Abstract
There is paucity of data on the genetic landscape of HIV-1 viruses circulating in the Limpopo Province of northeastern South Africa. Here, we examine the genetic diversity of viruses from Bela-Bela and Musina, two towns with high HIV prevalence. Between June 2007 and March 2008, blood samples were collected from antiretroviral-drug-naïve individuals. Viruses were analyzed for genetic subtypes and drug resistance mutations. All of the viruses in these samples were shown by phylogenetic analysis based on gag p17, gag p24, reverse transcriptase, protease and envelope C2-C3 gene regions to belong to HIV-1 subtype C. Two of 44 reverse transcriptase sequences (4.5%) contained N rather than the consensus K at position 103. The K103N mutation is normally associated with resistance to NNRTIs. No major mutations were observed in the protease gene. However, several polymorphisms and amino acid changes normally considered to be minor drug resistance mutations were observed in the protease sequences. These results suggest that HIV-1 subtype C remains the predominant variant responsible for the epidemic in northeastern South Africa and that the prevalence of drug-resistant viruses among the naïve population is low.
Introduction
Human immunodeficiency virus (HIV) presents a high degree of genetic variability, permitting the classification of isolates into types, groups, subtypes and recombinant forms. Genetic variability is attributed to the error-prone reverse transcriptase, which lacks proofreading functions (3′-5′ exonuclease activity), the diploid nature of the viral genome, the propensity for recombination, and the high rate of viral replication [1-3]. Phylogenetic analysis of different HIV type 1 strains from around the world has revealed three groups, namely M (main group), O (outliers) and N (non-M non-O viruses). Group M viruses are responsible for the global HIV pandemic and can be further subdivided into subtypes (A-D, F-H, J and K), sub-subtypes (A1/A2/A3, F1/F2), circulating recombinant forms (CRFs) and unique recombinant forms (URFs) [4-8]. The global distribution of HIV-1 subtypes is very heterogeneous, with subtype B largely responsible for the epidemic in the Americas and Europe. In the Central African region, all of the subtypes seem to be represented, while subtype C is driving the epidemic in India, China, Brazil, and in the Southern African region. In addition, subtype C accounts for more than 50% of infections globally [9].
South Africa has one of the highest numbers of HIV infections, with an estimated 5 million cases [10]. At the beginning of the epidemic in South Africa, HIV-1 subtype B viruses were identified among homosexual men who reported contacts in the United States [11]. Currently, HIV-1 subtype C viruses are responsible for the epidemic in South Africa [12, 13]. Subtypes A, D, and CRF01_AE and other recombinants viruses have also been identified, although in relatively small numbers [14-16]. As a result of viral diversity and redistribution of viral strains within countries and worldwide, regular monitoring of the genetic variants infecting individuals in a particular region is important because of its implications for diagnosis, treatment and prevention [17, 18].
The genetic diversity of HIV in several regions of South Africa has been documented [12, 14, 19, 20]. However, only two previous reports have provided genetic information from a highly endemic region of northeastern South Africa encompassing the Limpopo Province, using samples collected in 2001 from Bela Bela in the Waterberg district [21, 22]. The province is bordered to the north by Zimbabwe, to the northwest by Botswana, and to the east by Mozambique. Country reports indicate that in 2009 the prevalence of HIV was 14.3%, 23% and 12.2% for Zimbabwe, Botswana, and Mozambique, respectively [23]. The seroprevalence of HIV in Limpopo Province increased from 17.9% in 2000 to 21.4% in 2009 based on the national department of health’s antenatal sentinel HIV and syphilis prevalence survey [24]. Within South Africa, the province borders Gauteng province to the south, Mpumalanga province to the southeast, and North West province to the southwest, with HIV prevalence of 29.8%, 34.7% and 30% respectively.
The present study was carried out to update the genetic diversity profile of viruses in the Limpopo Province of South Africa, looking at viruses from two highly endemic areas. The diversity of viruses in one of two areas (Musina) has not been examined previously.
Materials and methods
Ethical considerations
The study protocol was approved by the Health, Safety and Research Ethics Committee of the University of Venda, South Africa. Approval to use public health institutions (clinics and hospitals) for the study was provided by the Limpopo Provincial Department of Health. Permission was also obtained from the authorities of the health establishments from which study participants were recruited. All of the study participants provided signed informed consent before the collection of demographic data and blood samples. Explanations of the study were provided in the local languages (Tshivenda and Sepedi) whenever there was a need.
Study areas, study population, and sample collection
Two study areas, Musina and Bela-Bela, were chosen for the study. Musina is the main border town between South Africa and Zimbabwe and also serves as a transit point for travelers and truckers going as far as Zambia and Malawi. The HIV seroprevalence in Musina is 30% in pregnant women according to the antenatal survey data of the National Department of Health [10], suggesting a relatively higher prevalence. No previous data are available on the genetic diversity of HIV from the Musina area. Study subjects from the Musina area were recruited from among individuals attending the Nancefield Voluntary Counseling and Testing (VCT) Center and the VCT service of the Madimbo clinic. Bela Bela is a tourist town comprising an informal settlement and an urban neighborhood. It is also a stopping point for long-distance truck drivers to and from Botswana, Zambia and Namibia. The seroprevalence of HIV in pregnant women in Bela-Bela was 32% in 2004 [10]. Subjects for this study were recruited from the VCT service of the HIV/AIDS Wellness Clinic run by the Bela-Bela HIV/AIDS Prevention Group. All study subjects were adults, with the exception of a 6-year-old boy.
Screening for HIV antibodies at the study sites was carried out according to the algorithm recommended by the South African Department of Health. The algorithm comprises the employment of a second rapid test of a different test principle for initially reactive samples. Doubly reactive individuals were considered HIV positive and were not tested further. Although the study participants were recruited from VCT services, the study questionnaire also sought to capture previous use, if any, of antiretrovirals by the self-reporting approach. None of the study participants were on antiretroviral therapy. A total of 150 individuals provided blood samples, 85 from the Musina area and 65 from Bela-Bela. The demographic data collected included age, sex, probable place and year of infection, probable route of infection, and WHO stage of disease. Recent infections were not determined by laboratory procedures. Samples were collected between June 2007 and May 2008. Five ml of venous blood was collection from each individual into EDTA vacutainer tubes. Plasma was subsequently extracted and stored in RNase- and DNAse-free tubes at −80°C until RNA isolation.
Viral RNA purification and RT-PCR
Total RNA was purified from plasma using a QIAGEN Viral RNA Mini Kit (QIAGEN GmbH, Germany) according to the manufacturer’s instructions. Amplification of the polymerase gene was performed by a one-tube reverse transcriptase PCR followed by nested PCR as described previously [21]. Briefly, a partial polymerase fragment of 1,400 bp was generated using the following primer pairs: RT-RV, 5′-TAT TTC AGC TAT CAA GTC TTT GAT GGG TCA-3′ and Pol1C 5′-GAA GGA CAC CAA TTG AAA GAC TGC AC-3′, for the RT-PCR step and GagP1,5′-CAA GGG GAG GCC AGG GAA TTT-3′, and Pol2R, 5′-TGA TGG GTC ATA ATA TAC TCC ATG-3′ for the nested PCR. The RT-PCR reaction was carried out in 50 μl of PCR mixture containing 5 μl of RNA, 5 μl of 10× buffer, 1 μl each of the RT-RV and Pol1C primers (10 pmol/μl), 0.5 μl of 10 mM dNTP mix, 0.25 μl each of Taq polymerase enzyme (5 U/μl), AMV RT (22 U/μl), RNase inhibitor (40 U/μl), 3 μl of MgCl2 (25 mM), and PCR-grade water to make up the final volume. The thermal cycling conditions for the RT-PCR step were as follows: 42°C for 60 min, then an initial step of 95°C for 3 min, followed by 30 cycles of 94°C for 1 min, 58°C for 1 min and 68°C for 2 min, and a final extension time of 10 min at 68°C.
The nested reactions were performed in 100 μl reaction mixture with 5 μl of the first amplification product, 10 μl of 10× buffer, 2 μl each of the second-round primers, 1 μl of 10 mM dNTP mix, 0.5 μl of Taq polymerase enzyme (5 U/μl), 6 μl of MgCl2, and water to make up the final volume. The thermal cycling conditions were as for the first round except for the RT step. A negative control was included in all PCR reactions to detect contamination. PCR products were examined for the expected band size after 1% gel electrophoresis followed by visualization under UV transillumination.
The p17 and p24 regions of the gag gene were generated as follows: cDNA was generated from 4 μl of RNA in a reaction mixture containing 1 μl each of dNTP mix (10 mM) and Gag D-rev primer (5′-AAT TCC TCC TAT CAT TTT TGG-3′) and placed in a heating block at 65°C for 5 min. Then, 4 μl of a master mixture of 2 μl of 5× cDNA buffer, 0.5 μl each of 0.1 M DTT, RNaseOUT (40 U/μl), Thermoscript (15 U/μl) and water of PCR grade was added to give a final volume of 10 μl. This was then incubated for 60 min at 50°C, and the reaction was terminated by heating to 85°C for 5 min. RNA template was removed with 1 μl (2 U/μl) E. coli RNase H and incubated on a heating block for 20 min at 37°C. The first-round PCR was performed using 1 μl each of Gag D-rev and Gag D-forw (5′-TCT CTA GCA GTG GCG CCC G-3′) targeting the entire gag region of 1220 bp in a reaction mixture of 5 μl of 10× buffer, 6 μl of 25 mM MgCl2, 4 μl of dNTP (200 μM), 0.125 μl of 5 U/μl Super-Therm Taq Pol, and 5 μl of the cDNA, with water making the final volume of 50 μl. The cycling conditions were as follows: 94°C for 2 min once and 94°C for 15 s, 55°C for 60 s and 72°C for 2 min for 35 cycles with a final extension time of 5 min at 72°C. The nested PCR was performed using 1 μl (10 pmol/μl) each of Gag A-forw (5′-CTC TCG AGC CAG GAC TCG GCT T-3′) and Gag C-rev (5′-TCT TCT AAT ACT GTA TCA GC-3′) in a reaction mixture containing 5 μl of 10× buffer, 2 μl of MgCl2 (25 mM), 4 μl of dNTP (200 μl), 0.125 μl of 5 U/μl Super-Therm Taq Pol and 5 μl of first-round product, with water to make a final volume of 50 μl. The cycling conditions were as follows: incubation at 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, 57°C for 45 s and 72°C for 1 min and a final extension time of 5 min at 72°C.
The C2-C3 region of the envelope gene was amplified using the following primer pairs: ED31, 5′-CCT CAGCCA ATT ACA CAG GCC TGT CCA AAG-3′, and ED33, 5′-TTA ACA GTA GAA AAA ATT CCC CTC-3′, for the first-round PCR and Env Bf, 5′-TAA CAC AAG CCT GCT CAA AGG T-3′, and Env Br, 5′-AAT TTC TAG GTC CCC TCC TGA-3′, for the nested PCR. The first round was carried out in a 50-μl reaction mixture consisting of 5 μl of 10× buffer, 2 μl of MgCl2 (25 mM), 4 μl of 10 mM dNTP mix, 1 μl each of the first-round primers (10 pmol/μl), 0.125 μl of 5 U/μl Super-Therm Taq Polymerase, and 5 μl of cDNA, which was generated in a manner like that of gag except that the ED33 primer was used instead of gag D-rev, with water to make up the final volume. The thermal cycling conditions were as follows: an initial 94°C for 2 min followed by 35 cycles of 94°C for 10 s, 50°C for 45 s, and 72°C for 1 min with a final extension time of 7 min at 72°C. The nested product of about 500 bp was generated using the second-round primers listed above. The 50-μl reaction mixture consisted of the following: 5 μl of 10× buffer, 4 μl of MgCl2 (25 mM), 4 μl of 10 mM dNTP mix, 1 μl each of the forward and reverse primers, 0.175 μl of 5 U/μl Super-Therm Taq Polymerase enzyme, and water to make the final volume 50 μl. The cycling conditions were as follows: an initial 94°C for 2 min followed by 35 cycles of 94°C for 15 sec, 57°C for 45 sec, and 72°C for 1 min, with a final extension time of 5 min at 72°C.
Sequencing and sequence compilation
Automated population-based sequencing was performed on both strands of viral DNA with the PCR nested primers using the dideoxynucleotide chain termination approach on an ABI PRISM Genetic Analyzer (ABI Prism310, Applied Biosystems, Foster City, CA). Forward and reverse nucleotide sequences were assembled, edited and translated into predicted amino acids with the SeqMan Pro and Seqbuilder programs included in the DNAStar software (DNASTAR, INC, Madison, Wisconsin USA).
Phylogenetic analysis
HIV-1 subtype assignment was done by phylogenetic analysis. Fifty-two gag p17, 47 p24, 43 RT, 29 PR and 22 C2-C3 env nucleotide sequences from 72 samples were available for analysis in this study. Sequences of some gene regions were not analyzed for all of the samples due either to a lack of amplification or a bad sequence that could not be reliably edited.
The nucleotide sequences generated from the test isolates and reference sequences obtained from the Los Alamos sequence database (http://hiv-web.lanl.gov) representing HIV-1 subtypes A-D, F-H, J and K were completely aligned using ClustalX software. Mean genetic distances were calculated according to the Kimura 2-parameter method assuming a constant evolutionary rate among the sequences. Evolutionary relatedness among the sequences was estimated by the neighbor-joining method, and phylogenetic trees were visualized with TreeView. Bootstrap re-sampling (1000 datasets) of the multiple alignments was performed in order to probe the statistical robustness of the tree.
Amino acid alignments
The predicted amino acid sequences of the gag p17, gag p24, RT, PR and env C2-C3 gene regions were analyzed for differences from the global consensus B and C sequences. Consensus amino acid sequences for the different gene regions under investigation were created and aligned with the global subtype B and C consensus sequences using BioEdit in order to determine the amino acid similarities or differences of test sequences to those of the global subtypes B and C.
Prediction of co-receptor usage
The V3 loop is a major determinant for viral tropism and coreceptor usage and also has a high degree of glycosylation. The predicted amino acid sequences of the V3 loop of the test isolates were submitted to webPSSM (http://indra.mullins.microbiol.washington.edu/pssm/webpssm3.pl), an online interactive program that predicts co-receptor usage of viruses by calculating the net charge of the V3 loop.
Analysis of drug-resistance mutations
Analysis of drug-resistance-related mutations in the RT and PR genes was performed using the Stanford HIV Drug Resistance Interpretation Algorithm (http://www.hivd.stanford.edu/hiv). This interactive program, based on the subtype B consensus sequence, compares codons of query sequences with resistance-encoding nucleotides contained in the database.
Results
Demographic and clinical profiles of the study population
The demographic characteristics of the study population were as follows: the mean age was 26.5 years, (range 6-52 years). Ninety percent (135) of the samples were obtained from women, and the main route of transmission was reported to be heterosexual contact, except in one case of perinatal transmission. The WHO clinical classification indicated that 60 of these patients were at stage 1, 65 at stage 2, 13 at stage 3 and 12 at stage 4. CD4 count and viral load assessment were not performed or available at the time of sample collection. None of the study participants thought they were infected within the previous six months at the time of sample collection.
Genetic subtypes, mean distances and immune pressure analysis
Of the 150 samples collected, viral DNA was obtained from 101, and 82 of the DNA samples were successfully sequenced (population-based sequencing) and edited reliably in one or more gene regions (57 for gag p17, 47 for gag p24, 35 for PR, 44 for RT, and 38 for env C2-C3). Two or more sequences were obtained for 58 viruses, and only one gene region was sequenced for 22 viruses. These sequences were used for HIV genetic diversity analysis. About 30% of the samples could not be amplified in any gene region.
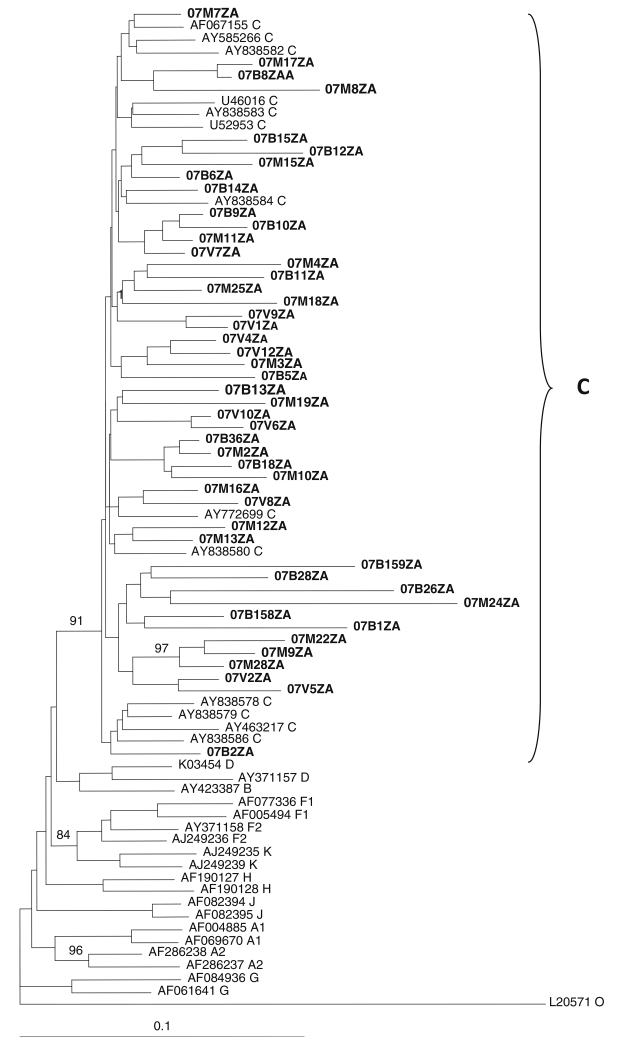
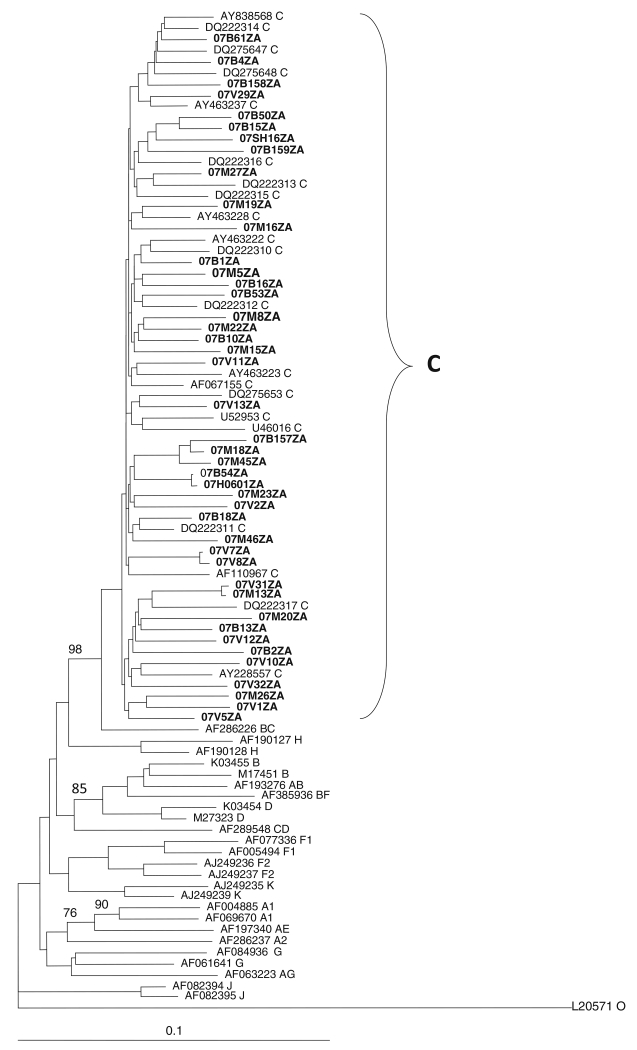
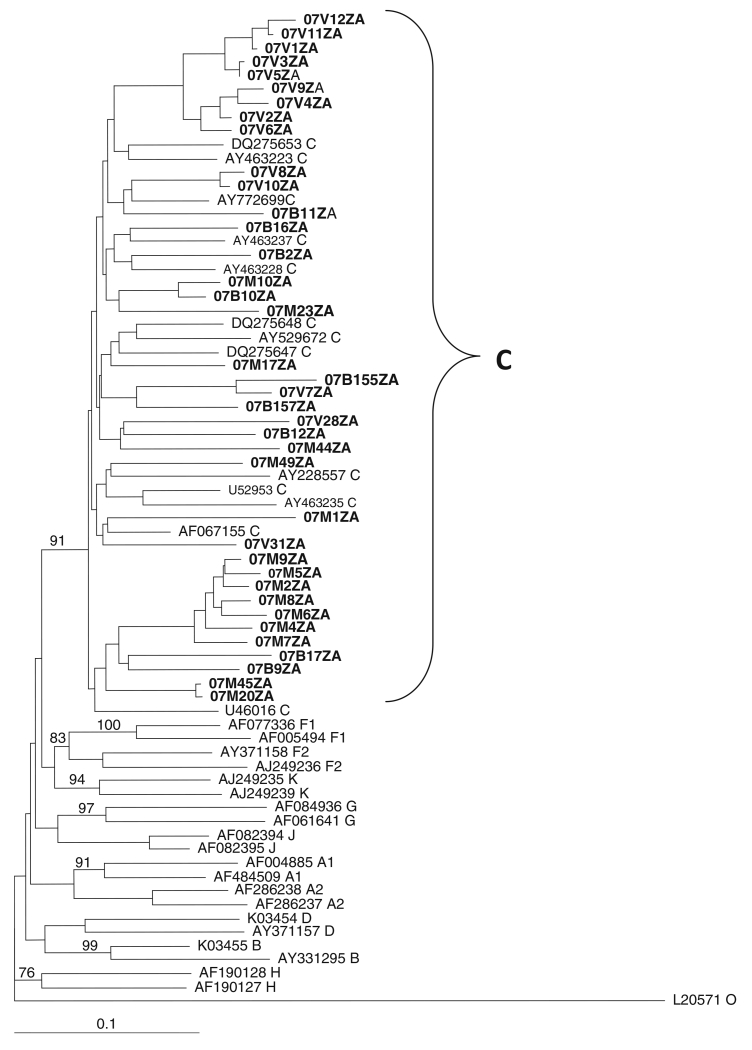
Phylogenetic analysis was performed on the gag p17, gag p24, protease, reverse transcriptase, and env C2-V3 envelope sequences. Neighbour-joining trees assigned the viruses to HIV-1 subtype C, as they all clustered and intermingle with subtype C reference sequences, with no monophyletic clustering of the test sequences observed in any of the regions. Figures 1, 2 and 3 show the phylogenetic relationships of the gap p24, RT and env C2-C3 sequences, respectively. A similar phylogenetic relationship was observed for the gag p17 and protease sequences (data not shown). The mean genetic distance between the gag p17 nucleotide sequences ranged from 0.0053 to 0.2929 (0.53% to 29.29%). All of the gag p17 amino acid sequences maintained the myristoylation site (http://www.bioafrica.net/proteomics/index.html). The mean genetic distance between the p24 nucleotide sequences ranged from 0.0550 to 0.2510 (5.3% to 25.10%). The amino acid alignments of the gag p17 and p24 showed no difference between their consensus sequences and the global subtype C consensus sequence, but they differed from the South African vaccine strain at seven positions (E11G, K15A, H27Q, I34L, M61I, K75R, and C111S) for the gag p17 and four positions (I91V, N125S, V129I and R151K) in the gag p24 region. Pairwise nucleotide distance analysis indicated a 3.4-9.6% divergence for the RT and 2.7-8.6% for the PR.
Fig. 1. Phylogenetic relationships of gag p24 nucleotide sequences from northeastern South Africa.
Sequences were aligned with reference HIV-1 subtype A-K sequences, and a phylogenetic tree was constructed by the neighbor-joining method implemented in ClustalX. Test sequences (in boldface) cluster and intermingle with subtype C reference sequences. Bootstraps values above 70% of 1000 replicates are indicated
Fig. 2. Phylogenetic relationship of partial reverse transcriptase nucleotide sequences from northeastern South Africa.
The sequences cluster and intermingle with subtype C reference sequences, indicating that they are HIV-1 subtype C. The tree was constructed by the neighbor-joining method as implemented in the ClustalX2 program. The validity of the branching orders was estimated with 1000 replicates. Bootstrap values above 70% of 1000 replicates are indicated
Fig. 3. Phylogenetic analysis of the env C2-C3 nucleotide sequences from northeastern South Africa.
Test sequences (in boldface) cluster and intermingle with HIV-1 subtype C reference sequences with a bootstrap value of 91% of 1000 replicates
In addition, the alignment of the 44 reverse transcriptase and 35 protease sequences with their respective subtype C global consensus sequence and their South African reference sequence showed no sequence variation between the global consensus C and the RT test sequences. However, differences were observed at six positions (I60V, G123F, V174A, K175Q, P277R and T286A) when compared to the South African reference sequence. Also, the consensus of the test sequences of the PR revealed one difference (T13I) between the global subtype C consensus and three positions (T19I, R20K and D35E) with regard to the South African reference sequence (figures not shown).
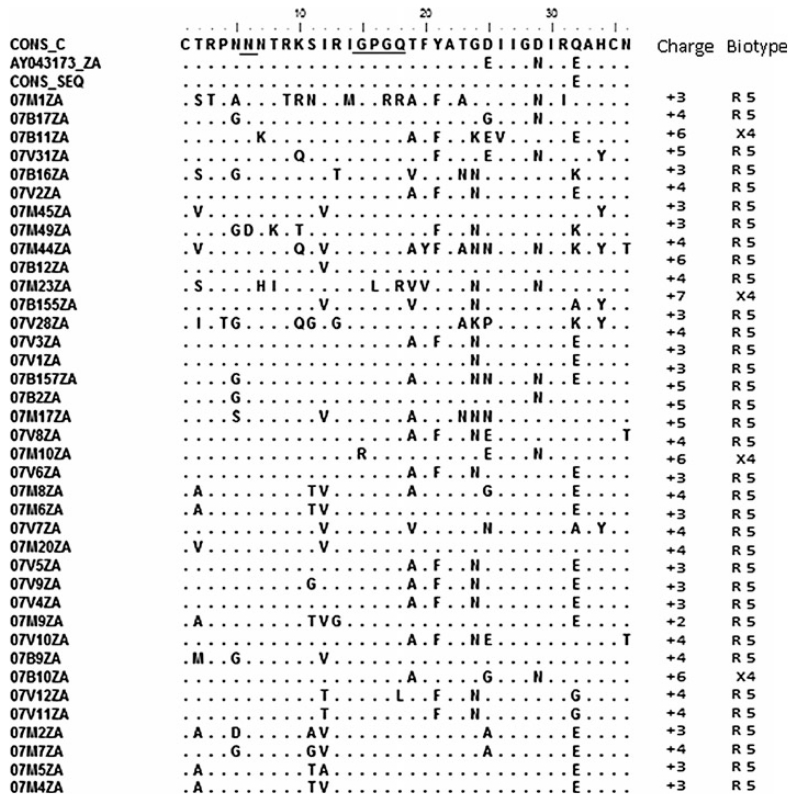
Comparison of the amino acid consensus sequence of the V3 loop showed it was identical in all positions except at Q32E to the global consensus sequences of subtypes C and B. The mean genetic distance ranged from 0.0354 to 0.2630 (3.54-26.30%). Analysis of the V3 loop of the 38 env amino acid sequences showed that all of the isolates had the GPGQ tetrapeptide, with the exception of 07M1ZA, 07M10ZA, and 07M23ZA, each of which had amino acid substitutions in this motif. Co-receptor usage prediction with webPSSM showed that all but 07B11ZA, 07B155ZA, and 07B10ZA (7.89%) were CCR5 viruses with net charges ranging from +2 to +7. The usual N-linked glycosylation sites at the start of the V3 loop were maintained in all of the test sequences except 07M49ZA and 07B155ZA (Fig. 4).
Fig. 4. Predicted V-3 loop amino acid sequences and biotypes of HIV from northeastern South Africa.
The multiple aligned sequences were compared to the consensus of the test sequences with those of the global subtype C consensus sequence and the South African vaccine strain AY043173_ZA. Underlined are the glycosylation sites and the GPGQ motif. Four viruses are X4 viruses, as predicted by webPSSM. The consensus of the test sequences showed close amino acid similarity to the global subtype C consensus sequence and the South African vaccine strain, except at position 32 for the consensus sequence and 25 and 29 for AY043173_ZA
Genetic evidence for drug resistance in the analyzed viruses
Nucleotide sequences were analyzed according to the Stanford HIV genotypic resistance interpretation algorithm in order to detect potential amino acid substitutions associated with resistance to PR and RT inhibitors. There were no primary substitutions expected to confer resistance in the PR sequences. However, substitutions in the protease that contribute to PI resistance when present with other substitutions were observed (Table 1). For the RT gene, the K103N substitution, which confers primary resistance to NNRTIs such as nevirapine was present in two of 44 sequences (4.5%) (samples 07M5ZA and 07V2ZA). In addition, secondary mutations were found in many samples, and some samples had combinations of substitutions (see Table 1).
Table 1.
Frequency of protease and reverse transcriptase resistance-related mutations from South African HIV-1 subtype C isolates
| Resistance-associated mutationsa |
Frequency (%) |
Coding nucleotideb |
|
|---|---|---|---|
| Wild type | Mutant | ||
| Protease (n = 29) | |||
| L10V | 2 (7%) | CTT | GTT |
| M36I | 26 (90%) | ATG | ATA/ATC |
| M36L | 3 (10%) | ATG | CTG |
| L60E | 4 (4%) | CTG/CTA/ CTC |
GAA |
| L63S | 3 (10%) | AGC | |
| L63P | 8 (27.5%) | CCC/CCT | |
| L63F | 1 (3%) | TTC | |
| L63T | 1 (3%) | ACT | |
| L63V | 3 (10%) | GTC | |
| A71T | 1 (3%) | GCT | ACT |
| T74S | 2 (6%) | ACC | TCC |
| V77I | 2 (6%) | GTT | ATT |
| V82I | 1 (3%) | GTT | ATT |
| I93L | 27 (94%) | ATT | GTT/CTC |
| Reverse transcriptase (n = 43) | |||
| V90I | 2 (4.6%) | GTT | ATT |
| A98S | 6 (13.9%) | GCC | TCC |
| V90I/K103N | 1 (2.3%) | ||
| K103N | 2 (4.6%) | AAA | AAC |
| V118I | 2 (4.6%) | GCT | ATC |
| E138A | 4 (9.3%) | GAA/GAG | GCA/ GCG |
| V118I/E138A | 1 (2.3%) | ||
| V118I/E138A/ V179D |
1 (2.3%) | ||
| V179I | 3 (6.9%) | GTT | ATC |
Resistance mutations were identified according to the Stanford Genotype Resistance Interpretation Algorithm
Nonsynonymous codons introducing substitutions at positions related to drug resistance. The wild-type reference is the HIV-1 subtype B consensus sequence
Sequence accession numbers
The nucleotide sequences analyzed here have been submitted to GenBank under the following accession numbers: Gag p17, GU201650—GU201706; Gag p24, GU201707—GU201753; Protease, GU201798—GU201826; Reverse transcriptase, GU201754—GU201797; Env C2-C3, GU201 612—GU201649.
Discussion and conclusion
Data on the genetic diversity of HIV in the Limpopo Province (northeastern South Africa) is limited, and the region has an HIV prevalence of 21.4% as estimated by the national HIV seroprevalence survey of 2009. Variation and evolution of HIV has implications for the efficacy of diagnostics, treatment and prevention strategies. Therefore, there is a need for periodic monitoring of the genetic landscape of the virus.
This study was undertaken to determine the HIV viral diversity in Bela-Bela and Musina, two areas in the northeastern South Africa with higher HIV prevalence. Partial gag, pol and env sequences were observed to be of HIV-1 subtype C by phylogenetic analysis, with no monophyletic clustering in any of the gene regions investigated. The predominance of subtype C viruses has been documented for the neighbouring provinces and other regions of South Africa [25-27]. Subtype C is also the predominant variant in the neighbouring southern African countries of Botswana, Mozambique, and Zimbabwe [28, 29], and there is a lack of evidence to suggest a clonal provenance of subtype C viruses in southern Africa [30, 31]. Also, alignments of the derived consensus amino acid sequences of the different gene regions against their respective subtype B and C global consensus sequences showed little variation between them and the subtype C consensus sequences. However, there were significant differences with the global subtype B consensus sequence, as expected. The inference is that the study population may be used as a clinical trial site for the evaluation of a subtype-C-based vaccine. Sequence analysis of the gag p17 revealed that all of the sequences retained the myristoylation motifs that are characteristic of these proteins (http://www.bioafrica.net/proteomics/index.html).
Before 2010, the South African national antiretroviral treatment guideline recommended a combination of stavudine, lamivudine, and efavirenz as the first-line regimen for adults with no prior exposure to antiretrovirals, with nevirapine replacing efavirenz for women of childbearing age. The second-line regimen consisted of zidovudine, didansone and lopinavir/ritonavir. In 2010, tenofovir replaced stavudine for those with poor tolerance of stavudine in the first-line regimen. Pregnant women and patients with HIV/TB co-infection with a CD4 count of less than 350/ml also became eligible for treatment in 2010 [10, 24].
Genetic characterization of the pol sequences (35 PR and 44 RT sequences), in addition to providing phylogenetic information, allowed us to evaluate the presence and prevalence of drug resistance-related mutations in these antiretroviral-naive individuals. There was no primary drug-resistant mutation in the protease sequences analyzed. However, several minor substitutions commonly associated with PI resistance were found, with several individuals harboring more than one such mutation. While mutations at these positions do not cause high-level drug resistance themselves, they contribute to drug resistance when present together with certain primary protease mutations and have been shown to compensate for the decrease in catalytic efficiency caused by PI-selected primary protease mutations [18, 32-36]. The secondary substitution was L10V in two viruses (7%), whereas K20R occurred in four (14%) of the 29 isolates. M36I occurred in 28 viruses (98%), T74S occurred in two viruses (7%), I93L occurred in 27 isolates with a prevalence of 93%, and V82I occurred in one virus (3.4%). These observations are similar to those of a previous report [22], with 93% prevalence for both M36I and I93L. However, that study reported 46% prevalence for K20R and 8% for V82I, which are higher than the findings in the current study. This very low prevalence of major resistance mutations in PR and high prevalence of secondary mutations are in agreement with other subtype C data obtained from drug-naïve individuals from a distinct geographical region [37]. These differences or discrepancies may be attributable to natural variation associated with founder effects rather than being representative of the transmission of drug-selected mutations, as PIs are not commonly used in South Africa or in most African countries [37].
Among the RT sequences, 2 of the 44 viruses (4.5%) harbored the K103N substitution, which confers resistance to nevirapine, and it also occurred in combination with V90I in one virus. The mutations occurred in two different women from the Musina area. Since the women were of childbearing age, it is possible that they could have taken nevirapine as part of a programme for prevention of mother-to-child transmission. Antiretrovirals were available at the Bela Bela site since 2004 for all eligible patients, and in 2006 in Musina. Our earlier study [22] did not find K103N among the study participants. However, others [12] have reported this mutation in epidemiologically linked patients in South Africa. We also found six viruses (13.9%) that harbored A98S and four (9.3%) with V179I, which is lower than the 15% and 23%, respectively, that were previously reported in the province [22]. One of the viruses harbored the combination V118I/E138A/V179D, while another had V118I/E138A. These mutations have been reported to cause low-level resistance when they occur alone and increased resistance when they occur in combination with other mutations like E44A/D (http://hivdb.stanford.edu/). Other substitutions that were found include V90I, E138A, and V179D, which are weakly associated with decreased NNRTI susceptibility, and V118I, which occurs in ~2% of untreated individuals and with increased frequency in those receiving multiple NRTIs. This latter substitution is known to cause low-level resistance to lamivudine and possibly to other NRTIs when present with E44A/D and/or one or more TAMs. The finding of K103N, which results in nevirapine resistance, in 4.5% of the study population is of importance, as nevirapine is used in the prevention of mother-to-child transmission of HIV.
Comparison of the global non-B RT consensus sequences to the subtype B RT global consensus sequence shows differences at 23 residues, of which position 179 is in a position related to subtype B drug resistance. V118I is a polymorphism in non-B RT that is associated with subtype B NRTI resistance [34]. Similar reports [21, 22] on viruses from Bela-Bela in a cross-sectional analysis of the pol gene showed that drug-resistance mutations may not be common among the drug-naïve population. However, it should be noted that ARVs were only recently introduced in Musina in 2006, two years prior to sample collection.
Genetic analysis of the V3 loop allowed us to evaluate the predicted co-receptor usage of the viruses. All but three of the viruses were predicted to be CCR5-utilizing variants, and this is in agreement with previous reports in which subtype C viruses were shown to use CCR5 preferentially. Should there be resistance to the available NRTIs and NNRTIs, which are commonly used in the management of patients, the newly approved entry inhibitor maraviroc, which is a CCR5 antagonist, will prove useful as an alternative therapy. However, the CXCR4 variants would pose a problem in management, as these viral variants are not susceptible to maraviroc [38-40].
This study has several limitations. The sample size is relatively small, and DNA amplification was not successful in about 30% of the samples. Lack of amplification could be attributed to low viral load, as the majority of the patients were in WHO clinical stage 1 or 2 of the disease. Also, some loss of RNA may have occurred during refrigeration of whole blood at 4°C in the outpost clinics for several days prior to RNA isolation. Partial gene regions were used to assign viral subtypes, potentially allowing recombinant viruses to be missed. The bulk sequencing employed may result in the lack of detection of minority-population viruses, which can lead to an underestimation of viral diversity and drug resistance mutations. Laboratory investigations of recent infections was not done, so it was difficult to correlate the probable infection dates as provided by the study subjects, at least for those who were in WHO stage 1 of the disease. Nevertheless, from the self-reporting questionnaire, none of the participants were thought to have been infected in the last 6 months preceding blood collection. This means that all of the infections were likely to have been chronic, again with the possibility of underestimating transmission of resistance mutations due to mutation reversion over time.
Regular monitoring of the HIV genetic landscape is important for diagnostic, treatment and prevention efforts. Given the enormous worldwide diversity of HIV variants, the design and choice of antigens for inclusion in candidate AIDS vaccines is a daunting challenge [18, 41-45]. Although full-length genomes are the ideal basis for viral subtype assignment, the present study has provided additional data on the molecular epidemiology of HIV in northeastern South Africa and has shown that the subtype distribution continues to be primarily dominated by HIV-1 subtype C viruses, at least in the highly endemic areas studied, with no distinct phylogenetic lineage. However, periodic monitoring for a virus as highly divergent as HIV continues to be necessary.
Acknowledgments
The South African AIDS Vaccine Initiative supported this study. Additional support was obtained from the National Research Foundation, South Africa. David Rekosh and Marie-Louise Hammarskjold were supported by the Myles H. Thaler and Charles Ross, Jr. Endowed Professorships at the University of Virginia. We thank the patients who participated in the study, and the nurses Ms. Cecile Manhaeve of the Bela Bela HIV Wellness Clinic, Ms. MS Nedzamba of Madimbo Clinic, and Ms. HS Nekhavhambe of the Musina HIV Information Centre for recruiting patients and blood sample collection. The opinions expressed here are those of the authors.
Contributor Information
Benson Chuks Iweriebor, AIDS Virus Research Laboratory, Department of Microbiology, University of Venda, PMB X5050, Thohoyandou 0950, South Africa.
Lufuno Grace Mavhandu, AIDS Virus Research Laboratory, Department of Microbiology, University of Venda, PMB X5050, Thohoyandou 0950, South Africa.
Tracy Masebe, AIDS Virus Research Laboratory, Department of Microbiology, University of Venda, PMB X5050, Thohoyandou 0950, South Africa.
David Rekosh, Department of Microbiology, Myles H. Thaler Center for AIDS and Human Retrovirus Research, University of Virginia, Charlottesville, VA 22908, USA.
Marie-Louise Hammarskjold, Department of Microbiology, Myles H. Thaler Center for AIDS and Human Retrovirus Research, University of Virginia, Charlottesville, VA 22908, USA.
Jeffrey M. Mphahlele, HIV and Hepatitis Research Unit, Department of Virology, University of Limpopo Medunsa Campus, Pretoria, South Africa
Pascal Obong Bessong, AIDS Virus Research Laboratory, Department of Microbiology, University of Venda, PMB X5050, Thohoyandou 0950, South Africa.
References
- 1.Moore MD, Hu WS. HIV-1 RNA dimerization: It takes two to tango. AIDS Rev. 2009;11(2):91–102. [PMC free article] [PubMed] [Google Scholar]
- 2.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev. 2009;73(3):451–480. doi: 10.1128/MMBR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon-Loriere E, Rossolillo P, Negroni M. RNA structures, genomic organization and selection of recombinant HIV. RNA Biol. 2011;8(2):280–286. doi: 10.4161/rna.8.2.15193. [DOI] [PubMed] [Google Scholar]
- 4.Powell RL, Zhao J, Konings FA, et al. Circulating recombinant form (CRF) 37_cpx: an old strain in Cameroon composed of diverse, genetically distant lineages of subtypes A and G. AIDS Res Hum Retroviruses. 2007;23(7):923–933. doi: 10.1089/aid.2007.0040. [DOI] [PubMed] [Google Scholar]
- 5.Brennan CA, Brites C, Bodelle P, et al. HIV-1 strains identified in Brazilian blood donors: significant prevalence of B/F1 recombinants. AIDS Res Hum Retroviruses. 2007;23(11):1434–1441. doi: 10.1089/aid.2007.0121. [DOI] [PubMed] [Google Scholar]
- 6.Gao F, Vidal N, Li Y, et al. Evidence of two distinct subsubtypes within the HIV-1 subtype A radiation. AIDS Res Hum Retroviruses. 2001;17(8):675–688. doi: 10.1089/088922201750236951. [DOI] [PubMed] [Google Scholar]
- 7.Van der Auwera G, Janssens W, Heyndrickx L, et al. Reanalysis of full-length HIV type 1 group M subtype K and subsubtype F2 with an MS-DOS bootscanning program. AIDS Res Hum Retroviruses. 2001;17(2):185–189. doi: 10.1089/08892220150217274. [DOI] [PubMed] [Google Scholar]
- 8.Castelbranco EP, da Silva Souza E, Cavalcanti AM, et al. Frequency of primary resistance to antiretroviral drugs and genetic variability of HIV-1 among infected pregnant women recently diagnosed in Luanda-Angola. AIDS Res Hum Retroviruses. 2010;26(12):1313–1316. doi: 10.1089/aid.2010.0111. [DOI] [PubMed] [Google Scholar]
- 9.Hemelaar J, Gouws E, Ghys PD, et al. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011;25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Department of Health . National HIV Seroprevalence survey of women attending antenatal clinics in South Africa. Directorate Health System Research and Epidemiology. Department of Health; Pretoria: 2004. [Google Scholar]
- 11.Williamson C, Englebrecht S, Lambrick M, et al. HIV-1 subtypes in different risk groups in South Africa. Lancet. 1995;346:782. doi: 10.1016/s0140-6736(95)91543-5. [DOI] [PubMed] [Google Scholar]
- 12.Gordon M, De Oliveria T, Bishop K, et al. Molecular characterization of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: implication for vaccine and antiretroviral control strategies. J Virol. 2003;77:2587–2599. doi: 10.1128/JVI.77.4.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs GB, Laten AD, Van Rensburg EJ, et al. Phylogenetic diversity and low level antiretroviral resistance mutations in HIV Type 1 treatment-naive patients from Cape Town, South Africa. AIDS Res Hum Retroviruses. 2008;24:1009–1012. doi: 10.1089/aid.2008.0028. [DOI] [PubMed] [Google Scholar]
- 14.Hemelaar J, Gouws E, Ghys PD, et al. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 15.Bredell H, Hunt G, Casteling A, et al. HIV-1 subtypes A, D, G, AG and unclassified sequences identified in South Africa. AIDS Res Hum Retroviruses. 2002;18:681–683. doi: 10.1089/088922202760019400. [DOI] [PubMed] [Google Scholar]
- 16.Papathanasopoulos MA, Vardas E, Wallis C, et al. Characterization of HIV type 1 genetic diversity among South African participants enrolled in the AIDS Vaccine Integrated Project (AVIP) study. AIDS Res Hum Retroviruses. 2010;26:705–709. doi: 10.1089/aid.2009.0281. [DOI] [PubMed] [Google Scholar]
- 17.Peeters M, Sharp PM. Genetic diversity of HIV-1: the moving target. AIDS. 2000;14(Suppl 3):S129–S140. [PubMed] [Google Scholar]
- 18.Lal RB, Sekher C, Chunfu Y. Impact of genetic diversity of HIV-1 on diagnosis, antiretroviral therapy & vaccine development. Indian J Med Res. 2005;121:287–314. [PubMed] [Google Scholar]
- 19.Bredell H, Williamson C, Sonnenberg P, et al. Genetic characterization of HIV type 1 from migrant workers in three South African gold mines. AIDS Res Hum Retroviruses. 1998;14:677–684. doi: 10.1089/aid.1998.14.677. [DOI] [PubMed] [Google Scholar]
- 20.Pillay C, Bredell H, McIntyre J, et al. HIV-1 subtype C reverse transcriptase sequences from drug naive pregnant women in South Africa. AIDS Res Hum Retroviruses. 2002;18:605–610. doi: 10.1089/088922202753747950. [DOI] [PubMed] [Google Scholar]
- 21.Bessong PO, Obi CL, Cilliers T, et al. Characterization of human immunodeficiency virus type 1 from a previously unexplored region of South Africa with a high HIV prevalence. AIDS Res Hum Retroviruses. 2005;21(1):103–109. doi: 10.1089/aid.2005.21.103. [DOI] [PubMed] [Google Scholar]
- 22.Bessong PO, Mphahlele J, Choge IA, et al. Resistance mutational analysis of HIV-1 subtype C among rural South African drug-naïve patients prior to large-scale availability of antiretrovirals. AIDS Res Hum Retroviruses. 2006;22(12):1306–1312. doi: 10.1089/aid.2006.22.1306. [DOI] [PubMed] [Google Scholar]
- 23.UNAIDS . AIDS epidemic update. Geneva: 2010. [Google Scholar]
- 24.Department of Health . National Antenatal Sentinel HIV and Syphilis Prevalence Survey in South Africa, 2009. 2010. [Google Scholar]
- 25.Jacobs GB, Loxton AG, Laten A, et al. Emergence and diversity of different HIV-1 subtypes in South Africa, 2000-2001. J Med Virol. 2009;81(11):1852–1859. doi: 10.1002/jmv.21609. [DOI] [PubMed] [Google Scholar]
- 26.Naidoo AF, Parboosing R, Gordon ML. Dual HIV infection uncommon in patients on antiretroviral therapy in a region with high HIV prevalence. AIDS Res Hum Retroviruses. 2009;25(12):1225–1230. doi: 10.1089/aid.2009.0095. [DOI] [PubMed] [Google Scholar]
- 27.Papathanasopoulos MA, Vardas E, Wallis C, et al. Characterization of HIV type 1 genetic diversity among South African participants enrolled in the AIDS Vaccine Integrated Project (AVIP) study. AIDS Res Hum Retroviruses. 2010;26(6):705–709. doi: 10.1089/aid.2009.0281. [DOI] [PubMed] [Google Scholar]
- 28.Bussmann H, Novitsky V, Wester W, et al. HIV-1 subtype C drug-resistance background among ARV-naive adults in Botswana. Antivir Chem Chemother. 2005;16(2):103–115. doi: 10.1177/095632020501600203. [DOI] [PubMed] [Google Scholar]
- 29.Bártolo I, Casanovas J, Bastos R, et al. HIV-1 genetic diversity and transmitted drug resistance in health care settings in Maputo, Mozambique. J Acquir Immune Defic Syndr. 2009;51(3):323–331. doi: 10.1097/qai.0b013e3181a24906. [DOI] [PubMed] [Google Scholar]
- 30.zur Megede J, Engelbrecht S, de Oliveira T, et al. Novel evolutionary analyses of full-length HIV type 1 subtype C molecular clones from Cape Town, South Africa. AIDS Res Hum Retroviruses. 2002;18(17):1327–1332. doi: 10.1089/088922202320886370. [DOI] [PubMed] [Google Scholar]
- 31.Bredell H, Martin DP, Van Harmelen J, et al. HIV type 1 subtype C gag and nef diversity in Southern Africa. AIDS Res Hum Retroviruses. 2007;23(3):477–481. doi: 10.1089/aid.2006.0232. [DOI] [PubMed] [Google Scholar]
- 32.Velaqueze-Campoy A, Todd MJ, Vega S, et al. Catalytic efficiency and vitality of HIV-1 protease from African viral subtype. Proc Natl Acad Sci USA. 2001;98:6062–6067. doi: 10.1073/pnas.111152698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soriano V, de Mendoza C. Genetic mechanisms of resistance to protease inhibitors and entry inhibitors. HIV Clin Trials. 2002;3:249–257. doi: 10.1310/hct.2002.3.3.009. [DOI] [PubMed] [Google Scholar]
- 34.Kantor R, Katzenstein D. Polymorphism in HIV-1 non-subtype B protease and reverse transcriptase and its potential impact on drug susceptibility and drug resistance evolution. AIDS Rev. 2003;5:25–35. [PubMed] [Google Scholar]
- 35.Holguin L, Paxinos E, Hertogs K, et al. Impact of frequent natural polymorphisms at the protease gene on the in vitro susceptibility to protease inhibitor in HIV-1 non-B subtypes. J Clin Virol. 2004;31:215–220. doi: 10.1016/j.jcv.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Wainberg MA. HIV-1 subtypes distribution and the problem of drug resistance. AIDS. 2004;18(suppl 3):S63–S68. doi: 10.1097/00002030-200406003-00012. [DOI] [PubMed] [Google Scholar]
- 37.Parreira R, Piedade J, Domingues A, et al. Genetic characterization of human immunodeficiency virus type 1 from Beira, Mozambique. Microbes Infect. 2006;8:2442–2451. doi: 10.1016/j.micinf.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Sayana S, Khanlou H. Maraviroc: a new CCR5 antagonist. Expert Rev Anti Infect Ther. 2009;7(1):9–19. doi: 10.1586/14787210.7.1.9. [DOI] [PubMed] [Google Scholar]
- 39.Reuter S, Braken P, Jensen B, et al. Maraviroc in treatment-experienced patients with HIV-1 infection—experience from routine clinical practice. Eur J Med Res. 2010;15(6):231–237. doi: 10.1186/2047-783X-15-6-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandekerckhove L, Verhofstede C, Demecheleer E, et al. Comparison of phenotypic and genotypic tropism determination in triple-class-experienced HIV patients eligible for maraviroc treatment. J Antimicrob Chemother. 2011;66(2):265–272. doi: 10.1093/jac/dkq458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tscherning C, Alaeus A, Fredriksson R, et al. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241:181–188. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 42.Thomson MM, Perez-Alvarez L, Najera R. Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy. Lancet Infect Dis. 2002;8:461–471. doi: 10.1016/s1473-3099(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 43.Gaschen B, Taylor J, Yusim K, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 44.Esparza J, Osmanvo S. HIV vaccines: a global perspective. Curr Mol Med. 2003;3:183–193. doi: 10.2174/1566524033479825. [DOI] [PubMed] [Google Scholar]
- 45.Stratov I, DeRose R, Purcell DF, et al. Vaccines and vaccine strategies against HIV. Curr Drug Targets. 2004;5:71–88. doi: 10.2174/1389450043490686. [DOI] [PubMed] [Google Scholar]