Abstract
Our objective was to characterize lipid profiles in cell models of adipocyte differentiation in comparison to mouse adipose tissues in vivo. A novel lipid extraction strategy was combined with global lipid profiling using direct infusion and sequential precursor ion fragmentation, termed MS/MSALL. Perirenal and inguinal white adipose tissue and interscapular brown adipose tissues from adult C57BL/6J mice were analyzed. 3T3-L1 preadipocytes, ear mesenchymal progenitor cells, and brown adipose-derived BAT-C1 cells were also characterized. Over 3000 unique lipid species were quantified. Principal component analysis showed that perirenal versus inguinal white adipose tissues varied in lipid composition of triacyl- and diacylglycerols, sphingomyelins, glycerophospholipids and, notably, cardiolipin CL 72:3. In contrast, hexosylceramides and sphingomyelins distinguished brown from white adipose. Adipocyte differentiation models showed broad differences in lipid composition among themselves, upon adipogenic differentiation, and with adipose tissues. Palmitoyl triacylglycerides predominate in 3T3-L1 differentiation models, whereas cardiolipin CL 72:1 and SM 45:4 were abundant in brown adipose-derived cell differentiation models, respectively. MS/MSALL data suggest new lipid biomarkers for tissue-specific lipid contributions to adipogenesis, thus providing a foundation for using in vitro models of adipogenesis to reflect potential changes in adipose tissues in vivo.
Keywords: Lipidomics, Mass Spectrometry, Phospholipids, Sphingolipids, Cardiolipin, Preadipocyte, MS/MSALL
Characterization of lipogenesis during the process of adipogenesis is fundamental to understanding the metabolism of adipogenic tissues in both normal metabolic responses to energy demands and in disease states. The field of lipidomics, or mass spectrometry-based unbiased global analysis of changes in lipid levels in cells and tissues, has become a critical area of metabolomic biomarker analysis. Many recent technological and software advances have made mass spectrometry (MS) ideally suited to lipid analysis. These advances have elevated the utility and reliability of shotgun lipidomic approaches for the analysis of lipid molecular species obtained directly from crude biological extracts of tissue or fluids. Accordingly, this has allowed for unbiased global analysis of complex biological and clinical specimens using a variety of MS-based approaches [Isaac, 2011; Wang and Han, 2014; Yang et al., 2009]. However, these more recent technical advances have not been fully exploited to characterize lipid changes in diverse biological contexts.
Unbiased lipid profiling is being applied to an ever-widening variety of human disease states. For example, lipidomic analysis of specimens from patients with nonalcoholic fatty liver disease, which includes steatosis, nonalcoholic steatohepatitis, and progression to cirrhosis, elucidate detailed differences in the proportions of glycerophospholipid (GPL) and diacylglycerol (DAG) species between normal and diseased human and murine liver extracts [Gorden et al., 2011]. The prospective study of patients with different stages of nonalcoholic fatty liver disease by profiling liver biopsies, blood plasma, and urine samples, with application of linear discriminant analysis, reveals novel lipid biomarkers of disease, including GPLs, sphingolipids, and sterols [Gorden et al., 2015]. Lipidomics studies have been applied in a variety of clinical areas, from diabetes [Lappas et al., 2015; Meikle et al., 2014], to cardiovascular disease [De Leon et al., 2015; Hinterwirth et al., 2014], prostate cancer [Min et al., 2011; Zhou et al., 2012] and clinical psychiatric areas such as the lipid profile responses to atypical antipsychotic drugs in cases of schizophrenia [McEvoy et al., 2013]. The effective interpretation of biological context-specific changes in lipid composition demands not only a firm basis in understanding lipid metabolism, but also the broader availability of diverse MS datasets that reveal the specific lipid changes that can be expected in normal homeostatic and disease-associated adipogenic states.
Important biological tools in understanding the molecular regulation of adipogenesis include in vitro cell models of adipocyte differentiation and mouse models of normal homeostatic and disease-altered lipid metabolism. One of the most widely used cell culture models, the mouse fibroblast-derived 3T3-L1 preadipocyte cell line, has been used as a model of adipogenesis since 1970 [Green and Kehinde, 1975]. Furthermore, the use of 3T3-L1 cells for lipidomics has provided information on lipid profiling [Sokol et al., 2013] and complex biological processes. These include identifying the role of peroxisomal fatty acid processing on the metabolic fate of fatty acids in the differentiating adipocyte [Su et al., 2004], clarifying the role of high density lipoprotein-associated phospholipids in circulating non-esterified fatty acid levels [Drew et al., 2011], providing new insight into retinoid control of adipocyte function [McIlroy et al., 2015], and the implication of critical upstream lipid-mediators of peroxisome proliferator-activated receptor γ (PPARγ) [Park et al., 2012], a key transcription factor in adipogenesis [Brun and Spiegelman, 1997; Wu et al., 1995]. Such studies not only shed light on the effect of lipids and correlative metabolic processes on adipogenesis, but provide a foundation for the comparative analysis of the lipidomes of nonadipose tissues [Cifkova et al., 2015; Kien et al., 2015] and cells [Arai et al., 2015; Sampaio et al., 2011]. However, the lipidomic characterization of other potentially useful cell culture models is relatively incomplete, thus underscoring the need for more diverse and interrelated cell culture-, tissue-, and organism-based data sets [Horn and Chapman, 2012; Layre and Moody, 2013; Zhang et al., 2015].
Emerging lipidomic data are starting to provide a foundation for understanding the differences in lipid metabolism between white and interscapular brown adipose tissues (WAT and BAT, respectively). Specific lipidomic signatures distinguish omental and subcutaneous adipose depots in humans [Jove et al., 2014]. Also, sex-specific lipidomic changes have been described in white and brown mouse adipose [Hoene et al., 2014], and in the BAT response to cold stress [Marcher et al., 2015]. These cell, tissue, and mouse model studies, as well as numerous clinical applications [Hyotylainen and Oresic, 2015; Wood, 2014; Zhao et al., 2014], demonstrate the utility of lipidomic analysis to address basic scientific and biomedical questions. However, there is a lack of clarity concerning the degree to which cell culture model lipidomes represent and correspond to the in vivo contexts that they are intended to model [Lamaziere et al., 2013].
Tandem mass spectrometry (MS) is well suited for the detailed characterization of lipid molecular species. MS profiling and quantitative analysis of mammalian lipids in blood plasma have characterized more than 500 lipid species [Quehenberger et al., 1987]. More recent high-throughput workflows allow lipids to be identified and quantified as specific molecular species [Jung et al., 2011], in which case specific lipid classes, including glycerophospholipids, glycerolipids, glycosphingolipids and sterol lipids, can be identified via: (i) their head group ions, (ii) their long-chain (e.g., sphingomyelin) bases, and (iii) the fatty acid acyl constituent ions. However, limited MS/MS data quality and laborious downstream analytical workflows have limited such strategies. An alternative approach utilizes important new features of the hybrid quadrupole time-of-flight technology that allow sequential precursor ion fragmentation (MS/MSALL, [Prasain et al., 2015; Simons et al., 2012; Sokol et al., 2015]). MS/MSALL employs sequential stepping through a pre-defined mass range in small increments that effectively isolates and fragments all ions within the user-defined mass range. In this workflow, precursor ions are selected in the Q1 quadrupole, at 1 mass unit (amu) resolution, in a step-wise fashion that covers the mass range of interest. Collision-induced dissociation is carried out in Q2 at high speed, while collecting more than a thousand MS/MS spectra that cover every precursor in the mass range of each cycle. Thus, the MS/MSALL approach is bias-free, and yields informative product ion spectra, even in the absence of an MS precursor ion signal, and is compatible with direct or “shotgun” infusion [Simons et al., 2012]. Such shotgun approaches are extremely efficient and have contributed to the evolution of MS methods in systems biology [Gross and Han, 2011]. Our goal was to use this approach in order to generate and compare a baseline lipid profile analysis of cell models of adipocyte differentiation with the lipid composition found in mouse white and brown adipose tissues.
Materials and Methods
Chemicals
Dichloromethane (Sigma-Aldrich), methanol and ammonium acetate (Fluka), and water (Burdick and Jackson) were LC-MS grade. Teflon coated caps and glass tubes were from VWR. 12 mm x 32 mm amber vials were from Thermo Scientific and 9 mm threaded caps were from Wheaton Inc. Internal standards (Avanti Polar Lipids, Inc.) were used for monitoring lipid class recoveries for: TAGs, D5-TAG (1,3 dipentadecanoyl-2-(9z-octadecanoyl)-sn-glycerol 2H5, Fw 810.34), GPLs, PC 17:0/14:1 (1-heptadecanoyl-2-(9z-tetradecanoyl)-sn-glycerol-phospholcholine, Fw 718.00), sphingosines, SM 18:1;2/17:0 (n-heptadecanoyl-D-erythro-sphingosylphosphocholine, Fw 717.07). Bovine heart extract (Avanti Polar Lipids, Inc.) at 0.25-2.5 ng/ml in methanol/dichloromethane/ammonium acetate infusion solvent was used for mass (MS and MS/MS) calibration.
Cell Culture
Three types of preadipocyte cultures were used in the study: 3T3-L1 mouse white preadipocytes (ZenBio), BAT-C1, brown adipose preadipocytes immortalized with SV40 large T antigen (gift of Bruce Spiegelman, Harvard University), and mouse ear-derived mesenchymal stem cells (MeMSCs). MeMSCs were isolated from C57BL/6J mice as previously described [Gawronska-Kozak, 2014; Rim et al., 2005]. Cells were cultured in 5% CO2 and maintained in DMEM/F-12 (Life Technologies) supplemented with 15% heat-inactivated FBS (Life Technologies), primocin (InvivoGen) and recombinant murine FGF-basic (10 ng/ml, Peprotech). The 3T3-L1 and BAT-C1 cell cultures were grown in DMEM with high glucose (Hyclone) supplemented with 10% fetal bovine serum (FBS, Hyclone) and 1% antibiotic-antimycotic (Gibco). Prior to lipid extraction, all cell pellets washed 5 times in 100x volume of phosphate-buffered saline containing 1 mM ethylenediaminetetraacetic acid (EDTA) to remove serum and medium lipid constituents and additives.
To induce white adipogenic differentiation, 3T3-L1 cells, grown to more than 90% confluence, were treated for 2 days in full medium supplemented with insulin (10 μg/mL), dexamethasone (1 μM), rosiglitazone (5 μM) and IBMX (0.5 mM), all reagents from Sigma-Aldrich. Two days later, the medium was aspirated, cells were washed and incubated for 5 days in full medium supplemented with insulin (10 μg/mL). After 7 totals days of chemical-induced differentiation, cells were collected by scraping in cold PBS supplemented with EDTA, and washed as described above.
For induction of brown adipogenesis, BAT-C1 cells at more than 90% confluency were treated for 2 days in full medium supplemented with insulin (0.1 μg/mL), rosiglitazone (5 μM), dexamethasone (1 μM), IBMX (0.5 mM), T3 hormone (1.5 nM) and indomethacin (125 μM), all reagents from Sigma-Aldrich. Two days later, the medium was aspirated, cells were washed and incubated for an additional 2 days in full medium supplemented with insulin (0.1 μg/mL) and T3 hormone (1.5 nM), and collected by scraping in cold PBS as noted above.
To induce adipogenic differentiation of MeMSCs, confluent cells were cultured for 48 hours in DMEM/F-12 (Life Technologies) supplemented with 5% heat-inactivated FBS (Life Technologies), primocin (InvivoGen), 10 μg/ml human insulin, 0.5 mM (IBMX) and 1 μM dexamethasone. On day 2, cells were cultured in DMEM/F-12 supplemented with 5% heat-inactivated FBS, primocin, 0.1 μg/ml human insulin and 2 μM rosiglitazone. Insulin, IBMX, dexamethasone and rosiglitazone were purchased from Sigma-Aldrich. Cells were harvested 3 days post-differentiation. Prior to analysis, cells were washed extensively in cold PBS supplemented with EDTA as described above.
Animal procedures
Mouse adipose tissues were collected from 5 week-old C57BL/6J male mice. Five independent samples from each anatomical site were collected. Mice were euthanized, and tissues were collected and immediately stored and frozen at −80°C prior to processing. From each mouse, white adipose tissue was collected from both the inguinal region and perirenally, dorsal to the kidney. Interscapular brown fat was also collected. All animals were obtained in compliance with federal, state and local laws and institutional regulations. Breeding, maintenance, and experimentation were conducted according to the NIH standards established in the Guidelines for the Care and Use of Experimental Animals. All studies were approved by the Maine Medical Center Institutional Animal Care and Use and Institutional Biosafety Committees.
Lipid extraction
Frozen tissue was cut into approximately 20 mg pieces, the wet mass noted, and subjected to homogenization in phosphate-buffered saline using a Kontes #19 glass homogenizer. For lipid analyses, 2×105 cells were pelleted in glass screw-cap tubes. Cell or tissue homogenate lipids were extracted into a dichloromethane-methanol-water monophase. Lipids were isolated using a modified Bligh and Dyer protocol [Bligh and Dyer, 1959] using dichloromethane and methanol. First, a dichloromethane/methanol/sample monophase was obtained, to which internal standards were added (e.g., odd-chain fatty acyl sphingomyelin, TAGs, and phosphatidylcholine species), and allowed to rest for 30 min to allow internal standards to equilibrate with sample lipids. Following the monophase step of the extraction procedure, aqueous and organic phases were separated by subsequent addition of dichloromethane and water, followed by centrifugation for 10 min at 300×g in a Heraeus Instruments Megafuge 2R. The lower dichloromethane organic phase was removed to a new glass tube and dried under nitrogen and a partial vacuum using a Visiprep manifold (Supelco, Bellefonte, PA). Lipid extracts were dissolved in methanol/dichloromethane (50:50, v/v, containing 5 mM ammonium acetate) for MS analysis.
Mass Spectrometry
Unbiased MS/MSALL shotgun lipidomic analyses were performed by direct infusion in 50% dichloromethane, 50% methanol, and 5 mM ammonium acetate, using the loading pump stage of a ThermoFischer/Dionex RSLC U3000 and autosampler onto a Sciex 5600 TripleTOF mass spectrometer. Lipids were analyzed using a bias-free global lipid profiling acquisition technique, termed MS/MSALL, employing sequential precursor ion fragmentation. Minimum signal-to-noise and minimum percent intensities for MS and MS/MS were: TAGs (MS 5/1 and MSMS, 10/1, 2%); SMs (MS 5/1 and MS/MS, 5/1, 2%); DAGs (MS 5/1 and MS/MS, 2/1, 2%); and GPLs (MS 5/2 and MS/MS, 10/1, 2%). Otherwise, MS/MSALL parameters were as previously published [Simons et al., 2012].
Statistical Analysis
For sample comparisons, a t-test was applied using MarkerView software (Sciex) essentially as described [Ivosev et al., 2008] . The calculated t-value was subsequently converted to a P value, yielding the probability that any difference between the two respective sample groups is statistically significant, defined as P<0.05. Briefly, MarkerView calculates a separate t-value for each peak. For a given spectral ion peak, the resultant square of the t-value for comparing group 1 to group 2 is defined as:
and <R1> and <R2> are the average responses for the spectral peaks for group 1 and group 2, n1 and n2 are the number of samples for the groups, and σ1 and σ2 are the standard deviations of the responses [Ivosev et al., 2008]. The P value calculated from the t-test was measured for each possible pair-wise comparison, and each group was also compared to all other samples. Additionally, the first acquired samples were compared to the following fourth sample to help detect potential drift for variables. Samples with a drift of greater than 10% were excluded.
Principle component analyses (PCA) utilized Pareto autoscaling, where each value is subtracted from the average and divided by the square root of the standard deviation of the value, as described previously [Ivosev et al., 2008]. PCA was guided using the sample groupings for each cell line, differentiated or undifferentiated, and adipose tissue samples. Normalization of profiles based on total deisotoped peak areas was used when comparing similar samples, e.g., adipose tissues, to better identify relative differences in lipid composition. Normalization was not used for comparing sample types exhibiting large changes in lipid composition and total amounts, e.g., between undifferentiated and differentiated cells, or between cells and adipose tissues.
Results
Mouse white and brown adipose tissues are distinguished by many lipid classes
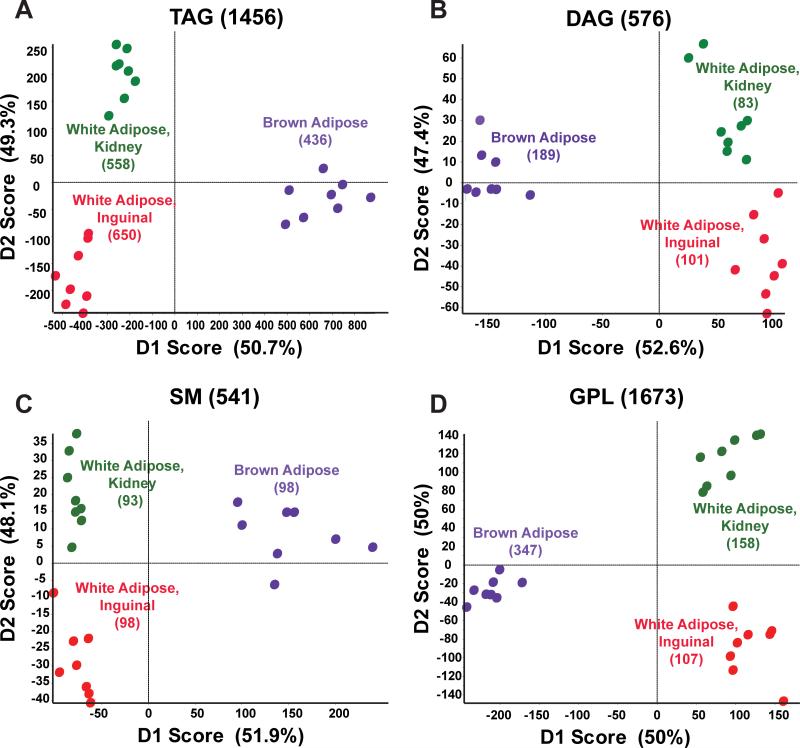
Samples of mouse white adipose tissue were taken from inguinal fat (WIF), perirenal fat (WKF), and interscapular brown adipose tissue (BAT). MS/MSALL mass spectra were obtained for four lipid class groupings: 1) triacylglycerides (TAGs), 2) diacylglycerides (DAGs) and monoalkyldiacylglycerides (MADAGs), 3) sphingomyelin (SM), ceramides (Cer), ceramide phosphates (CerP), cholesteryl esters (CE), monohexosylceramides (HexCer) and dihexosylceramides (Hex2Cer), and 4) glycerophospholipids (GPL), cardiolipins (CL), free fatty acids (FFA), phosphatidic acids (PA), phosphatidylcholines (PC), phosphatidylethanolamines (PE), phosphatidyleinositols (PI), phosphatidyleglycerols (PG), and phosphatidyleserines (PS). For all lipid constituents, the deisotoped, corrected peak intensities were used for principle component analysis as previously described [Ejsing et al., 2006]. For adipose tissue, a total of 3172 distinct molecular species were detected, with 112, 574, and 728 distinct molecular species identified as different between WKF versus WIF, BAT versus WKF and WIF, respectively (P<0.05, Table 1). Principle component analysis (PCA) indicated that although the two WAT depots clustered more tightly together with certain lipid classes, all three depots were readily resolved by any of the four lipid groupings (Fig. 1A-D).
Table 1.
Number of lipid molecular species observed and differences between groups.
| Group | Total molecular species detected | Group comparisons | Number molecular species different between comparison groups (P<0.05) |
|---|---|---|---|
| BAT | 2445 | WKF vs. WIF | 112 |
| WKF | 1810 | BAT vs. WKF | 574 |
| WIF | 1893 | BAT vs. WIF | 728 |
| 3T3-L1* | 5411 | BAT-C1 cells, undiff vs. diff | 1524 |
| BAT-C1 | 5270 | MeMSC, undiff vs. diff | 1123 |
| MeMSC | 3092 | 3T3-L1 cells, undiff vs. diff | 1114 |
Each tissue or cell population was analyzed, and total lipid species detected per group is listed. For group comparisons, the number of significantly different molecular species is indicated.
Total molecular species for the cell populations include both undifferentiated (undiff) and differentiated (diff) states.
Figure 1. Principle component analysis with Pareto scaling on MS/MSALL adipose lipids.
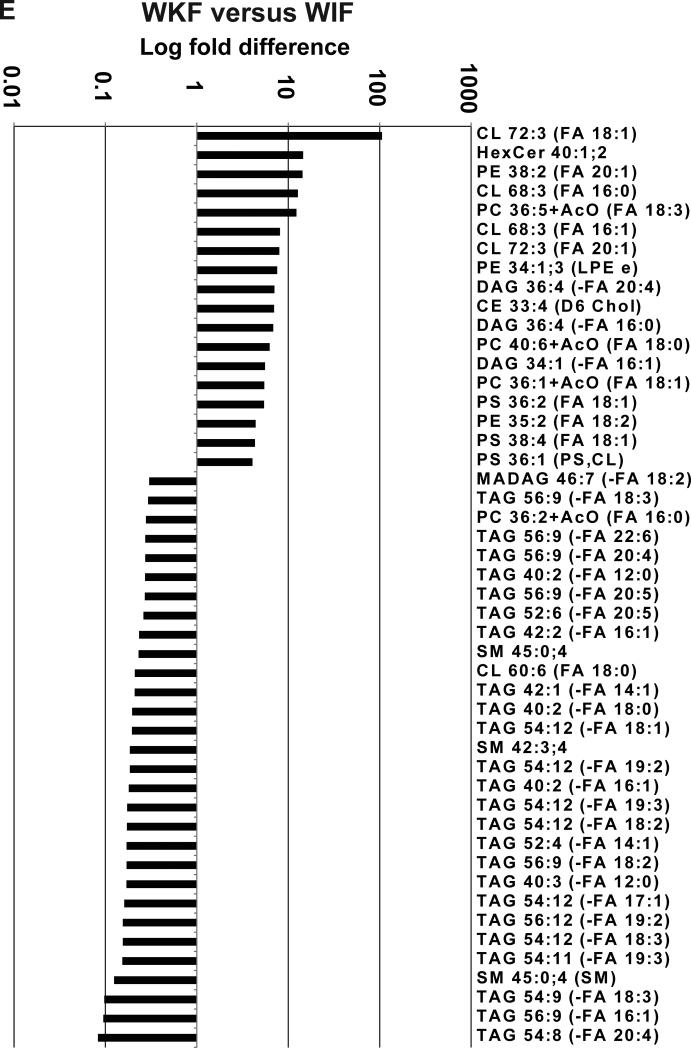
(A-D) Directed principle component analysis (PCA) discrimination of mouse adipose-derived lipid MS/MSALL spectra was conducted as described [Ivosev et al., 2008]. PCA of mass spectra data for triacylglycerols (1456 TAGs), diacylglycerols and monoalkyldiacylglycerols (576 DAGs), the SM grouping, including sphingomyelins; cholesteryl esters, ceramides, ceramide phosphates, monohexosylceramides, and dihexosylceramides (541 SMs), and the glycerophospholipid grouping, including cardiolipins, free fatty acids, phosphatidic acid, phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, phosphatidylglycerol, phosphatidylserine, lysophosphatidylcholine, and lysophosphatidylinositol (1673 GPLs). These data were derived from white adipose (inguinal and perirenal) and interscapular brown adipose depots (n=4 from each depot, analyzed in duplicate). (E, F) Shown are lipids that most differentiated between (E) perirenal and inguinal white adipose and (F) brown adipose and combined white adipose samples (P<0.05).
Data were analyzed to identify the greatest differences in lipids species between the different types of adipose tissues. Although the number of lipids that were significantly different between comparisons was extensive, examples are shown of highly quantitatively different lipid molecules when comparing white adipose tissue from perirenal versus inguinal depot (Fig. 1E) and when comparing brown adipose tissue to the combined white adipose tissues (Fig. 1F). Perirenal versus inguinal white adipose differed most from each other in that perirenal WAT had more pronounced levels of a specific cardiolipin, monohexosylceramide, and phosphatidylethanolamine (Fig. 1E, CL 72:3, HexCer 40:1;2, and PE 38:2, respectively) whereas inguinal WAT had more pronounced levels of several highly unsaturated TAGs (Fig. 1E, TAGs 54:9, 56:9, and 54:8).
Interscapular BAT lipids were similarly characterized in comparison to both WAT depots. Interestingly, interscapular BAT showed strong elevation of dihexosylceramides and sphingomyelins (Fig. 1F, Hex2Cer 34:0;3 and Hex2Cer 36:2;3, SM 38:0;2, and SM 40:1;2). Whereas longer chain 56:2 TAGs were also more abundant in BAT, less saturated, shorter acyl chain TAGs and sphingomyelin (Fig. 1F, TAG 50:5 and TAG 42:2 and SM 40:2;4) were the most predominant lipid species that significantly distinguished interscapular BAT from both WAT depots.
Cell culture models differ in the most prominent lipids upregulated during differentiation
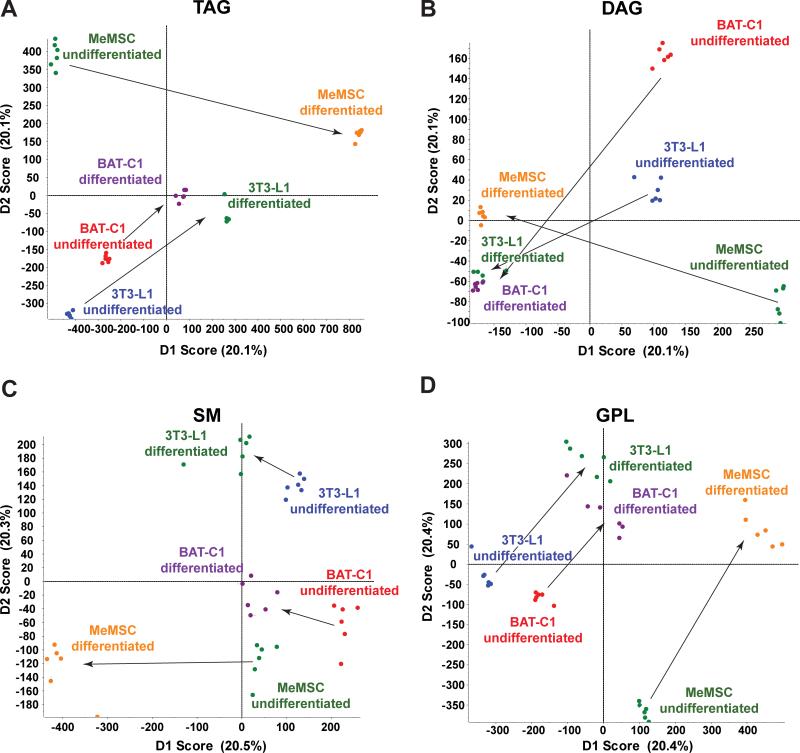
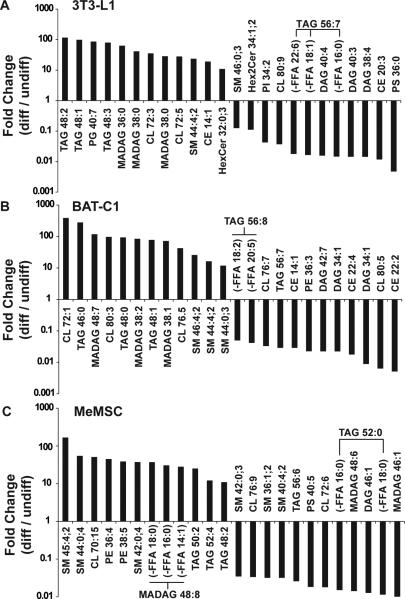
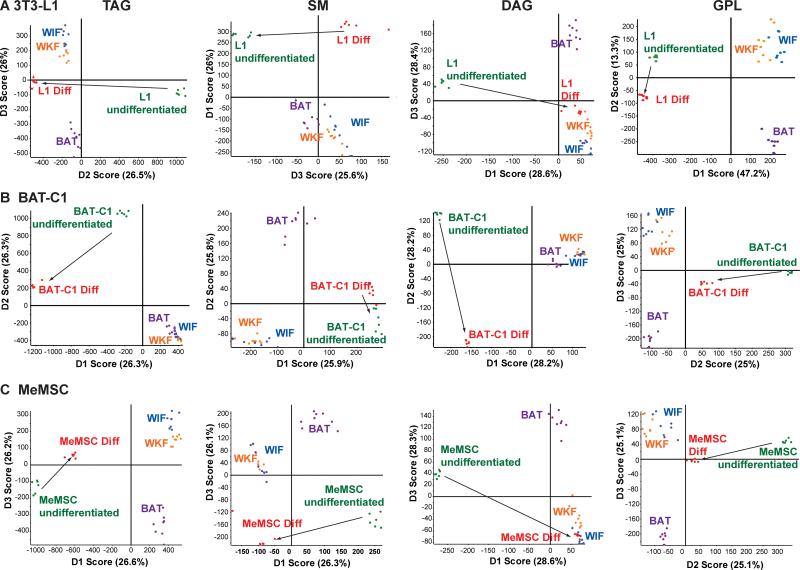
We next analyzed the lipid profiles of 3T3-L1, BAT-C1, and MeMSC cell culture models of adipocyte differentiation. 3T3-L1 cells were chosen as a long-established murine preadipocyte cell model of lipogenesis during differentiation to an adipocyte-like cell [Mackall et al., 1976]. Brown adipose-derived BAT-C1 cells [Uldry et al., 2006] were selected as a preadipocyte cell model for BAT adipogenesis. Murine ear mesenchymal preadipocyte progenitor cells (MeMSCs) were used as a primary progenitor cell that can be chemically differentiated into adipocytes with concomitant lipogenesis [Rim et al., 2005]. For 3T3-L1, BAT-C1, and MeMSC cells, we detected more total molecular lipid species compared to the in vivo tissues (Table 1). In addition, each of these cell populations had numerous lipid species that were significantly changed following induction of differentiation (Table 1, >1000 regulated in each cell population). As shown by the PCA in Fig. 2A-D, all four groupings of analyzed lipid classes readily discriminated their pre- and post-differentiated lipid compositions, indicating strong differences between these cell lines both before and after their differentiation to adipocytes. As noted above, these data were used to generate lists of the most distinctive lipid markers of cell model differentiation, using a P-value of 0.05 or less as the criteria for significance (Fig. 3A-C). Many interesting lipid molecular distinctions were evident. For example, specific sphingomyelin molecular species stood out as a potential marker of MeMSC-to-adipocyte differentiation (Fig. 3C, SM45:4;2, SM44:0;4 and SM42:0;4), whereas sphingomyelins were not the most characteristic lipid class distinguishing 3T3-L1 from BAT-C1 adipogenic differentiation (Fig. 3A-B). Specific TAGs were a major feature of 3T3-L1 adipogenic differentiation, with TAGs 48:2, 48:1, and 48:3 predominating. In contrast, longer chain TAGs appeared diminished in 3T3-L1 and BAT-C1 adipocytes (TAG 56:7 and 56:8, Fig. 3A-B). BAT-derived BAT-C1 cells, when chemically-induced to differentiate, revealed upregulated TAGs signatures. However, CL 72:1 and longer-chain cardiolipins (CL 80:3 and 76:5), as well as additional sphingomyelin species, differed from those seen in differentiated 3T3-L1 cells (Fig. 3B, SMs 46:4;2, 44:4;2, and 44:0;3 versus SMs 38:0;2 and 40:1;2, Fig. 1F). Finally, MeMSC progenitor primary cells were distinct in their differentiation-dependent upregulation of sphingomyelins and phosphatidylethanolamines (Fig. 3C). In addition, TAG 52 species were more desaturated, and the TAG 48:n class appeared less desaturated with BAT-C1 adipogenic differentiation. All three cell models showed upregulation of ether-linked monoalkyldiacylglycerols (MADAGs). However, differentiated MeMSCs differed from differentiated 3T3-L1 cells as evidenced by increasing longer-chain MADAGs, whereas BAT-C1 cells showed evidence of both shorter- and longer-chain MADAGs.
Figure 2. Lipid profiling of adipocyte cell differentiation models.
MS/MSALL mass spectra were obtained from non-differentiated and differentiated 3T3-L1, BAT-C1, and MeMSC adipocyte cell cultures models (n=3 independent differentiation experiments, analyzed in duplicate). (A-D) PCA of mass spectra for TAG, DAG, SM group and GPL group, as described above. Arrows highlight the transition from the undifferentiated to the differentiated state.
Figure 3. Characterization of lipids in cell models of adipogenic differentiation.
3T3-L1 (A), BAT-C1 (B), and MeMSC (C) were analyzed before and after induction of differentiation.Shown are lipid species that were significantly different (P <0.05) and showing greater than 10-fold change. Fold changes represent the levels of lipids between differentiated (diff) versus undifferentiated (undiff) cells.
Restricted subsets of distinct lipids relate different adipogenesis models to adipose tissue
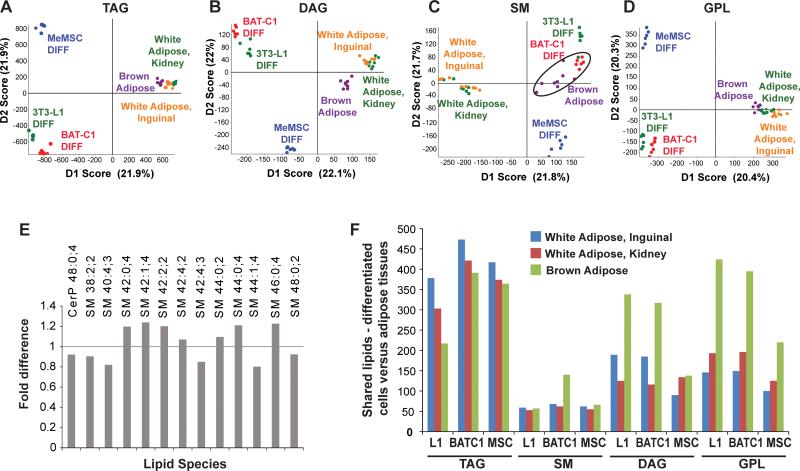
Lipid class PCA data were assessed for each cell model of differentiation in comparison to WAT and BAT, to ascertain the lipids that best related cell culture models of adipogenesis to adipose tissue. Each panel in Figure 4 shows the cell differentiation PCA versus adipose tissue for the four lipid class groupings. This survey suggests that TAGs and DAG/MADAGs (Fig. 4A, 4C), but neither SMs, CERs, CEs, nor GPLs as a group (Fig. 4B, 4D), most related 3T3-L1 cell adipogenic differentiation to WAT. In terms of BAT-C1 differentiation, no markers showed strong correlation with WAT. However, the PCA of GPL-class changes of expression in BAT-C1 during differentiation showed moderate correspondence to BAT (Fig. 4H, arrow). For MeMSC adipocyte progenitor cells, most lipid classes indicated that MeMSC adipogenic differentiation was toward a WAT-like profile (Fig. 4I, 4K, 4L). In particular, and consistent with 3T3-L1 cells, changes in MeMSC DAG/MADAGs during adipogenic differentiation were strongly characteristic of both WAT depots (Fig. 4C, 4K).
Figure 4. Principle component analysis of cell models and adipose tissue.
MS/MSALL spectra for undifferentiated and differentiated cells for (A) 3T3-L1, (B) BAT-C1, and (C) MeMSC (n=3 each, analyzed in duplicate) were compared with data from inguinal, perirenal, and brown adipose tissue (n=4 each, analyzed in duplicate)..
To gain further insight into the identities of specific lipids that relate each cell model to adipose depot lipids, and because the undifferentiated cells of these three cell models reflect disparate cell types and tissue sources, we restricted comparison of the differentiated cell types to adipose tissue. As shown in Fig. 5A-D, all differentiated cell culture models were highly distinct from WAT and BAT. As noted above, MS/MSALL data resolve inguinal and perirenal white adipose tissue depots from each other and from BAT, though these components appear compressed when viewed on the same scale as the differentiated cell culture models. However, BAT-C1 adipocytes showed striking overlap with BAT when the SM-group classes were sampled (Fig. 5C, circled). The highest lipids that contributed most strongly to BATC1:BAT similarities were ceramide 48:0 and a broad range of specific sphingomyelins (Fig. 5E). In contrast, differentiated 3T3-L1 and MeMSCs were less similar from the three lipid depots as seen by global lipid profiling. Thus, an overall count of detected lipids that were similar between the differentiated cell models and adipose tissues indicated large numbers of overlapping species of lipids. These lipid species included sphingomyelins in BAT-C1 cells showing overlap with interscapular BAT, many TAG species were shared between all three differentiated cell culture models and adipose sources, with 3T3-L1 cells showing the greatest similarity to TAG expression in inguinal WAT, and strongly shared DAG and GPL signatures in all three cell models overlapping with BAT (Fig. 5F). Interestingly, despite these overlapping similarities, the expression of numerous lipids was restricted to the adipogenic cell culture models, and not among the prominent lipids found in adipose tissue. These data suggest that the adipogenic cell culture models, in comparison to white and brown adipose tissues, have many distinctive lipid features that include both overlapping and distinct species, and knowing which specific lipids fall into each group is critical for insights into which potential metabolic control points may be the most relevant to processes of interest such as tissue-precursor-specific adipogenesis.
Figure 5. Analysis of differentiated cell models and adipose tissues.
(A-D) Principle component analysis of lipids from differentiated 3T3-L1, BAT-C1, and MeMSC compared with adipose tissues. Circled points in panel C indicate concordance between differentiated BAT-C1 cells and brown adipose tissue. (E) Shown are lipids that are the most similar between differentiated BAT-C1 cells and brown adipose tissue. (F) The number of shared lipids detected in the cell models and adipose tissues, for major lipid categories.
Discussion
By combining a dichloromethane/methanol modified Bligh and Dyer extraction procedure and tandem MS using an MS/MSALL [Prasain et al., 2015; Simons et al., 2012; Sokol et al., 2015] direct infusion, or “shotgun”, workflow, we obtained detailed lipid profiles of three models of preadipocyte-to-adipocyte differentiation and ascertained the relationships of lipid species of these widely used (3T3-L1) and more recently developed (BAT-C1 and MeMSC) mouse adipogenic cell culture models, and mouse white and brown adipose tissues. We undertook this study to generate a lipidomic baseline comparing the lipid expression patterns of mouse cell model adipogenesis to adipose tissue, obtained using the MS/MSALL workflow, with future applications including the study of mouse models of adipose tissue homeostasis and disease.
In our protocol, internal standards were primarily used for confirmation and normalization of the recovery of lipids. However, for our analyses, normalization of results to internal standards yielded no significant differences in the relative levels observed for lipids (data not shown). This observation persisted regardless of whether software-based normalization of the data via MarkerView software processing was used. Thus, our data suggest that the MS/MSALL workflow and downstream data analysis based on principle component analyses and analysis of significant changes, is robust and reproducible, with the primary utility of single-lipid-class internal standardization being for sample recovery adjustments and high precision lipid quantitation that is needed for the resolution of subtle differences in absolute lipid levels. Under these experimental conditions, absolute lipid quantitation is not possible due to differential efficiency of fragmentation based on acyl chain length and the degree of unsaturation.
Detailed global lipid data are consistent with previous studies indicating strong lipid differences based on anatomical location and the specific type of adipose. For example, Hoene et al., [Hoene et al., 2014] used tandem LC-MS to profile mouse brown and white adipose tissue. Moreover, our data (Fig. 1F) showing SM 38:0;2, SM 40:1;2 and PE 38:2 increase in interscapular BAT versus WAT depots, are in agreement with their finding that sphingomyelins and phosphatidylethanolamines are among the lipids most enriched in interscapular BAT versus WAT. In addition, our method revealed many significant differences between the two WAT depots (perirenal and inguinal) in this study. Though Hoene et al., [Hoene et al., 2014] did not find many differences between subcutaneous and inguinal white fat, they did observe a significant decrease in HexCer in inguinal WAT relative to subcutaneous WAT, thus agreeing with our data, which suggests that HexCer (40:1;2) was significantly increased in perirenal WAT versus inguinal WAT depots (Fig. 1E).
Our PCA data indicate that perirenal WAT was distinct from inguinal WAT in terms of their respective lipid profiles. Moreover, the strongest lipid contributions between groups, such as the differences between perirenal and inguinal WAT, were determined by chi-square and associated P values with respect to their fold differences. These data indicated that inguinal white fat is richer in TAGs whereas the perirenal adipose depot is rich in hexosylceramides, cardiolipins, and phosphatidyl species.
At this level of analysis, there appeared to be many more differences than similarities when comparing the detailed molecular composition of lipids present in various adipose depots. This is most obvious in the number of lipids with significant expression levels (greater than 100 counts, with a P<0.05) that were restricted to only one adipose depot. Thus, the data shown in Table 2 suggested that perirenal WAT contains substantial levels of unique cardiolipins, phosphatidylserines and phosphatidylcholine, as compared to inguinal WAT. In contrast, though the lipids unique to inguinal WAT included cardiolipin and phosphatidylcholine, this depot is predominantly distinguished from perirenal WAT by its content of medium acyl chain length TAGs (Table 2). When each WAT depot was compared to the other two combined depots (in order to determine which lipids were unique to each adipose tissue depot), inguinal WAT was unique only with respect to the expression of certain TAG species (Table 3). In contrast, perirenal WAT was uniquely distinguished due to its content of cardiolipin CL 64:3. When this selection criteria (e.g., greater than 100 counts, P<0.05) was applied to BAT as compared to the two WAT depots (perirenal and inguinal), we observed that cardiolipins and specific glycerophosphatides, as well as TAGs with fewer but longer acyl chains, were unique to BAT (Table 3).
Table 2.
Pair-wise comparison: Lipids unique to one white adipose tissue depot.
| Depot | Lipid | Counts |
|---|---|---|
| WKF | CL 72:3 (FA 18:0) | 267 |
| PS 36:1 (PS,CL) | 237 | |
| PC 40:6 (FA 18:0) | 231 | |
| PS 38:4 (FA 18:1) | 198 | |
| CL 68:3 (FA 16:1) | 190 | |
| WIF | TAG 40:0 (-FA 12:0) | 316 |
| TAG 54:12 (-FA 18:2) | 234 | |
| TAG 40:1 (-FA 12:0) | 232 | |
| PC 36:2 (FA 16:0) | 230 | |
| TAG 44:3 (-FA 18:2) | 213 | |
| CL 68:3 (FA 16:0) | 212 | |
| TAG 40:0 (-FA 16:0) | 206 | |
| TAG 48:5 (-FA 18:2) | 172 | |
| TAG 40:1 (-FA 18:1) | 159 | |
| TAG 50:8 (-FA 16:0) | 159 |
For white adipose tissue, molecular species are listed that are restricted to that depot when comparing WKF to WIF (P<0.05). Counts listed are all at least two-fold above background.
Table 3.
Three-way comparisons: Lipids unique to a single adipose tissue.
| WIF vs. (WKF and BAT) | Counts | P value |
|---|---|---|
| TAG 40:1 (-FA 12:0) | 316 | 1.82E-06 |
| TAG 44:3 (-FA 18:2) | 232 | 2.09E-05 |
| TAG 40:0 (-FA 16:0) | 213 | 2.19E-06 |
| TAG 48:5 (-FA 18:2) | 206 | 1.65E-05 |
| TAG 40:1 (-FA 18:1) | 172 | 1.00E-04 |
| TAG 50:8 (-FA 16:0) | 159 | 1.40E-04 |
| TAG 58:9 (-FA 22:6) | 159 | 1.00E-04 |
| TAG 44:3 (-FA 16:1) | 145 | 0.01123 |
| TAG 40:0 (-FA 14:0) | 141 | 2.35E-06 |
| TAG 48:5 (-FA 18:3) | 131 | 0.01099 |
| TAG 44:3 (-FA 18:3) | 111 | 2.71E-03 |
| TAG 58:8 (-FA 22:6) | 110 | 2.77E-05 |
| TAG 40:1 (-FA 12:0) | 103 | 0.01079 |
| WKF vs. (WIF and BAT) | Counts | P value |
|---|---|---|
| CL 64:3 (FA 16:1) | 102 | 0.04239 |
| BAT vs. (WKF and WIF) | Counts | P value |
|---|---|---|
| CL 70:7 (FA 18:2) | 3887 | 3.65E-06 |
| SM 38:1;2 | 2552 | 0.00027 |
| CL 50:2 (FA 18:2) | 1093 | 3.74E-05 |
| PA 38:3 (FA 18:1) | 1010 | 0.0002 |
| PE 35:3 (FA 18:2) | 674 | 0.00014 |
| PC 33:2 (FA 18:2) | 519 | 0.01202 |
| SM 38:2;2 | 435 | 1.84E-05 |
| PG 40:6 (FA 18:1) | 286 | 2.47E-05 |
| PC 30:1 (FA 18:0) | 280 | 0.0433 |
| PA 36:3 (FA 18:2) | 234 | 0.01095 |
| PE 39:7 (FA 22:6) | 215 | 0.00259 |
| SM 40:2;3 | 212 | 0.01204 |
| DAG 32:0 (-FA 16:0) | 201 | 2.58E-05 |
| PE 38:6 (FA 18:1) | 191 | 2.44E-06 |
| CL 50:1 (FA 18:1) | 174 | 0.00091 |
| TAG 56:2 (-FA 18:1) | 162 | 0.04197 |
| TAG 56:2 (-FA 18:2) | 160 | 2.83E-05 |
| PE 38:6 (FA 20:5) | 135 | 0.0001 |
| PE 38:1;1 | 126 | 2.36E-06 |
| PE 34:4 (FA 18:3) | 119 | 7.97E-06 |
| PS 31:5 | 117 | 4.66E-06 |
| PI 26:7 | 116 | 2.67E-06 |
Shown are lipids that were restricted to the tissue type (P<0.05), above 100 counts, or greater than two-fold above background.
Lipids that appeared to be the most similar between the two WAT depots were predominantly longer acyl chain length TAGs, and relatively fewer glycerophosphatides (PC, PS) and a MADAG, suggesting that these WAT and BAT depots are predominantly distinguished by their specific cardiolipins and glycerophosphatides (Table 4).
Table 4.
Lipids most similar between adipose tissues.
| Depot | Lipid | Fold difference |
|---|---|---|
| WKF and WIF | PE 35:2 (FA 18:2) | 4.4 |
| PS 38:4 (FA 18:1) | 4.3 | |
| PS 36:1 (PS,CL) | 4.1 | |
| MADAG 46:7 (-FA 18:2) | 0.3 | |
| TAG 56:9 (-FA 18:3) | 0.3 | |
| PC 36:2 (FA 16:0) | 0.3 | |
| TAG 56:9 (-FA 22:6) | 0.3 | |
| TAG 56:9 (-FA 20:4) | 0.3 | |
| TAG 40:2 (-FA 12:0) | 0.3 | |
| TAG 56:9 (-FA 20:5) | 0.3 | |
| TAG 52:6 (-FA 20:5) | 0.3 | |
| BAT and WAT (WKF and WIF) | PC 36:4 (FA 18:2) | 1.9 |
| SM 45:4;2 | 1.9 | |
| PE 38:4 (FA 20:4) | 1.9 | |
| SM 48:2;2 | 0.4 | |
| TAG 50:3 (-FA 20:3) | 0.4 |
Shown are those lipids that were least changed between the indicated tissues, detected at least two-fold above background, and less than 5-fold different.
While our data sets provide detailed lipid composition profiles of adipose depots, these data are not specific to adipocytes per se. WAT and BAT are complex tissues containing many different cell types, including stromal cells and blood vessel-associated endothelial and smooth muscle cells, whose distinctive lipid profiles contribute to our sample complexity. Thus, in vitro cell culture systems are important tools for understanding and comparing preadipocyte contributions to the synthesis or degradation of specific lipids during the process of adipogenic differentiation.
Although the cell culture models showed profound visual accumulation of waxy lipid droplets and gross changes in morphology (data not shown), these cell models exhibited overlapping yet highly distinctive patterns of both lipid accumulation and loss with the chemical induction of adipocyte differentiation. Thus, in vitro modeling of the metabolic changes that occur during adipogenic differentiation, as evidenced by unbiased lipid expression profiling, provides a potentially rich source of information regarding the specific metabolic processes that accompany adipogenesis. First, our studies sought to compare the changes in lipid content for different preadipocyte cell models of adipogenic differentiation. Using a chemical-induced differentiation approach, we determined that the lipid expression profiles of our three cell models differed with respect to the lipids most characteristic of their respective adipogenic transitions. For example, differentiation of the extensively utilized 3T3-L1 cell model resulted in accumulation of TAGs as expected. Interestingly, the most predominant TAGs were lower carbon number and more saturated TAGs 48:(1-3), whereas longer, more extensively desaturated TAG 56:7s were not as highly expressed (Fig. 3). This result suggests utilization of longer chain unsaturated C18-C20 fatty acids during the process of 3T3-L1 preadipocyte-to-adipocyte differentiation. This trend in fatty acyl chain length utilization during 3T3-L1 differentiation was accompanied by a change in hexosylceramides, sphingomyelins, and cholesteryl esters (e.g., HexCer32:0;3 versus Hex2Cer34:1;2; SM44:4;2 versus 46:0;3; and CE14:1 vs. CE20:3, respectively), which suggests that these species are important to the metabolic demands of 3T3-L1 differentiation. Finally, our data suggested that diacylglycerides were reduced and monoalkyldiacylglycerides appeared increased during 3T3-L1 preadipocyte-to-adipocyte differentiation. This result is reminiscent of a previous study in which comparison of the lipids characteristic of rat keratinocytes and intact epidermis suggests that the lipid profile of cultured keratinocytes was consistent with monoalkyldiacylglyceride accumulation [Oku et al., 1993].
The changes in lipids seen during BAT-derived BAT-C1 cell differentiation showed both overlapping, but also strongly distinctive trends as compared to 3T3-L1 cells. Consistent with 3T3-L1 adipogenic differentiation, induction of BAT-C1 differentiation yielded MADAGs, and longer and more unsaturated TAGs (e.g., TAGs 56:8 and TAG 56:7) decreased, while the saturated TAGs 46:0 and 48:0, the monounsaturated TAG 48:1, and cardiolipin CL 72:n were increased. However, BAT-C1 differentiation was distinct from 3T3-L1 differentiation in that longer chain species of cardiolipins (CL 76:5,and CL 80:3, versus CL 72:n) and sphingomyelins (e.g., SM 44:0 and 46:0) were upregulated, in addition to SM 44:4;2, which was a shared feature of 3T3-L1 and BAT-C1 differentiation. Moreover, an apparent shift toward saturation of longer acyl chain cardiolipins (e.g., CL 80:3 from CL 80:5) was only observed during BAT-C1 adipogenic differentiation.
As with BAT-C1 cells, our primary cell model, MeMSCs, has not been previously characterized with respect to its lipid expression profile concomitant to adipogenic differentiation. For these cells, chemical induction of adipogenesis yielded sphingomyelins, including an odd chain length species (e.g., SM 45:4;2, SM 44:0;4, and SM 42:0;4) and phosphatidylethanolamine species (PE 36:4 and PE 38:5, Fig. 3C), as well as the reduced predominance of TAGs and shorter chain sphingomyelins; furthermore, MADAG 46:1 appeared to be a distinctive lipid feature of MeMSCs following chemical induction of its adipogenic differentiation. Interestingly, this result is consistent with previous studies that show differentiated 3T3-L1 cells acquiring the capacity to alpha oxidize unbranched fatty acids, leading to the presence of odd-chain-length fatty acids in all major lipid classes [Su et al., 2004]. Thus, our MeMSC data corroborated this oxidative trend seen in 3T3-L1 cells by showing that the expression of odd-chain fatty acids may not simply be a feature of 3T3-L1 adipogenic differentiation, but can occur in primary cells as well.
Finally, comparison of the differentiated cell models with adipose tissue revealed many differences between our in vitro models and mouse perirenal and inguinal WAT, or interscapular BAT. For example, because changes in 3T3-L1 cell TAG and DAG MS/MSALL profiles show correlative behavior similar to WAT (Fig. 4C), these cells, as well as MeMSCs as a primary cell model (Fig. 4K), may be better suited for the study of DAG/TAG metabolism in WAT. Similarly, our data suggested that MeMSCs (given the specific conditions of its culture and differentiation, Materials and Methods) are a broadly applicable cell model to WAT but not BAT. Finally, BAT-derived BAT-C1 cells appeared to proceed on a trajectory toward the BAT lineage, where sphingomyelin-class lipids and glycerophosphatide lipids are considered markers of brown adipogenic differentiation (Fig. 4F, 4H, Fig. 5C). Thus, in the context of distinguishing lipid expression in our cell models versus adipose tissues, specific lipid classes were determined and characterized that may better correlate specific cell models of adipogenic differentiation to anatomically-targeted adipose tissues.
The MS/MSALL workflow provides a platform for comprehensive lipidome profiling across a wide range of adipogenic cell models and adipose tissues that will facilitate functional studies to elucidate the role of the progression of specific lipids from preadipocyte progenitor to diverse adipose types, and will also enable the identification of specific lipids that may serve as diagnostic or prognostic disease biomarkers. Based on these insights, our work may guide subsequent investigations that seek to leverage the biologically relevant metabolic aspects of in vitro cell culture models of adipogenesis in comparison to specific anatomical and metabolic adipose tissue.
Acknowledgments
Funding: American Heart Association GRNT20460045 (CPHV), National Institutes of General Medical Science 8P30GM103392 (REF); NIH Grants and 5R01HL10965 (LL), and Maine Medical Center.
Abbreviations
- WAT
white adipose tissue
- WIF
white inguinal fat
- WKF
white perirenal kidney fat
- BAT
interscapular brown adipose tissue
- TAG
triacylglycerol
- DAG
diacylglycerol
- MADAG
monoalkyldiacylglycerol
- SM
sphingomyelin
- CE
cholesteryl ester
- Cer
ceramide
- CerP
ceramide phosphate
- HexCer
monohexosylceramide
- Hex2Cer
dihexosylceramide
- GPL
glycerophospholipid
- CL
cardiolipin
- FFA
free fatty acid
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PG
phosphatidylglycerol
- PS
phosphatidylserine
- LPC
lysophosphatidylcholine
- LPI
lysophosphatidylinositol
- PCA
principle component analysis
- MeMSC
mouse ear mesenchymal progenitor cell
- BAT-C1
brown adipose tissue derived cell line-1
References
- Arai Y, Sampaio JL, Wilsch-Brauninger M, Ettinger AW, Haffner C, Huttner WB. Lipidome of midbody released from neural stem and progenitor cells during mammalian cortical neurogenesis. Front Cell Neurosci. 2015;9:325. doi: 10.3389/fncel.2015.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–7. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brun RP, Spiegelman BM. PPAR gamma and the molecular control of adipogenesis. J Endocrinol. 1997;155:217–8. doi: 10.1677/joe.0.1550217. [DOI] [PubMed] [Google Scholar]
- Cifkova E, Holcapek M, Lisa M, Vrana D, Melichar B, Student V. Lipidomic differentiation between human kidney tumors and surrounding normal tissues using HILIC-HPLC/ESI-MS and multivariate data analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2015;1000:14–21. doi: 10.1016/j.jchromb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- De Leon H, Boue S, Szostak J, Peitsch MC, Hoeng J. Systems Biology Research into Cardiovascular Disease: Contributions of Lipidomics-based Approaches to Biomarker Discovery. Curr Drug Discov Technol. 2015;12:129–54. doi: 10.2174/1570163812666150702123319. [DOI] [PubMed] [Google Scholar]
- Drew BG, Carey AL, Natoli AK, Formosa MF, Vizi D, Reddy-Luthmoodoo M, Weir JM, Barlow CK, van Hall G, Meikle PJ, Duffy SJ, Kingwell BA. Reconstituted high-density lipoprotein infusion modulates fatty acid metabolism in patients with type 2 diabetes mellitus. J Lipid Res. 2011;52:572–81. doi: 10.1194/jlr.P012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem. 2006;78:6202–14. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- Gawronska-Kozak B. Preparation and differentiation of mesenchymal stem cells from ears of adult mice. Methods Enzymol. 2014;538:1–13. doi: 10.1016/B978-0-12-800280-3.00001-3. [DOI] [PubMed] [Google Scholar]
- Gorden DL, Ivanova PT, Myers DS, McIntyre JO, VanSaun MN, Wright JK, Matrisian LM, Brown HA. Increased diacylglycerols characterize hepatic lipid changes in progression of human nonalcoholic fatty liver disease; comparison to a murine model. PLoS One. 2011;6:e22775. doi: 10.1371/journal.pone.0022775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorden DL, Myers DS, Ivanova PT, Fahy E, Maurya MR, Gupta S, Min J, Spann NJ, McDonald JG, Kelly SL, Duan J, Sullards MC, Leiker TJ, Barkley RM, Quehenberger O, Armando AM, Milne SB, Mathews TP, Armstrong MD, Li C, Melvin WV, Clements RH, Washington MK, Mendonsa AM, Witztum JL, Guan Z, Glass CK, Murphy RC, Dennis EA, Merrill AH, Jr., Russell DW, Subramaniam S, Brown HA. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J Lipid Res. 2015;56:722–36. doi: 10.1194/jlr.P056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- Gross RW, Han X. Lipidomics at the interface of structure and function in systems biology. Chem Biol. 2011;18:284–91. doi: 10.1016/j.chembiol.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinterwirth H, Stegemann C, Mayr M. Lipidomics: quest for molecular lipid biomarkers in cardiovascular disease. Circ Cardiovasc Genet. 2014;7:941–54. doi: 10.1161/CIRCGENETICS.114.000550. [DOI] [PubMed] [Google Scholar]
- Hoene M, Li J, Haring HU, Weigert C, Xu G, Lehmann R. The lipid profile of brown adipose tissue is sex-specific in mice. Biochim Biophys Acta. 2014;1842:1563–70. doi: 10.1016/j.bbalip.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Horn PJ, Chapman KD. Lipidomics in tissues, cells and subcellular compartments. Plant J. 2012;70:69–80. doi: 10.1111/j.1365-313X.2011.04868.x. [DOI] [PubMed] [Google Scholar]
- Hyotylainen T, Oresic M. Analytical Lipidomics in Metabolic and Clinical Research. Trends Endocrinol Metab. 2015;26:671–3. doi: 10.1016/j.tem.2015.08.006. [DOI] [PubMed] [Google Scholar]
- Isaac G. Electrospray ionization tandem mass spectrometry (ESI-MS/MS)-based shotgun lipidomics. Methods Mol Biol. 2011;708:259–75. doi: 10.1007/978-1-61737-985-7_16. [DOI] [PubMed] [Google Scholar]
- Ivosev G, Burton L, Bonner R. Dimensionality reduction and visualization in principal component analysis. Anal Chem. 2008;80:4933–44. doi: 10.1021/ac800110w. [DOI] [PubMed] [Google Scholar]
- Jove M, Moreno-Navarrete JM, Pamplona R, Ricart W, Portero-Otin M, Fernandez-Real JM. Human omental and subcutaneous adipose tissue exhibit specific lipidomic signatures. FASEB J. 2014;28:1071–81. doi: 10.1096/fj.13-234419. [DOI] [PubMed] [Google Scholar]
- Jung HR, Sylvanne T, Koistinen KM, Tarasov K, Kauhanen D, Ekroos K. High throughput quantitative molecular lipidomics. Biochim Biophys Acta. 2011;1811:925–34. doi: 10.1016/j.bbalip.2011.06.025. [DOI] [PubMed] [Google Scholar]
- Kien CL, Bunn JY, Fukagawa NK, Anathy V, Matthews DE, Crain KI, Ebenstein DB, Tarleton EK, Pratley RE, Poynter ME. Lipidomic evidence that lowering the typical dietary palmitate to oleate ratio in humans decreases the leukocyte production of proinflammatory cytokines and muscle expression of redox-sensitive genes. J Nutr Biochem. 2015;26:1599–606. doi: 10.1016/j.jnutbio.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaziere A, Farabos D, Wolf C, Quinn PJ. The deficit of lipid in cultured cells contrasted with clinical lipidomics. Mol Nutr Food Res. 2013;57:1401–9. doi: 10.1002/mnfr.201200741. [DOI] [PubMed] [Google Scholar]
- Lappas M, Mundra PA, Wong G, Huynh K, Jinks D, Georgiou HM, Permezel M, Meikle PJ. The prediction of type 2 diabetes in women with previous gestational diabetes mellitus using lipidomics. Diabetologia. 2015;58:1436–42. doi: 10.1007/s00125-015-3587-7. [DOI] [PubMed] [Google Scholar]
- Layre E, Moody DB. Lipidomic profiling of model organisms and the world's major pathogens. Biochimie. 2013;95:109–15. doi: 10.1016/j.biochi.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackall JC, Student AK, Polakis SE, Lane MD. Induction of lipogenesis during differentiation in a “preadipocyte” cell line. J Biol Chem. 1976;251:6462–4. [PubMed] [Google Scholar]
- Marcher AB, Loft A, Nielsen R, Vihervaara T, Madsen JG, Sysi-Aho M, Ekroos K, Mandrup S. RNA-Seq and Mass-Spectrometry-Based Lipidomics Reveal Extensive Changes of Glycerolipid Pathways in Brown Adipose Tissue in Response to Cold. Cell Rep. 2015;13:2000–13. doi: 10.1016/j.celrep.2015.10.069. [DOI] [PubMed] [Google Scholar]
- McEvoy J, Baillie RA, Zhu H, Buckley P, Keshavan MS, Nasrallah HA, Dougherty GG, Yao JK, Kaddurah-Daouk R. Lipidomics reveals early metabolic changes in subjects with schizophrenia: effects of atypical antipsychotics. PLoS One. 2013;8:e68717. doi: 10.1371/journal.pone.0068717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy GD, Tammireddy SR, Maskrey BH, Grant L, Doherty MK, Watson DG, Delibegovic M, Whitfield PD, Mody N. Fenretinide mediated retinoic acid receptor signalling and inhibition of ceramide biosynthesis regulates adipogenesis, lipid accumulation, mitochondrial function and nutrient stress signalling in adipocytes and adipose tissue. Biochem Pharmacol. 2015 doi: 10.1016/j.bcp.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle PJ, Wong G, Barlow CK, Kingwell BA. Lipidomics: potential role in risk prediction and therapeutic monitoring for diabetes and cardiovascular disease. Pharmacol Ther. 2014;143:12–23. doi: 10.1016/j.pharmthera.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Min HK, Lim S, Chung BC, Moon MH. Shotgun lipidomics for candidate biomarkers of urinary phospholipids in prostate cancer. Anal Bioanal Chem. 2011;399:823–30. doi: 10.1007/s00216-010-4290-7. [DOI] [PubMed] [Google Scholar]
- Oku H, Urahashi A, Yagi N, Nagata J, Chinen I. Fatty acid and lipid composition in vitro and in vivo of rat epidermis. Comp Biochem Physiol B. 1993;105:293–9. doi: 10.1016/0305-0491(93)90232-t. [DOI] [PubMed] [Google Scholar]
- Park BH, Kim DS, Won GW, Jeon HJ, Oh BC, Lee Y, Kim EG, Lee YH. Mammalian ste20-like kinase and SAV1 promote 3T3-L1 adipocyte differentiation by activation of PPARgamma. PLoS One. 2012;7:e30983. doi: 10.1371/journal.pone.0030983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasain JK, Wilson L, Hoang HD, Moore R, Miller MA. Comparative Lipidomics of Caenorhabditis elegans Metabolic Disease Models by SWATH Non-Targeted Tandem Mass Spectrometry. Metabolites. 2015;5:677–96. doi: 10.3390/metabo5040677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quehenberger O, Koller E, Jurgens G, Esterbauer H. Investigation of lipid peroxidation in human low density lipoprotein. Free Radic Res Commun. 1987;3:233–42. doi: 10.3109/10715768709069788. [DOI] [PubMed] [Google Scholar]
- Rim JS, Mynatt RL, Gawronska-Kozak B. Mesenchymal stem cells from the outer ear: a novel adult stem cell model system for the study of adipogenesis. FASEB J. 2005;19:1205–7. doi: 10.1096/fj.04-3204fje. [DOI] [PubMed] [Google Scholar]
- Sampaio JL, Gerl MJ, Klose C, Ejsing CS, Beug H, Simons K, Shevchenko A. Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci U S A. 2011;108:1903–7. doi: 10.1073/pnas.1019267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons B, Kauhanen D, Sylvanne T, Tarasov K, Duchoslav E, Ekroos K. Shotgun Lipidomics by Sequential Precursor Ion Fragmentation on a Hybrid Quadrupole Time-of-Flight Mass Spectrometer. Metabolites. 2012;2:195–213. doi: 10.3390/metabo2010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol E, Almeida R, Hannibal-Bach HK, Kotowska D, Vogt J, Baumgart J, Kristiansen K, Nitsch R, Knudsen J, Ejsing CS. Profiling of lipid species by normal-phase liquid chromatography, nanoelectrospray ionization, and ion trap-orbitrap mass spectrometry. Anal Biochem. 2013;443:88–96. doi: 10.1016/j.ab.2013.08.020. [DOI] [PubMed] [Google Scholar]
- Sokol E, Ulven T, Faergeman NJ, Ejsing CS. Comprehensive and quantitative profiling of lipid species in human milk, cow milk and a phospholipid-enriched milk formula by GC and MS/MS. Eur J Lipid Sci Technol. 2015;117:751–759. doi: 10.1002/ejlt.201400575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Han X, Yang J, Mancuso DJ, Chen J, Bickel PE, Gross RW. Sequential ordered fatty acid alpha oxidation and Delta9 desaturation are major determinants of lipid storage and utilization in differentiating adipocytes. Biochemistry. 2004;43:5033–44. doi: 10.1021/bi035867z. [DOI] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–41. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Wang M, Han X. Multidimensional mass spectrometry-based shotgun lipidomics. Methods Mol Biol. 2014;1198:203–20. doi: 10.1007/978-1-4939-1258-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood PL. Mass spectrometry strategies for clinical metabolomics and lipidomics in psychiatry, neurology, and neuro-oncology. Neuropsychopharmacology. 2014;39:24–33. doi: 10.1038/npp.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Xie Y, Bucher NL, Farmer SR. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 1995;9:2350–63. doi: 10.1101/gad.9.19.2350. [DOI] [PubMed] [Google Scholar]
- Yang K, Cheng H, Gross RW, Han X. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal Chem. 2009;81:4356–68. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Chen S, Liang X, Zhang H. Development of a mass-spectrometry-based lipidomics platform for the profiling of phospholipids and sphingolipids in brain tissues. Anal Bioanal Chem. 2015;407:6543–55. doi: 10.1007/s00216-015-8822-z. [DOI] [PubMed] [Google Scholar]
- Zhao YY, Cheng XL, Lin RC. Lipidomics applications for discovering biomarkers of diseases in clinical chemistry. Int Rev Cell Mol Biol. 2014;313:1–26. doi: 10.1016/B978-0-12-800177-6.00001-3. [DOI] [PubMed] [Google Scholar]
- Zhou X, Mao J, Ai J, Deng Y, Roth MR, Pound C, Henegar J, Welti R, Bigler SA. Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS One. 2012;7:e48889. doi: 10.1371/journal.pone.0048889. [DOI] [PMC free article] [PubMed] [Google Scholar]