Synopsis
Late endocytic membrane trafficking delivers target materials and newly synthesized hydrolases into lysosomes and is critical for maintaining an efficient degradation process and cellular homoeostasis. Although some features of late endosome–lysosome trafficking have been described, the mechanisms underlying regulation of this event remain to be elucidated. Our previous studies showed that Snapin, as a SNAP25 (25 kDa synaptosome-associated protein)-binding protein, plays a critical role in priming synaptic vesicles for synchronized fusion in neurons. In the present study, we report that Snapin also associates with late endocytic membranous organelles and interacts with the late endosome-targeted SNARE (soluble N-ethylmaleimide-sensitive factor-attachment protein receptor) complex. Using a genetic mouse model, we further discovered that Snapin is required to maintain a proper balance of the late endocytic protein LAMP-1 (lysosome-associated membrane protein-1) and late endosomal SNARE proteins syntaxin 8 and Vti1b (vesicle transport through interaction with target SNAREs homologue 1b). Deleting the snapin gene in mice selectively led to the accumulation of these proteins in late endocytic organelles. Thus our present study suggests that Snapin serves as an important regulator of the late endocytic fusion machinery, in addition to its established role in regulating synaptic vesicle fusion.
Keywords: late endocytic organelle, late endosome, lysosome-associated membrane protein-1 (LAMP-1), membrane fusion, membrane trafficking, soluble N-ethylmaleimide-sensitive factor-attachment protein receptor (SNARE)
INTRODUCTION
Membrane fusion is an obligatory event at each vesicular trafficking step along the secretory and endocytic pathways, allowing the delivery of quantal packages of cargo between different membrane compartments. Endosomes are major cargo-sorting junctions that employ complex protein- and cargo-recognition mechanisms to differentiate between distinct vesicle trafficking pathways [1]. Lysosomes are acidic organelles containing hydrolytic enzymes, which receive and degrade macromolecules and organelles from these pathways [2,3]. Studies using time-lapse confocal microscopy in living cells have suggested that direct fusion events contribute to the mixing of the contents of late endosomes and lysosomes in hybrid organelles for degradation [4].
Membrane fusion depends on the pairing of proteins known as v-SNAREs [vesicle SNAREs (soluble N-ethylmaleimide-sensitive factor-attachment protein receptors)] and t-SNAREs (target SNAREs), which contribute to the specificity of membrane trafficking [5]. Heterotypic fusion between late endosomes and lysosomes requires a trans-SNARE complex, which consists of t-SNAREs [syntaxin 7, syntaxin 8 and Vti1b (vesicle transport through interaction with target SNAREs homologue 1b)] and v-SNAREs [VAMP7 (vesicle-associated membrane protein 7) or VAMP8] [6–8]. SNARE-mediated membrane fusion is highly regulated, as suggested by the study of synaptic vesicle exocytosis [9]. However, it has not been shown whether the formation of the late endocytic trans-SNARE complex on its own is sufficient to mediate fusion or whether additional proteins are required to regulate proper fusion between late endosomes and lysosomes.
Snapin was first identified as a SNAP25 (25 kDa synaptosome-associated protein)-binding protein that enhances the association of the Ca2+-sensor synaptotagmin I with the neuronal SNARE complex [10,11]. Using snapin knockout mice in combination with genetic rescue experiments, our previous work provided evidence that Snapin plays a critical role in priming large dense-core vesicles for fusion in chromaffin cells [12] and in facilitating synchronized fusion of synaptic vesicles in neurons [13]. In addition to its association with synaptic vesicles, Snapin is also present in both the cytosol- and peripheral-membrane-associated fractions and interacts with non-neuronal SNAP23 and other protein trafficking machineries, suggesting a broader role for Snapin in intracellular membrane trafficking [14–25]. However, most of these interactions were identified via yeast two-hybrid screening, and the physiological relevance of these diverse interactions with Snapin must be critically evaluated using the snapin-deficient mouse model.
In the present study, we discovered a new role for Snapin as a component of the late endocytic SNARE machinery. First, we show that the deletion of the snapin gene in mice results in a significant increase in the late endocytic marker LAMP-1 (lysosome-associated membrane protein-1) and the late endosomal SNARE proteins syntaxin 8 and Vti1b. Second, Snapin is enriched in the late endocytic compartments. Furthermore, Snapin associates with the late endocytic trans-SNARE complex (syntaxin 7–syntaxin 8–Vti1b–VAMP8) through a direct interaction with syntaxin 8, which corresponds to the C-terminal half of SNAP25. Altogether, our present study suggests a novel role for Snapin in late endocytic membrane fusion.
MATERIALS AND METHODS
Materials
The rabbit polyclonal antibody against N-terminal mouse Snapin has been described previously [12] and was further purified with a His–Snapin affinity Sepharose column. Sources of other antibodies or reagents are as follows: mouse monoclonal anti-(syntaxin 8), anti-Vti1a, anti-Vti1b, anti-p115, anti-(cytochrome c) and anti-Rab11 antibodies (BD Biosciences); rat monoclonal anti-LAMP-1 antibody (Developmental Studies Hybridoma Bank); mouse monoclonal and rabbit polyclonal anti-HA (haemagglutinin) antibodies (Covance); rabbit polyclonal anti-(syntaxin 7), anti-VAMP3 and anti-VAMP8 antibodies (Synaptic Systems); mouse monoclonal anti-(syntaxin 13) antibody and rabbit polyclonal anti-calnexin antibody (Stressgen); goat polyclonal anti-EEA1 (early endosome antigen 1) antibody (Santa Cruz Biotechnology); rabbit polyclonal anti-(syntaxin 4) antibody (Alomone Labs); rabbit polyclonal anti-SNAP23 antibody (Affinity BioReagents); Alexa Fluor® 488- and 546-conjugated secondary antibodies (Invitrogen); goat anti-(rat IgG) (Fc fragment specific) (Jackson ImmunoResearch Laboratories).
Immunocytochemistry of MEFs (mouse embryonic fibroblasts) and transfection of COS7 cells
All animal experiments were conducted in accordance with the National Institutes of Health Animal Use Guidelines. MEFs from snapin wild-type and mutant mice were obtained from E13.5 (embryonic day 13.5) or E14.5 embryos. Each embryo was minced and trypsinized, and then the cells were dispersed and incubated for 1 or 2 days with high-glucose DMEM (Dulbecco’s modified Eagle’s medium), containing sodium pyruvate, l-glutamine, supplemented with 10% FBS (fetal bovine serum) and penicillin/streptomycin (1×; Invitrogen), until the cells became confluent. Primary cells (at passage below 7) were used for the experiments. COS7 cells cultured in 100-mm diameter dishes were maintained in DMEM with 10% FBS and 0.5% l-glutamine and were transfected with 15 µg of cDNA using Lipofectamine 2000 (Invitrogen). After 48 h, the cells were harvested and solubilized in TBS (Tris-buffered saline) (50 mM Tris/HCl, pH 7.5, and 140 mM NaCl) with 1% Triton X-100 and protease inhibitors (1 mM PMSF, 10 mg/ml leupeptin and 2 mg/ml aprotinin). Cell lysates were centrifuged at 15 500 g for 20 min at 4°C, and the supernatants were used for immunoprecipitation studies.
Fusion-protein preparation, in vitro-binding and immunoprecipitation experiments
Full-length syntaxin 7, syntaxin 8 and Vti1b were added into the GST (glutathione transferase)-fusion vector pGEX-4T (GE Healthcare) and the His-tagged vectors pcDNA His (Invitrogen). GST- and His-tagged fusion proteins were prepared as crude bacterial lysates by mild sonication in PBS containing 1% Triton X-100, 1 mM PMSF, 10 µg/ml leupeptin and 2 µg/ml aprotinin. In vitro-binding experiments were performed as described previously [12]. Briefly, GST-fusion proteins were bound to glutathione–Sepharose beads (GE Healthcare) in TBS with 0.1% Triton X-100 and protease inhibitors, incubated at 4°C for 1 h with constant agitation, and washed with TBS with 0.1% Triton X-100 to remove unbound protein. Glutathione–Sepharose beads coupled to 1 µg of GST-fusion protein were added to Hisfusion proteins or mouse liver homogenates, and then incubated with gentle mixing for 3 h at 4 °C. The beads were washed three times with TBS with 0.1% Triton X-100, and the bound proteins were eluted and processed for SDS/PAGE and immunoblotting. Co-precipitated proteins were detected with antibodies against Snapin and GST (GE Healthcare). HRP (horseradish peroxidase)-conjugated secondary antibodies and enhanced chemiluminescence reagents (GE Healthcare) were used to visualize detected proteins. For multiple detection with different antibodies, blots were stripped in a solution of 62.5 mM Tris/HCl, pH 7.5, 20 mM dithiothreitol and 1% SDS for 20 min at 50°C with agitation, and washed twice with TBS with 0.1% Tween 20 for 15 min each time.
For immunoprecipitation experiments, solubilized crude membrane fractions from embryonic mouse liver or transfected COS7 cell lysates were incubated with polyclonal anti-Snapin antibody or control rabbit IgG (Sigma) in 200 µl of TBS with 0.1% Triton X-100, 1 mM Ca2+ and protease inhibitors, and incubated on a microtube rotator at 4°C overnight. Protein A–Sepharose CL-4B resin (2.5 mg; GE Health Sciences) was added to each sample, and the incubation was allowed to continue for an additional 3 h, followed by three washes with TBS with 0.1% Triton X-100. The immobilized protein complexes were processed for SDS/PAGE and immunoblotting.
Light membrane preparation and immuno-isolation of late endocytic organelles
Liver tissue from E17–E18 snapin wild-type or knockout embryos was dissected out and homogenized in homogenization buffer (10 mM Hepes, pH 7.4, 1 mM EDTA, 0.25 M sucrose, and protease inhibitors). The homogenate was centrifuged at 750 g for 10 min and the supernatant was collected. The pellet was re-suspended in homogenization buffer by using a glass rod with 3 to 4 gentle strokes of the pestle of the 30 ml Dounce homogenizer and re-centrifuged at 750 g for 10 min. The combined first and second supernatant was centrifuged at 3500 g for 10 min and the supernatant was collected for high-speed centrifugation at 23 000 g for 20 min. The pellet was then re-suspended in homogenization buffer and subjected to the subsequent immuno-isolation assay. Immuno-isolation was performed with tosylated superparamagnetic beads (M-500 Dynabeads, subcellular; Dynal) as described previously [12,26]. Briefly, goat anti-rat IgG (Fc fragment specific, linker) was incubated for 24 h at 37°C on a rotator with M-500 Dynabeads at a ratio of 7 mg of linker per 107 beads in 0.1 M borate buffer (100 mM H3BO3, pH 9.5) at a final concentration of 4 × 108 beads/ml. For this and all subsequent steps, beads were collected with a magnetic device (MPC; Dynal). The linker-coated beads were washed twice, 5 min each, in PBS (pH 7.4) with 0.1% BSA at 4°C on a rotator, and incubated for 20 h in Tris blocking buffer (0.2 M Tris, pH 8.5, and 0.1% BSA) at room temperature (25°C). After washing once for 5 min in PBS (pH 7.4) with 0.1% BSA at 4°C, the linker-coated beads (1.4 mg) were incubated with 1 mg of anti-LAMP-1 monoclonal antibody or control IgG overnight at 4°C on a rotator. After incubation, the beads were washed four times (5 min each) in PBS (pH 7.4) with 0.1% BSA at 4°C, and then re-suspended in incubation buffer containing PBS, pH 7.4, 2 mM EDTA and 5% FBS. Light membrane fractions (~150 µg) from snapin wild-type or knockout embryonic liver were mixed with incubation buffer containing beads (final reaction volume, 1 ml) and incubated overnight at 4°C on a rotator. After incubation, the beads were collected with a magnetic device and washed five times with the incubation buffer and two times with PBS for 10 min each. After the final wash, the beads, as bound fractions, were resolved by 4–12% Bis-Tris PAGE and then sequentially detected by Western blot with antibodies used on the same membranes after stripping between each application of the antibodies.
RESULTS
Loss of snapin selectively alters protein levels of the late endocytic marker LAMP-1 and late endosomal SNAREs
Our previous study showed that snapin deficiency in mice results in defective priming of large dense-core vesicles for fusion in chromaffin cells [12], and a reduced and unsynchronized release of synaptic vesicles in neurons [13]. However, impaired synaptic transmission is not sufficient to explain why the homozygous deletion of snapin causes mouse perinatal death. In addition to its association with synaptic vesicles and binding to neuronal SNAREs, Snapin is also widely present in cytosol and membrane-associated fractions of non-neuronal cells and interacts with non-neuronal SNAP23 [14], and is co-purified as a component of BLOC-1 (biogenesis of lysosome-related organelle complex-1) [16]. These studies highlight a possibility for Snapin as having multiple effects on broader intracellular membrane trafficking events; the perinatal death of snapin homozygous mouse might be attributed to the defects of these trafficking processes.
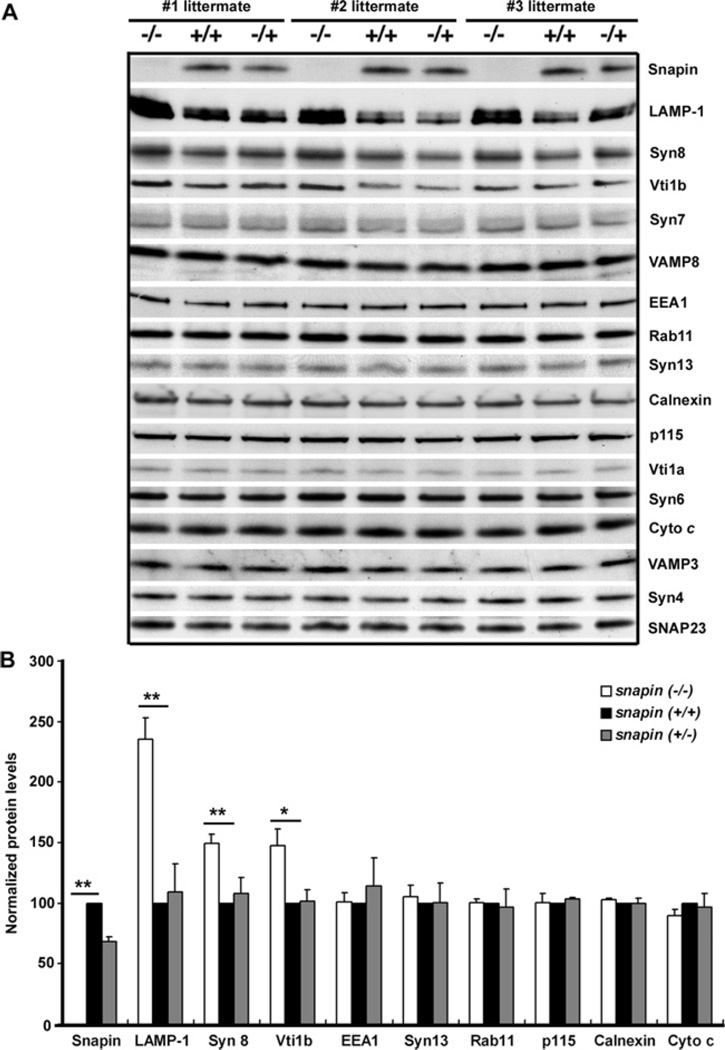
To examine whether Snapin homozygous deletion and heterozygous mutants affect expression of a large variety of proteins involved in intracellular membrane trafficking, we performed immunoblot analysis of the liver homogenates of snapin wild-type and deficient littermates. Deleting snapin in mouse significantly increased protein levels of LAMP-1 (P = 0.002) and late endosomal SNARE proteins syntaxin 8 (P = 0.003) and Vti1b (P = 0.024) relative to the wild-type littermates (Figure 1). LAMP-1 is a membrane protein located in both late endosomes and lysosomes [27,28]. However, snapin deficiency has no detectable effect on the intensity of the markers and SNAREs that are specific for other membrane organelles, including early and recycling endosomes (EEA1, Rab11 and syntaxin 13), ER (endoplasmic reticulum) (calnexin), Golgi (p115), trans-Golgi (Vti1a and syntaxin 6), mitochondria (cytochrome c) and plasma membrane SNAREs (syntaxin 4).
Figure 1. Deletion of the snapin gene in mice selectively increases late endosomal SNAREs and LAMP-1.
(A) Immunoblot analysis of SNAREs and markers specific for various intracellular membrane organelles. Equal amounts of liver homogenates (30 µg) from three littermates of E18.5 embryos of all snapin genotypes were sequentially detected with antibodies, as indicated in the same membranes after stripping between applications of each antibody. (B) Relative protein levels from the snapin +/+, +/− and −/− mouse livers. Protein intensity was normalized by p115 intensity in the same littermate and averaged from three littermates. A two-tailed Student’s t test for paired data was used and error bars indicate S.E.M.; *P < 0.05, **P < 0.01. Note that deleting snapin in mouse significantly increases LAMP-1 (late endocytic marker), syntaxin 8 (Syn8) and Vti1b (late endocytic SNARE proteins) without detectable changes in the markers of early and recycling endosomes [EEA1, Rab11 and syntaxin 13 (Syn13)], ER (calnexin), Golgi (p115), trans-Golgi [Vti1a and syntaxin 6 (Syn6)], mitochondria [cytochrome c (Cyto c)] and plasma membrane SNAREs [syntaxin 4 (Syn4)].
Our previous analysis of brain homogenates showed that Snapin homozygous deletion and heterozygous mutants did not affect expression of a large variety of proteins involved in synaptic vesicle exocytosis in brain homogenates [12,13]. These included SNAP25, syntaxin 1A/B, synaptobrevin/VAMP2, synaptotagmin 1, synapsin 1, Rab3a, NSF (N-ethylmaleimide-sensitive factor), α-SNAP, Munc-18, complexin I/II, Munc13-1 and CAPS (Ca2+-dependent activator protein for secretion). Selectively altered levels of the late endocytic marker LAMP-1, and syntaxin 8 and Vti1b, two SNARE proteins involved in late endocytic trafficking, suggest that Snapin may play a critical role in the late endocytic membrane trafficking. Deleting snapin in mice may lead to the accumulation of LAMP-1 due to defective late endocytic trafficking or result in apparent compensatory changes in the expression of late endosomal SNAREs to bypass snapin deficiency.
Snapin associates with late endocytic compartments
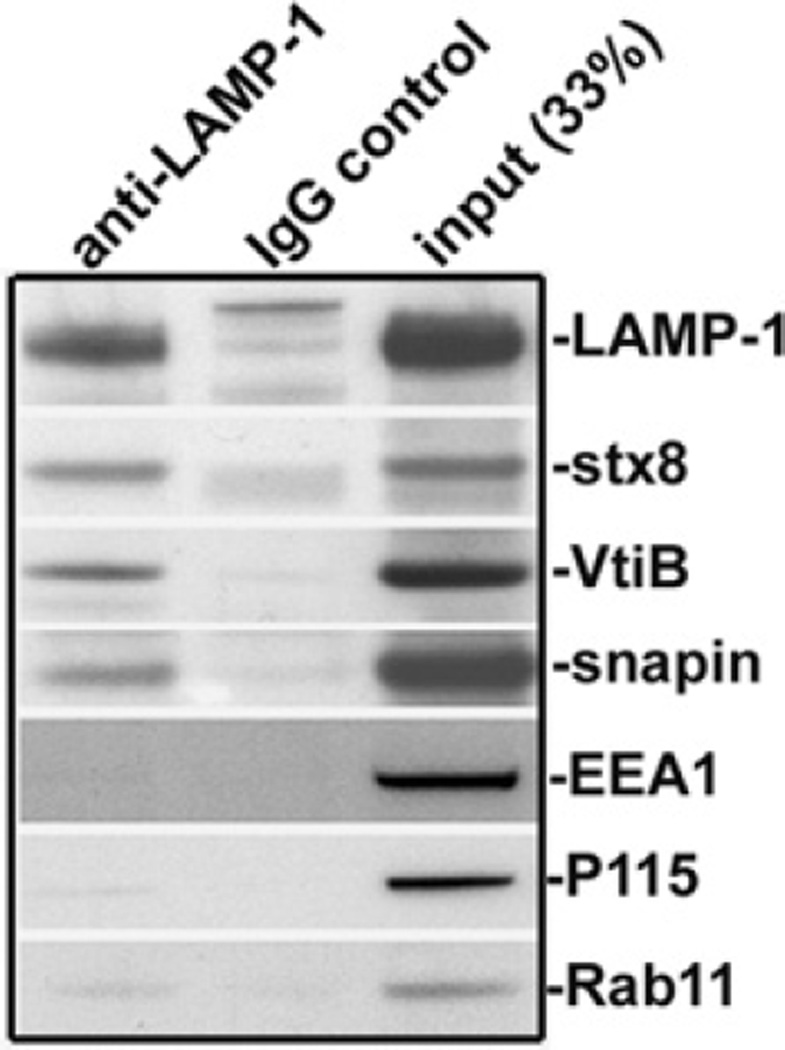
Next, we asked whether Snapin associates with the late endocytic membrane. Given that Snapin is present in both cytosolic and peripheral membrane-associated fractions [14], immunostaining of cells is not an appropriate means to determine specific attachment of Snapin to late endocytic organelles. Instead, we performed immuno-isolation of the LAMP-1-containing organelles from embryonic mouse liver membrane fractions using magnetic beads coated with the antibody against LAMP-1. The purified membranous organelles were assessed by sequential immunoblots on the same membrane. Snapin was detected along with LAMP-1, as well as late endosomal SNARE proteins, including syntaxin 8 and Vti1b (Figure 2). None of these proteins was isolated with the magnetic beads coated with IgG control. The relative purity was further confirmed by the absence of markers for other membranous organelles including EEA1 (early endosomes), p115 (Golgi) and Rab11 (recycling endosomes).
Figure 2. Snapin associates with late endocytic compartments.
LAMP-1-associated membranous organelles were immuno-isolated from light membrane fractions of mouse livers with magnetic Dynabeads coated with anti-LAMP-1 antibody or normal IgG as control. The bead-bound organelles were solubilized and resolved by PAGE, and sequentially detected with antibodies in the same membrane as indicated. The relative purity of the isolated organelles was assessed by detecting the markers for late endocytic compartments [LAMP-1, syntaxin 8 (stx8) and Vti1b (VtiB)] and the markers for early endosomes (EEA1), recycling endosomes (Rab11) and Golgi (p115). Note that Snapin, along with late endocytic SNAREs, was detected in the LAMP-1-containing membrane organelles.
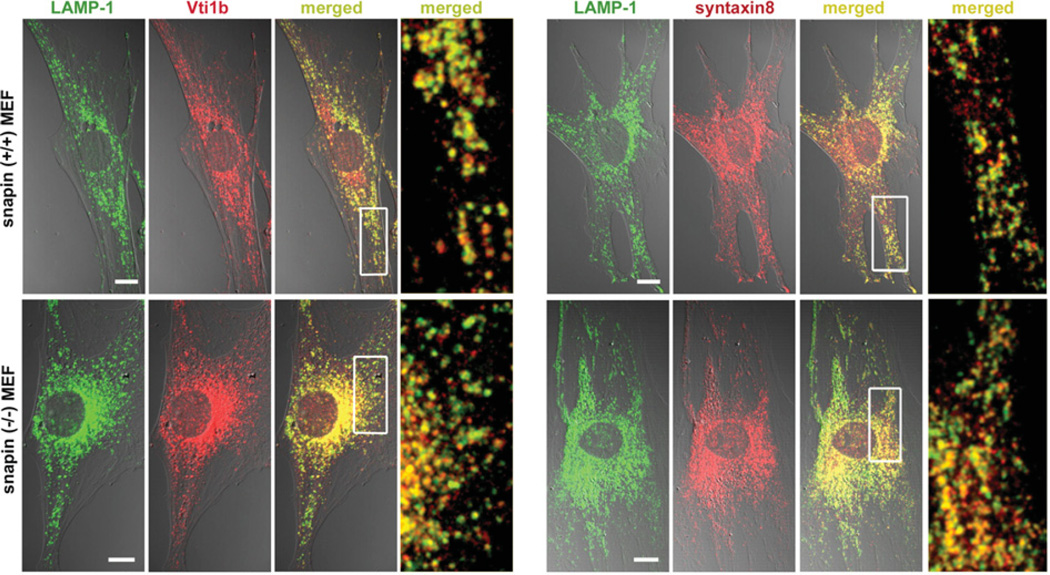
Second, we further explored whether snapin deficiency would alter the sorting and targeting of late endosomal SNARE proteins that may affect late endocytic trafficking. We performed immunostaining of MEFs from snapin wild-type and mutant E13.5 or E14.5 embryos. MEFs have larger cell bodies, allowing us to analyse the late endocytic organelles in more detail. Confocal microscopy analysis showed no detectable changes in the co-localization of the LAMP-1-labelled late endocytic organelles with syntaxin 8 or Vti1b in snapin-deficient MEFs (Figure 3).
Figure 3. Intracellular localization of late endosomal/lysosomal SNAREs.
Representative images showing the co-localization of LAMP-1 with Vti1b or syntaxin 8 were captured by confocal microscopy in snapin +/+ and −/− MEFs. The colour images are shown in DIC (differential interference contrast) and the co-localization is indicated in yellow in the merged images. The boxed regions are shown at a higher magnification. Scale bar, 10 µm.
Snapin interacts with late endosomal SNAREs
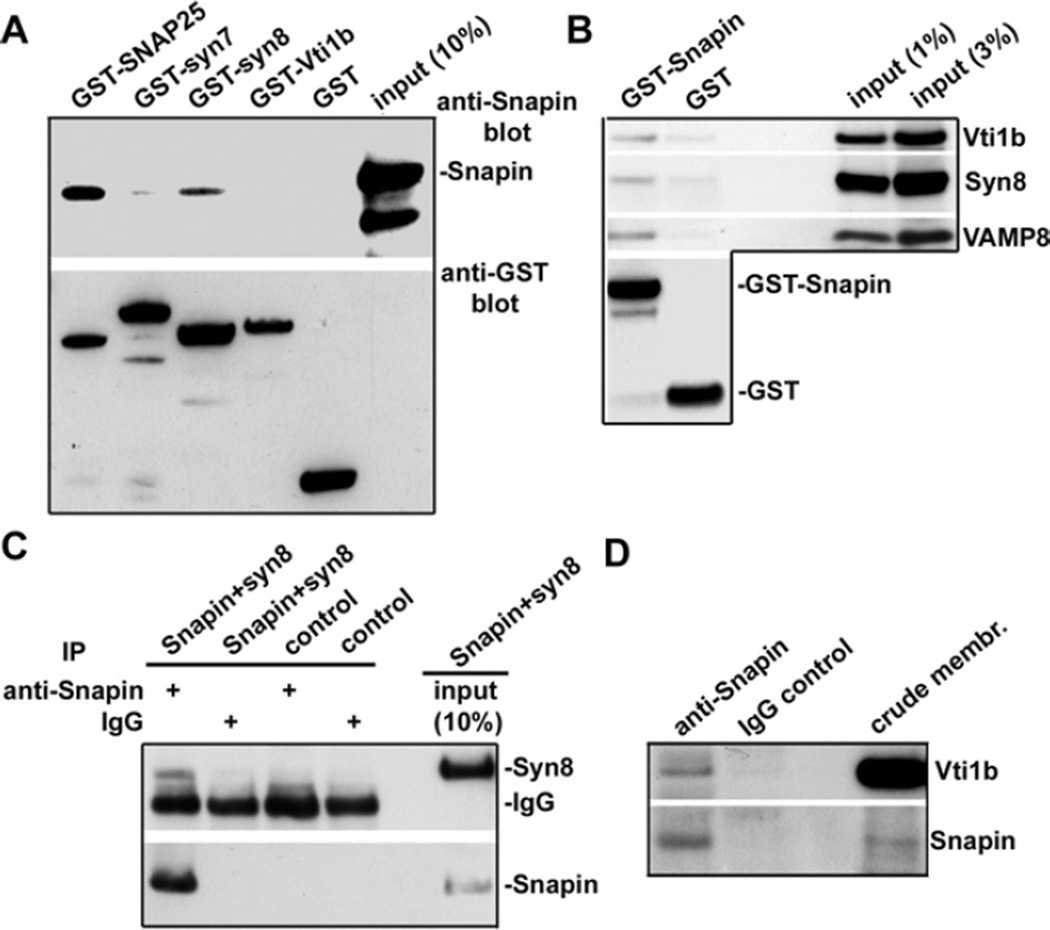
The resolved crystal structure of the late endosomal SNARE core complex containing syntaxin 7, syntaxin 8, Vti1b and VAMP8 is remarkably similar to those of the neuronal SNARE complex, where syntaxin 8 corresponds to the C-terminal coiled-coil domain of SNAP25 [7]. To determine whether Snapin interacts with late endosomal SNAREs, we conducted four lines of biochemical analysis. First, in vitro-binding assays showed that His-tagged Snapin selectively interacts with GST–syntaxin 8 and GST–SNAP25, a neuronal SNARE protein identified as the first Snapin-binding partner [10] (Figure 4A). No direct interaction was detected for Snapin with syntaxin 7 and Vti1b. Second, we conducted a pull-down study using liver homogenates. Consistently, GST–Snapin, but not GST, pulled down the native late endosomal SNAREs Vti1b, syntaxin 8, and VAMP8 (Figure 4B). Furthermore, an anti-Snapin antibody co-immunoprecipitated syntaxin 8 from co-transfected COS7 cell lysates (Figure 4C) and Vti1b from solubilized mouse liver crude membrane fractions (Figure 4D). Thus our analysis provides biochemical evidence that Snapin associates with late endocytic compartments, probably through its interaction with late endosomal SNARE proteins. By interacting with SNAP-25 or syntaxin 8, Snapin may regulate both synaptic vesicle exocytosis and late endocytic membrane fusion.
Figure 4. Snapin interacts with late endosomal SNAREs.
(A) Snapin binds to syntaxin 8 in vitro. GST or GST-tagged fusion proteins were immobilized on glutathione–Sepharose beads and then incubated with His-tagged Snapin. Bound protein complexes were sequentially immunoblotted with antibodies against Snapin and GST tag on the same membrane. Note that Snapin selectively binds to both neuronal SNAP25 and late endosomal syntaxin 8 (syn8), but not to syntaxin 7 (syn7) and Vti1b under the same conditions. (B) GST–Snapin pulls down the native late endosomal SNARE complex. GST or GST-tagged Snapin was immobilized on glutathione–Sepharose beads followed by incubation with mouse liver homogenates. Bound complexes were sequentially blotted with antibodies against late endosomal SNARE proteins and GST. (C) Snapin interacts with syntaxin 8 in COS7 cells. COS7 cells were co-transfected with Snapin and syntaxin 8. The Snapin–syntaxin 8 complex was immunoprecipitated (IP) by an anti-Snapin antibody or control IgG followed by sequential immunoblotting with antibodies against syntaxin 8 and Snapin on the same membrane after stripping between the applications of each antibody. (D) Immunoprecipitation of late endosomal SNARE protein Vti1b with Snapin. The Vti1b–Snapin complex was immunoprecipitated from mouse liver crude membrane fractions (crude membr.) with an anti-Snapin antibody or normal rabbit IgG, followed by sequentially blotting with antibodies against Vti1b and Snapin.
DISCUSSION
The endocytic pathway is a highly dynamic system that co-ordinates multiple trafficking routes. The lysosome is generally considered to be the end point of the endocytic pathway and most of the biosynthetic and endocytic proteins are targeted to this degradative compartment. Lysosomes are also required for the digestion of cytosolic components and organelles that are segregated during the process of autophagy [29–31]. Although the mechanism underlying the transfer of endocytosed materials from endosomes to lysosomes remains controversial, the direct and complete fusion of late endosomes with lysosomes and their transient fusion (‘kiss and run’) mode have been proposed as two major mechanisms for producing mature lysosomes [3]. However, the mechanisms regulating endosome–lysosome fusion and lysosomal biogenesis remain unclear.
In the present study, we reveal that Snapin associates with late endocytic membranous organelles and interacts with the late endosome-targeted SNARE complex, thus highlighting its potential role in late endocytic membrane trafficking. By using a genetic mouse model, we further discovered that Snapin is required to maintain a proper balance of the late endocytic protein LAMP-1 and late endosomal SNAREs. Deleting the snapin gene in mice significantly and selectively accumulates these proteins in the late endocytic pathway or results in apparent compensatory changes in the expression of late endosomal SNAREs, suggesting a regulatory role for Snapin in the late endocytic membrane fusion process.
Similar to synaptic vesicle fusion events, late endocytic membrane fusion is considered to have three sequential steps: initial tethering, the formation of a trans-SNARE complex that bridges across the two organelles and final membrane fusion. Live-cell imaging, together with studies in cell-free systems and transfected cells, has established the role of late endocytic SNAREs in the fusion of late endosomes with lysosomes [6–8]. The fusion of late endosomes and lysosomes requires the presence of Q-SNAREs (glutamine-SNAREs; syntaxin 7, Vti1b and syntaxin 8) and R-SNAREs (arginine-SNAREs; VAMP8 or VAMP7). The structure of the four-helix bundle for the endocytic SNARE complex shares similar structural features with the neuronal SNARE complex [7]. Although synaptic vesicle fusion with the presynaptic plasma membrane is tightly primed or regulated by a number of SNARE-binding proteins [9], it has not been proven whether the formation of the trans-SNARE complex on its own is sufficient for late endosome–lysosome fusion. Our biochemical experiments revealed the presence of Snapin in late endocytic organelles, its direct binding to syntaxin 8 and its association with the late endosomal SNARE complex both in vitro and in vivo. Our previous study [12] using snapin mutant mice illustrated that Snapin plays a critical role in priming the release-ready vesicle pool and facilitates synaptic vesicle fusion by enhancing the structural coupling of the Ca2+-sensor synaptotagmin I with the neuronal SNARE complex. Evidence indicates that secretory lysosomes are able to release their contents into the extracellular milieu via exocytosis [32,33], a process that occurs via the plasma membrane-targeted SNARE fusion machinery and the Ca2+-sensor synaptotagmin-VII [34]. Thus it is necessary to determine whether Snapin can facilitate the endsosome–lysosome membrane fusion through a similar priming mechanism, although the candidate Ca2+ sensor specific for endosome–lysosomal fusion has not yet been identified. Alternatively, it is possible that Snapin may participate in secretory lysosomal fusion with the plasma membrane by regulating the SNARE–synaptotagmin-VII interaction. Future studies using time-lapse imaging in live cells will provide direct evidence of whether Snapin can facilitate the interaction between late endosomes and lysosomes or between late endosomes/secretory lysosomes and the plasma membrane.
Although melanosomes and platelet-dense granules are not endocytic vesicles themselves, they belong to the exocytic group of LROs (lysosome-related organelles). LROs co-exist with conventional lysosomes in some specific cell types, but are functionally, morphologically and compositionally distinct from lysosomes. Snapin was co-purified as a component of BLOC-1 [16]. Mutation of the BLOC-1 genes in mice displays HPS (Hermansky–Pudlak syndrome)-like phenotypes which are characterized by specific defects in melanosomes and platelet-dense granules [35–38]. It has been reported that BLOC-1 deficiency selectively altered the targeting of the late endocytic SNARE proteins syntaxin 7 and syntaxin 8, and Vti1b [39], and that BLOC-1 acts as the endosomal trafficking machinery to facilitate the traffic from early endosomes toward LROs [40,41]. The evidence from our genetic study suggests that loss of snapin has no detectable effect on the targeting of these SNAREs to the late endocytic organelles (Figure 3). In addition, snapin deficiency does not change the levels of early endosomal markers EEA1, Rab11 and syntaxin 13 (Figure 1). Notably, the snapin mice with homozygous mutations are neonatal lethal, whereas mice deficient in BLOC-1 components are fertile and fully viable [42,36]. It is therefore conceivable that BLOC-1-mediated SNARE protein trafficking is likely to be independent of Snapin. Identification of Snapin as a component of late endocytic fusion machinery will provide new avenues for understanding these diverse cellular processes. Future studies using the snapin mouse model will provide molecular and cellular details of how Snapin regulates late endocytic membrane trafficking, an essential process for controlling protein and organelle turnover or clearance during development and degeneration.
Acknowledgments
We thank the following people for their help: H. Arnheiter and J. Bonifacino and the members of the Sheng laboratory for helpful discussions; C. Gerwin for critical reading of the manuscript and for mouse maintenance; and H. Arnheiter for preparation of MEFs. The anti-LAMP-1 monoclonal antibody developed by D. Messner and J. T. August was obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the National Institute of Child Health and Development and maintained by the University of Iowa.
FUNDING
This work was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, National Institutes of Health (to Z.-H.S.) L.L. is a graduate student of the National Institutes of Health–Shanghai JiaoTong University Joint PhD Program in Neuroscience, supported by the National Institutes of Health Intramural Research Program.
Abbreviations used
- BLOC
biogenesis of lysosome-related organelle complex
- DMEM
Dulbecco’s modified Eagle’s medium
- E13.5
embryonic day 13.5 etc.
- EEA
early endosome antigen
- ER
endoplasmic reticulum
- FBS
fetal bovine serum
- GST
glutathione transferase
- LAMP
lysosome-associated membrane protein
- LRO
lysosome-related organelle
- MEF
mouse embryonic fibroblast
- NSF
N-ethylmaleimide-sensitive factor
- SNAP25
25 kDa synaptosome-associated protein
- SNARE
soluble N-ethylmaleimide-sensitive factor-attachment protein receptor
- TBS
Tris-buffered saline
- t-SNARE
target SNARE
- VAMP
vesicle-associated membrane protein
- v-SNARE
vesicle SNARE
- Vti1b
vesicle transport through interaction with t-SNAREs homologue 1b
REFERENCES
- 1.Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 2.Mullins C, Bonifacino JS. The molecular machinery for lysosome biogenesis. Bioessays. 2001;23:333–343. doi: 10.1002/bies.1048. [DOI] [PubMed] [Google Scholar]
- 3.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 4.Bright NA, Gratian MJ, Luzio JP. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr. Biol. 2005;15:360–365. doi: 10.1016/j.cub.2005.01.049. [DOI] [PubMed] [Google Scholar]
- 5.Hong W. SNAREs and traffic. Biochim. Biophys. Acta. 2005;1744:493–517. [PubMed] [Google Scholar]
- 6.Mullock BM, Bright NA, Fearon CW, Gray SR, Luzio JP. Fusion of lysosomes with late endosomes produces a hybrid organelle of intermediate density and is NSF dependent. J. Cell Biol. 1998;140:591–601. doi: 10.1083/jcb.140.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonin W, Fasshauer D, Becker S, Jahn R, Schneider TR. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 2002;9:107–111. doi: 10.1038/nsb746. [DOI] [PubMed] [Google Scholar]
- 8.Pryor PR, Mullock BM, Bright NA, Lindsay MR, Gray SR, Richardson SC, Stewart A, James DE, Piper RC, Luzio JP. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;5:590–595. doi: 10.1038/sj.embor.7400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jahn R, Scheller RH. SNAREs – engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 10.Ilardi JM, Mochida S, Sheng ZH. Snapin: a SNARE-associated protein implicated in synaptic transmission. Nat. Neurosci. 1999;2:119–124. doi: 10.1038/5673. [DOI] [PubMed] [Google Scholar]
- 11.Chheda MG, Ashery U, Thakur P, Rettig J, Sheng ZH. Phosphorylation of Snapin by PKA modulates its interaction with the SNARE complex. Nat. Cell Biol. 2001;3:331–338. doi: 10.1038/35070000. [DOI] [PubMed] [Google Scholar]
- 12.Tian JH, Wu ZX, Unzicker M, Lu L, Cai Q, Li C, Schirra C, Matti U, Stevens D, Deng C, et al. The role of Snapin in neurosecretion: snapin knock-out mice exhibit impaired calcium-dependent exocytosis of large dense-core vesicles in chromaffin cells. J. Neurosci. 2005;25:10546–10555. doi: 10.1523/JNEUROSCI.3275-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan PY, Tian JH, Sheng Z-H. Snapin facilitates the synchronization of synaptic vesicle fusion. Neuron. 2009;61:412–424. doi: 10.1016/j.neuron.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buxton P, Zhang XM, Walsh B, Sriratana A, Schenberg I, Manickam E, Rowe T. Identification and characterization of Snapin as a ubiquitously expressed SNARE-binding protein that interacts with SNAP23 in non-neuronal cells. Biochem. J. 2003;375:433–440. doi: 10.1042/BJ20030427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thakur P, Stevens DR, Sheng ZH, Rettig J. Effects of PKA-mediated phosphorylation of Snapin on synaptic transmission in cultured hippocampal neurons. J. Neurosci. 2004;24:6476–6481. doi: 10.1523/JNEUROSCI.0590-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starcevic M, Dell’Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J. Biol. Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- 17.Rüder C, Reimer T, Delgado-Martinez I, Hermosilla R, Engelsberg A, Nehring R, Dörken B, Rehm A. EBAG9 adds a new layer of control on large dense-core vesicle exocytosis via interaction with Snapin. Mol. Biol. Cell. 2005;16:1245–1257. doi: 10.1091/mbc.E04-09-0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukui K, Yang Q, Cao Y, Takahashi N, Hatakeyama H, Wang H, Wada J, Zhang Y, Marselli L, Nammo T, et al. The HNF-1 target collectrin controls insulin exocytosis by SNARE complex formation. Cell Metab. 2005;2:373–384. doi: 10.1016/j.cmet.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Zissimopoulos S, West DJ, Williams AJ, Lai FA. Ryanodine receptor interaction with the SNARE-associated protein snapin. J. Cell Sci. 2006;119:2386–2397. doi: 10.1242/jcs.02936. [DOI] [PubMed] [Google Scholar]
- 20.Talbot K, Cho DS, Ong WY, Benson MA, Han LY, Kazi HA, Kamins J, Hahn CG, Blake DJ, Arnold SE. Dysbindin-1 is a synaptic and microtubular protein that binds brain snapin. Hum. Mol. Genet. 2006;15:3041–3054. doi: 10.1093/hmg/ddl246. [DOI] [PubMed] [Google Scholar]
- 21.Krapivinsky G, Mochida S, Krapivinsky L, Cibulsky SM, Clapham DE. The TRPM7 ion channel functions in cholinergic synaptic vesicles and affects transmitter release. Neuron. 2007;52:485–496. doi: 10.1016/j.neuron.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki F, Morishima S, Tanaka T, Muramatsu I. Snapin, a new regulator of receptor signaling, augments α1A-adrenoceptor-operated calcium influx through TRPC6. J. Biol. Chem. 2007;282:29563–29573. doi: 10.1074/jbc.M702063200. [DOI] [PubMed] [Google Scholar]
- 23.Mistry AC, Mallick R, Fröhlich O, Klein JD, Rehm A, Chen G, Sands JM. The UT-A1 urea transporter interacts with snapin, a SNARE-associated protein. J. Biol. Chem. 2007;282:30097–30106. doi: 10.1074/jbc.M705866200. [DOI] [PubMed] [Google Scholar]
- 24.Granata A, Watson R, Collinson LM, Schiavo G, Warner TT. The dystonia-associated protein torsina modulates synaptic vesicle recycling. J. Biol. Chem. 2007;283:7568–7579. doi: 10.1074/jbc.M704097200. [DOI] [PubMed] [Google Scholar]
- 25.Bao Y, Lopez JA, James DE, Hunziker W. Snapin interacts with the Exo70 subunit of the exocyst and modulates GLUT4 trafficking. J. Biol. Chem. 2008;283:324–331. doi: 10.1074/jbc.M706873200. [DOI] [PubMed] [Google Scholar]
- 26.Cai Q, Gerwin C, Sheng ZH. Syntabulin-mediated anterograde transport of mitochondria along neuronal processes. J. Cell Biol. 2005;170:959–969. doi: 10.1083/jcb.200506042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen JW, Murphy TL, Willingham MC, Pastan I, August JT. Identification of two lysosomal membrane glycoproteins. J. Cell Biol. 1985;101:85–95. doi: 10.1083/jcb.101.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis V, Green SA, Marsh M, Vihko P, Helenius A, Mellman I. Glycoproteins of the lysosomal membrane. J. Cell Biol. 1985;100:1839–1847. doi: 10.1083/jcb.100.6.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn WA., Jr Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 30.Levine B, Klionsky DJ. Development by self-digestion; molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 31.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrews NW. Regulated secretion of conventional lysosomes. Trends Cell Biol. 2000;10:316–321. doi: 10.1016/s0962-8924(00)01794-3. [DOI] [PubMed] [Google Scholar]
- 33.Blott EJ, Griffiths GM. Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 2002;3:122–131. doi: 10.1038/nrm732. [DOI] [PubMed] [Google Scholar]
- 34.Rao SK, Huynh C, Proux-Gillardeaux V, Galli T, Andrews NW. Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J. Biol. Chem. 2004;279:20471–20479. doi: 10.1074/jbc.M400798200. [DOI] [PubMed] [Google Scholar]
- 35.Dell’Angelica EC. The building BLOC(k)s of lysosomes and related organelles. Curr. Opin. Cell Biol. 2004;16:458–464. doi: 10.1016/j.ceb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Li W, Rusiniak ME, Chintala S, Gautam R, Novak EK, Swank RT. Murine Hermansky–Pudlak syndrome genes: regulators of lysosome-related organelles. Bioessays. 2004;26:616–628. doi: 10.1002/bies.20042. [DOI] [PubMed] [Google Scholar]
- 37.Di Pietro SM, Dell’Angelica EC. The cell biology of Hermansky–Pudlak syndrome: recent advances. Traffic. 2005;6:525–533. doi: 10.1111/j.1600-0854.2005.00299.x. [DOI] [PubMed] [Google Scholar]
- 38.Raposo G, Marks MS, Cutler DF. Lysosome-related organelles: driving post-Golgi compartments into specialisation. Curr. Opin. Cell Biol. 2007;19:394–401. doi: 10.1016/j.ceb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salazar G, Craige B, Styers ML, Newell-Litwa KA, Doucette MM, Wainer BH, Falcon-Perez JM, Dell’Angelica EC, Peden AA, Werner E. BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 cargoes. Mol. Biol. Cell. 2006;17:4014–4026. doi: 10.1091/mbc.E06-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Pietro SM, Falcón-Pérez JM, Tenza D, Setty SR, Marks MS, Raposo G, Dell’Angelica EC. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol. Biol. Cell. 2006;17:4027–4038. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Setty SR, Tenza D, Truschel ST, Chou E, Sviderskaya EV, Theos AC, Lamoreux ML, Di Pietro SM, Starcevic M, Bennett DC. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol. Biol. Cell. 2007;18:768–780. doi: 10.1091/mbc.E06-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nazarian R, Falcón-Pérez JM, Dell’Angelica EC. Biogenesis of lysosome-related organelles complex 3 (BLOC-3): a complex containing the Hermansky–Pudlak syndrome (HPS) proteins HPS1 and HPS4. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8770–8775. doi: 10.1073/pnas.1532040100. [DOI] [PMC free article] [PubMed] [Google Scholar]