Abstract
Background
Epigenetic events mediated by methylation and histone modifications have been associated with the development of metastasis in patients with uveal melanoma. The role of epigenetic events mediated by microRNA (miR) is less clear. Tumor and plasma miR expression was examined in patients with primary uveal melanoma with tumor monosomy-3, a predictor of metastasis.
Results
miR profiling of tumors by microarray found six miRs over-expressed and 19 under-expressed in 33 tumors with monosomy-3 compared to 22 without. None of the miRs differentially expressed in tumors with and without monosomy-3 was differentially expressed in tumors with and without tumor infiltrating lymphocytes. Tumors manifesting monosomy-3 were also characterized by higher levels of TARBP2 and DDX17 and by lower levels of XPO5 and HIWI, miR biogenesis factors. miR profiling of plasma by a quantitative nuclease protection assay found elevated levels of 11 miRs and reduction in four in patients with tumor monosomy-3. Only three miRs differentially expressed in the tumor arrays were detectable in plasma. miRs implicated in uveal melanoma development were not differentially expressed. Elevated plasma levels in patients with tumor monosomy-3 of miR-92b, identified in the tumor array, and of miR-199-5p and miR-223, identified in the plasma array, were confirmed by quantitative real-time polymerase chain reaction. Levels were also higher in patients compared to normal controls.
Conclusions
These results support a role for epigenetic mechanisms in the development of metastasis in patients with uveal melanoma and the analysis of miRs as biomarkers of metastatic risk. They also suggest that potentially useful blood miRs may be derived from the host response as well as the tumor.
Keywords: Prognosis, Biomarkers, miR-92b, miR-199-5p, miR-223
Background
Uveal melanoma is a rare cancer that leads to metastatic death in up to half of patients. That loss of chromosome 3 in tumors is associated with the development of metastasis is well established, and a variety of techniques are being used to test tumors for monosomy-3 [1]. Gene expression profiling (GEP) has also been effectively applied to characterize tumors with a high risk of metastasis, “class 2,” and tumors with a low risk, “class 1” [2]. Epigenetic events have also been implicated in uveal melanoma metastasis. When independently analyzed for global DNA methylation profiles, primary uveal melanomas cluster into two groups that are identical to the class-2 and class-1 groups identified by GEP [3]. Expression levels of a number of histone-modifying genes and polycomb family members are significantly lower in uveal melanoma with monosomy-3/class-2 GEP [4]. Although epigenetic events mediated by microRNA (miR) have been implicated in uveal melanoma development [5–7], a role for miRs in the metastatic process has not been established. Worley et al. found six miRs to be upregulated in 12 tumors expressing high-risk, class-2 GEP and 68 to be upregulated in 12 tumors expressing low-risk, class-1 GEP [8]. The most significant discriminators of class 2 were upregulation of let-7b and miR-199a. In contrast, Larsen et al. found no association between miR-expression profiles and histopathological features, staging, metastasis, or survival in a study of 20 patients [9].
Obtaining uveal melanoma tumors for genotyping can be problematic. There is a need for blood biomarkers [10]. miRs are very stable in blood due in part to their incorporation into microparticles and exosomes, and serum and plasma levels of miRs are also under investigation as diagnostic and prognostic biomarkers in cancer and other diseases [11]. Blood miR levels have not been previously reported in uveal melanoma. We used tumor monosomy-3 as well as tumor and plasma miR profiling as guides as to develop miR-based prognostic blood biomarkers for patients with primary uveal melanoma. We also examined the expression of miR biogenesis factors. Differential expression of miRs and miR biogenesis factors were identified.
Results
Tumor array
miR and gene expression profiles of 33 enucleated uveal tumors with monosomy-3 and 22 without were obtained. This analysis identified 26 miRs as discriminators; 19 were down-regulated >2.0-fold and six were upregulated >2.0-fold (Table 1). In this data set, 13 patients, all with tumor monosomy-3, had manifested metastatic disease clinically on follow-up. Eight of the 26 miRs identified were also differentially expressed in these patients (Table 1). The strongest associations with monosomy-3 were observed for under-expression of X-linked miRs. Neither X-linked miRs nor other any miRs were differentially expressed by tumors from the 31 males compared to the 24 females studied. Twenty-seven of the tumors evaluated were considered to have TILs, a potential source of miRs; 28 were not. None of the miRs differentially expressed in tumors with and without monosomy-3 was differentially expressed in tumors with and without TILs. Gene expression of 12 miR biogenesis factors were also profiled by microarray in the 55 enucleated uveal melanoma tumors. Tumors manifesting monosomy-3 were characterized by higher levels of TARB2 and DDX17 and lower levels of XPO5 and HIWI (Fig. 1).
Table 1.
Tumor miRs differentially expressed
| miR | Chr | Monosomy-3 vs. disomy-3a fold difference | Monosomy-3 vs. disomy-3a P | Metastatic vs. nonmetastaticb P | Plasma levelsc | |
|---|---|---|---|---|---|---|
| Monosomy-3 | Disomy-3 | |||||
| Over-expressed in tumors with monosomy-3 | ||||||
| hsa-miR-135a* | 3 | 80 | 0.0003 | NS | ND | ND |
| hsa-miR-624 | 14 | 7.6 | 0.0003 | 0.0004 | ND | ND |
| hsa-miR-449b | 5 | 5.9 | 0.0005 | NS | ND | ND |
| hsa-miR-142-5p | 17 | 7.5 | 0.0006 | NS | 1236 | 1059 |
| hsa-miR-92b | 1 | 2.9 | 0.0009 | NS | 688 | ND |
| hsa-miR-628-5p | 15 | 4.8 | 0.001 | NS | ND | ND |
| Under-expressed in tumors with monosomy-3 | ||||||
| hsa-miR-509-3-5p | X | 615 | 0.00000008 | 0.0002 | ND | ND |
| hsa-miR-508-3p | X | 4672 | 0.0000003 | 0.0004 | ND | ND |
| hsa-miR-514 | X | 2887 | 0.0000009 | NS | ND | ND |
| hsa-miR-506 | X | 1308 | 0.000001 | 0.0008 | ND | ND |
| hsa-miR-513a-5p | X | 1674 | 0.000002 | 0.001 | ND | ND |
| hsa-miR-507 | X | 61 | 0.000003 | NS | ND | ND |
| hsa-miR-509-3p | X | 818 | 0.000004 | 0.0008 | ND | ND |
| hsa-miR-513b | X | 75 | 0.00003 | 0.00002 | ND | ND |
| hsa-miR-876-3p | 9 | 81 | 0.00003 | NS | ND | ND |
| hsa-miR-378* | 5 | 5.1 | 0.0002 | NS | ND | ND |
| hsa-miR-935 | 19 | 4.1 | 0.0003 | 0.001 | ND | ND |
| hsa-miR-181a | 9 | 7.4 | 0.0004 | NS | 546 | ND |
| hsa-miR-99a | 21 | 5.6 | 0.0009 | NS | ND | ND |
| hsa-miR-194 | 1 | 4.5 | 0.001 | NS | ND | ND |
| hsa-miR-592 | 7 | 4.1 | 0.001 | NS | ND | ND |
| hsa-miR-1296 | 10 | 15 | 0.001 | NS | ND | ND |
| hsa-miR-624* | 14 | 7.5 | 0.002 | NS | ND | ND |
| hsa-miR-140-5p | 16 | 10 | 0.002 | NS | ND | ND |
| hsa-miR-651 | X | 6.1 | 0.002 | NS | ND | ND |
NS not significant, ND not detectable
aMonosomy-3, n = 33; disomy-3, n = 22
bMetastatic, n = 13; nonmetastatic, n = 42
cAverage signal intensity
Fig. 1.
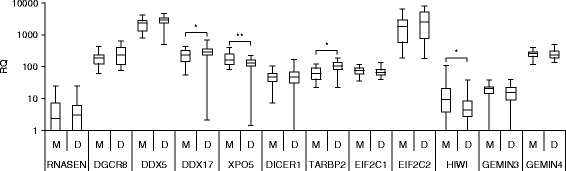
miR biogenesis factor expression by gene expression array in enucleated tumors with (M), n = 33, and without (D), n = 22, monosomy-3. The box represents the 25th and 75th percentiles, the horizontal lines represent the median, and the whiskers represent the minimum and maximum. Brackets with an asterisk above indicate statistical significance P <0.05 , **P < 0.01, Wilcoxon rank-sum test
Plasma array
Plasma miR profiles of pooled samples from 10 patients with monosomy-3 and 10 without were analyzed using quantitative nuclease protection assay (qNPA). Of the 674 human miRs assayed, 96 were detectable in plasma. Compared to patients without, 11 miRs were elevated >2.0-fold and four were reduced >2.0-fold in patients with tumor monosomy-3 (Table 2). None of the miRs that was discriminatory in the tumor array met the level of discrimination set for the plasma array. Only two of the miRs over-expressed in the tumor array were quantifiable in plasma, miR-92b and miR-142-5p (Table 1). The 1.6-fold increase in plasma miR-92b was statistically significant (P <0.02); the 1.2-fold increase in plasma miR-142-5p was not (P <0.5). The only other miR measurable in blood was miR-181a, levels of which were increased in plasma while being under-expressed in tumors in the presence of tumor monosomy-3.
Table 2.
Plasma miRs differentially expressed
| miR | Chr | Monosomy-3a | Disomy-3b | P |
|---|---|---|---|---|
| Increased in patients with tumor monosomy-3 | ||||
| hsa-miR-191 | 3 | 7456 | 1760 | 0.0000001 |
| hsa-miR-93 | 7 | 3344 | 836 | 0.000001 |
| hsa-miR-221 | X | 10411 | 3449 | 0.00006 |
| hsa-miR-342-3p | 14 | 962 | ND | 0.00007 |
| hsa-miR-19b | 13 | 2385 | 1017 | 0.0002 |
| hsa-miR-199a-5p | 19 | 1977 | ND | 0.0003 |
| hsa-miR-25 | 7 | 1490 | ND | 0.0009 |
| hsa-miR-27a | 19 | 5182 | 1993 | 0.0009 |
| hsa-miR-23a | 19 | 4566 | 1886 | 0.001 |
| hsa-miR-15b | 3 | 1195 | 530 | 0.001 |
| hsa-miR-223 | X | 10286 | 3413 | 0.002 |
| Decreased in patients with tumor monosomy-3 | ||||
| hsa-miR-1227 | 19 | 1686 | 10791 | 0.0000008 |
| hsa-miR-663 | 20 | 2196 | 16206 | 0.00001 |
| hsa-miR-654-5p | 14 | 420 | 1148 | 0.00008 |
| hsa-miR-1238 | 19 | 1561 | 6172 | 0.0001 |
aAverage signal intensity, n = 10
bAverage signal intensity, n = 10
ND not detectable
Plasma miR quantification
Plasma levels of select miRs increased in the tumor and in the pooled plasma arrays in the presence of tumor monosomy-3 were then examined by quantitative real-time polymerase chain reaction (qRT-PCR) in the individual patients tested, again 10 with tumor monosomy-3 and 10 without. The focus was on the two miRs that were over-expressed in the tumor array that were measurable in plasma, miR-92b and miR-142-5p, and three miRs elevated in the plasma array, miR-191, miR-199a-5p, and miR-223. Three miRs previously reported to be upregulated in uveal melanoma tumors compared to normal choroid, miR-20a, miR-21, and miR-106a, that were not differentially expressed in either the tumor or plasma array, were also assessed [5]. Differential expression in plasma as assessed by qRT-PCR paralleled the qNPA results (Fig. 2). miR-92b, miR-199a-5p, and miR-223 were significant higher in both the qNPA and the qRT-PCR analysis. miR-191 tended to be higher in the qRT-PCR analysis, but increases did not reach the level of significance (P < 0.10), as it did in the qNPA analysis. miR-142-5, miR-20a, miR-21, and miR-106a were not differentially expressed. Levels of the three miRs that were significantly different were then examined in another set of patients with primary uveal melanoma in which tumor chromosome 3 status was obtained on fine needle aspiration (FNA) biopsies. Levels of these miRs were also compared to those of 26 healthy donor controls. Plasma levels of miR-92b, 199a-5p, and 223 were significantly higher in patients with monosomy-3 when compared to patients with disomy; levels of all three were also higher when compared to levels of normal controls (Fig. 3).
Fig. 2.
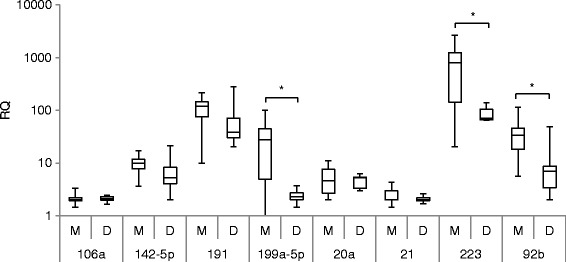
Plasma miR quantification by qRT-PCR in patients with enucleated tumors with (M), n = 10, and without (D), n = 10, tumor monosomy-3. The box represents the 25th and 75th percentiles, the horizontal lines represent the median, and the whiskers represent the minimum and maximum. Brackets with an asterisk above indicate statistical significance P < 0.05, Wilcoxon rank-sum test
Fig. 3.
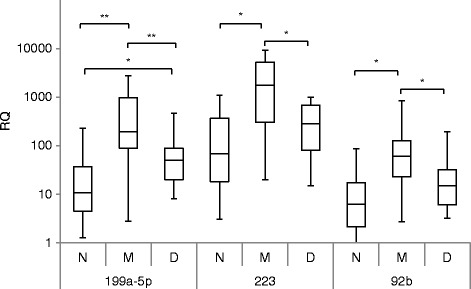
Plasma miR quantification by qRT-PCR in patients with (M), n = 33, and without (D), n = 32, monosomy-3 in which tumor chromosome 3 status was obtained on FNA biopsies. Also displayed are plasma levels of normal controls (N), n = 26. The box represents the 25th and 75th percentiles, the horizontal lines represent the median, and the whiskers represent the minimum and maximum. Brackets with an asterisk above indicate statistical significance P < 0.05, **P < 0.01, Kruskal-Wallis test
Discussion
Tumor and plasma miR profiling of patients with primary uveal melanoma was applied to investigate the role of epigenetic mechanisms in the metastatic process with an overall goal to develop blood biomarkers that could potentially help guide adjuvant therapy decisions and follow-up. Of 858 miRs assessed in tumors manifesting monosomy-3, an accurate predictor of the development of metastasis, 6 were found to be over-expressed and 20, under-expressed. The over-expressed miRs associated with monosomy-3 were analyzed by DIANA mirPath (Multiple microRNA Analysis), a web-based miR pathway analysis application [12]. The top three pathways potentially regulated were actin cytoskeleton, adherens junctions, and TGF-beta signaling, pathways implicated in metastasis, including in uveal melanoma [13–15]. Of note, whereas the target genes of most miRs are enriched, for example, on chromosomes 6, 16, 17, 19, and 22, miR target genes are not enriched on chromosome 3 [16]. None of the miRs we found to be differentially expressed in tumors with monosomy-3 was differentially expressed in tumors studied by Worley et al. [8], who used the class-2 GEP as a surrogate for metastasis. The most significant discriminators in our study were under-expression of miRs of the 506-514 cluster, which has been implicated in initiating melanocyte transformation and promoting melanoma growth and invasiveness [17, 18].
miR production is a complicated process requiring a large number of molecular events to be coordinated. None of the major miR biogenesis factors are encoded on chromosome 3. Tumors manifesting monosomy-3 were characterized by alterations in miR processing factors, which have been associated with the development of metastasis in several types of cancer. DDX17 and TARBP2 were upregulated, and XPO5 and HIWI were down-regulated. DDX17 (22q13.1), a nuclear endonuclease that produces 60 to 70 nucleotide pre-miRs, was identified as a metastasis-associated gene in renal cell carcinoma [19]. A decrease in exportin 5 (XPO5; 6p21.1), which transports pre-miRs into the cytoplasm, has been associated with prognosis in head and neck and in lung cancers [20, 21]. Upregulation of TARBP2 (12q12-q13), a cytoplasmic endonuclease which cleaves pre-miRs into 21 to 22 nucleotide mature miRs in conjunction with Dicer (DICER1), has been associated with metastasis in breast cancer [22]. Down regulation of HIWI (12q24.33), which is integrated into the silencing complex, has been associated with metastasis in pancreatic cancer [23]. Alterations in Dicer, Drosha, and Gemin4, which have been observed in cutaneous melanoma, were not observed [24]. Most of the miRs identified that were discriminatory in tumors with monosomy-3 were down-regulated. How the alterations in miR biogenesis factors influenced this observation will require further study.
None of the miRs that we found to be discriminatory in the tumor array was found to be discriminatory in the plasma array. The plasma miRs most significantly increased was miR-191, which has been implicated in several oncogenic processes [25]. We were able to confirm using qRT-PCR that specific miRs differentially expressed in the arrays were increased in the plasma of patients with tumor monosomy-3 and significantly increased when compared to levels in normal donors. These included one miR over-expressed in the tumor array, miR-92b, and two increased in the plasma array, miR-199a-5p and miR-223. These miRs have also been implicated in several cellular processes. All three have been implicated in regulating genes that promote metastasis [26–28]. All three also regulate host responses. Of note, miR-92b belongs to a cluster of miRs that regulate T cells, including regulatory T cells [29], miR-199a-5p promotes regulatory T cells [30], and miR-223 regulates myeloid suppressor cells [31]. Regulatory T cells and myeloid suppressor cells have been implicated in uveal melanoma progression [32, 33]. Plasma levels of miRs reported to be upregulated in uveal melanoma tumors when compared to normal choroid, miR-20a, miR-21, and miR-106a, were also measured to examine the possibility that increases may represent circulating uveal melanoma cells, a potential predictor of metastasis [5, 34]. Levels of these miRs were not increased in the plasma of patients with tumor monosomy-3.
Virtually, all of the miRs discriminatory in the tumor array were not quantifiable in the plasma array. In contrast to tumor where more miRs were differentially under-expressed, more miRs were differentially increased in the plasma in patients with tumor monosomy-3 compared to without. That miR-expression patterns of tumor differ from those of blood has been previously reported [35]. Several mechanisms may generate blood miRs, including passive leakage from apoptotic or necrotic cells and active secretion of miR-containing microparticles and exosomes. These can occur in malignant cells but also in nonmalignant cells with a short half-life, such as blood cells, or upon tissue damage. There is evidence that most circulating miRs are blood-cell derived [36]. At least 100 different miRs have been shown to circulate in the blood of healthy donors, including most of the miRs we found to be differentially increased, miR-19b, miR-191, miR-199a-5p, miR-25, miR-23a, miR-223, and miR-93 [37, 38]. Several of the differentially expressed miRs identified have been previously reported to be elevated in the plasma or serum of patients with cancer, including miR-92b in prostate [39]; miR-223 in lung, esophageal, and hepatocellular [40–42]; and miRs-199a-5p, miRs-19b, miRs-15b, and miRs-25 in lung [40, 43, 44].
In addition to chromosome 3, abnormalities in chromosomes 1, 6, and 8 have also been associated with metastasis in uveal melanoma. Only three of the 26 tumor miRs and two of the 18 plasma miRs differentially expressed are located on these chromosomes. One of the miRs over-expressed in the tumor array, miR-135a*, localizes to chromosome 3. miR levels are regulated by several transcriptional and post transcription mechanisms as well as poorly understood degradation pathways [45]. miR-135a levels are regulated by Wnt/beta-catenin signaling [46]. Wnt/beta-catenin signaling has been implicated in uveal melanoma development [47]. The miR-506-514 cluster maps to the human X chromosome, which contains approximately 10 % of all miRs detected in the human genome. miR-223 and miR-221, which were increased in plasma, are also X-linked. Although the role of most has not yet been described, several X-linked miRs have been shown to have important functions in cancer as well as in immunity [48]. Nonrandom abnormalities have been previously observed on the sex chromosomes in uveal melanoma, but a consensus regarding their prognostic significance has not been established [49–51]. Although males manifest a slightly higher incidence and mortality rate, gender is not considered to play a major role in uveal melanoma predisposition or prognosis [52].
Conclusions
These results, which derive from the largest number of uveal melanoma samples reported to date, support a role for epigenetic events mediated by miRs in uveal melanoma metastasis and further analysis of miRs as biomarkers of metastatic risk. They also suggest that potentially useful blood miRs may be derived from the host response as well as the tumor. Tumor monosomy-3 and class-2 GEP, although accurate predictors, are surrogate endpoints of metastatic death. Larger scale, prospective studies with clinical endpoints, including early compared to late metastases, will be necessary. The use of blood miR levels in conjunction with imaging studies as part of systemic surveillance for metastasis also merits study.
Methods
Patients
Tumors that had been cryopreserved from patients with uveal melanoma treated by enucleation at the Cleveland Clinic Cole Eye Institute between 2004 and 2010 were analyzed. Starting in 2009, blood was also collected from patients treated with enucleation and from patients undergoing FNA biopsy at the time of plaque radiotherapy. Computed tomography scans of the chest, abdomen, and pelvis were initially performed to rule out metastatic disease. Chromosome 3 status in the enucleation specimen was assessed by single nucleotide polymorphism array and in the FNA biopsies by fluorescent in situ hybridization as previously described [53]. Standard clinical and histologic characteristics were also assessed. This included the presence or absence of significant TILs, which was defined as being more than 100 lymphocytes in 20 high power (40×) fields [54]. All patients underwent scheduled surveillance for the development of metastases with clinical evaluation with liver imaging.
Tumor miR array
Total RNA was extracted from 25 mg of cryopreserved tumor tissue using Trizol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions and further purified using the miRNeasy Mini Kit (Qiagen, Valencia, CA). RNA quality was assessed using the Agilent 2100 Bioanalyzer, and concentration was measured using Nanodrop 1000 (Thermo Fisher Scientific, Waltham, MA). The total RNA (400 ng) was hybridized to the Illumina MicroRNA Profiling BeadChip, containing 858 mature human miR probes and 287 hypothetical small RNA probes according to the standard protocol.
Tumor miR biogenesis factors
RNA was isolated from snap-frozen primary uveal melanoma tissue isolated from enucleated eyes. Purity and concentration were determined using a NanoDrop ND-1000 Spectrometer. Quality RNA was subsequently hybridized using a direct hybridization array kit (Illumina). Each RNA sample was hybridized using the HumanHT-12 BeadChip array (Illumina) in a multiple-step procedure; the chips were washed, dried, and scanned on the BeadArray Reader (Illumina). Raw microarray data were generated using BeadStudio v3.0 (Illumina). Microarray data analysis and quality control were performed using BeadArray R package v1.0.0. After background subtraction (using median background method), the data were normalized using quantile normalization and log-transformed.
Plasma miR array
Plasma samples were forwarded to the HTG Molecular Diagnostics, Inc. (Tucson, AZ) for miR profiling using qNPA. The expression of 674 human miRs was analyzed using Whole Transcript miRNA Microarray Version 11. Each sample was tested in duplicate.
Plasma miR quantification
The total RNA was isolated from plasma using the miRNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. With the exceptions of miR-142-5p and miR-92b, reverse transcription reactions were performed using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. qRT-PCR was performed using the reverse transcription reaction product, TaqMan MicroRNA Assay kit, and TaqMan Universal PCR Master Mix (Applied Biosystems) according to the manufacturer’s instructions. TaqMan MicroRNA Assay kits for human miRs were used. Reactions were loaded onto a 96-well plate and run in duplicate on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems). The reactions were incubated at 50 °C for 20 s and 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, then 1 min of annealing/extension at 60 °C. The ΔΔCT method was used to determine relative number of copies (RQ) of miR. Data were normalized to a Caenorhabditis elegans synthetic miR sequence, cel-miR-39 (Qiagen), which was spiked in as a control during RNA isolation. The miScript PCR System from Qiagen (Valencia, CA) was used for quantification of miR-142-5p and miR-92b. miRs were isolated as described previously; 5 μL of isolated template RNA were used for subsequent reverse transcription reactions which were performed using the miScript II RT Kit according to the manufacturer’s instructions. Real-time PCR was performed using 2× QuantiTect SYBR Green PCR Master Mix, 10× miScript Universal Primer, 10× miScript Primer Assay, and template cDNA from reverse transcription; all reaction volumes suggested by the manufacturer were doubled to perform reactions in duplicate.
Statistical analysis
Significance analysis of microarrays (http://statweb.stanford.edu/~tibs/SAM/) was used to identify miRs differentially expressed between monosomy- and disomy-3 tumors. Normalization by median centering and t test statistic were used in the analysis. In order not to miss subtly expressed miRs in tumors that may be measureable in plasma, the false discovery rate threshold for the tumor array was set at <0.01. Expression of tumor miR biogenesis factors was evaluated by Wilcoxon rank-sum tests. The miR array of plasma from patients with monosomy- and disomy-3 was analyzed by HTG Molecular Diagnostics, Inc. Data were normalized to the total signal for each microarray. Results are reported as average signal intensities. miRs were considered quantifiable if the average signal intensity was >526 in plasma from monosomy-3 donors and >531 in plasma from disomy-3 donors. Differentially expressed plasma miRs were selected using random variance t test, P <0.05, and absolute fold change >2.0. Differential expression of plasma RQ of specific miRs was assessed by Wilcoxon rank-sum test for comparison between two groups or Kruskal-Wallis test for comparison between multiple groups. All tests were two-sided with P < 0.05 considered significant.
Abbreviations
FNA, fine needle aspiration; GEP, gene expression profiling; miR, microRNA; qNPA, quantitative nuclease protection assay; qRT-PCR, quantitative real-time polymerase chain reaction; RQ, relative number of copies
Acknowledgements
The authors would also like to acknowledge the contributions of Dr. Raymond R. Tubbs and the Genomics Core of Cleveland Clinic’s Lerner Research Institute.
Funding
This work was supported in part by RO1CA136776 from the National Cancer Institute, National Institutes of Health, Bethesda, MD, and by the Falk Medical Research Trust, Chicago, IL.
Availability of data and materials
Data are uploaded to the Gene Expression Omnibus (GEO) under accession number GSE44297.
Authors’ contributions
PLT is responsible for the concept, design, and conduct of the study and manuscript preparation. SA and WA processed the tumor and plasma and performed the miR-expression assays. YS assisted with the tumor profiling studies. JWC and ADS provided clinical materials and assisted in the analysis. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The studies were approved by the Cleveland Clinic Institutional Review Board and adhered to the Declaration of Helsinki, and all patients provided written informed consent. Plasma was also obtained from healthy donors without ocular disease, also on an approved study.
Contributor Information
Pierre L. Triozzi, Phone: (336) 716-5772, Email: ptriozzi@wakehealth.edu
Susan Achberger, Email: achbers@ccf.org.
Wayne Aldrich, Email: aldricw@ccf.org.
John W. Crabb, Email: crabbj@ccf.org
Yogen Saunthararajah, Email: saunthy@ccf.org.
Arun D. Singh, Email: singha@ccf.org
References
- 1.Damato B, Coupland SE. Translating uveal melanoma cytogenetics into clinical care. Arch Ophthalmol. 2009;127:423–9. doi: 10.1001/archophthalmol.2009.40. [DOI] [PubMed] [Google Scholar]
- 2.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12:461–8. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landreville S, Agapova OA, Harbour JW. Emerging insights into the molecular pathogenesis of uveal melanoma. Future Oncol. 2008;4:629–36. doi: 10.2217/14796694.4.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herlihy N, Dogrusöz M, van Essen TH, Harbour JW, van der Velden PA, van Eggermond MC, et al. Skewed expression of the genes encoding epigenetic modifiers in high-risk uveal melanoma. Invest Ophthalmol Vis Sci. 2015;56:1447–58. doi: 10.1167/iovs.14-15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang C, Wei W. The miRNA expression profile of the uveal melanoma. Sci China Life Sci. 2011;54:351–8. doi: 10.1007/s11427-011-4149-y. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Wang J, Shen H, Lu J, Li C, Hu DN, et al. Epigenetics, microRNAs, and carcinogenesis: functional role of microRNA-137 in uveal melanoma. Invest Ophthalmol Vis Sci. 2011;52:1193–9. doi: 10.1167/iovs.10-5272. [DOI] [PubMed] [Google Scholar]
- 7.Yan D, Zhou X, Chen X, Hu DN, Dong XD, Wang J, et al. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Invest Ophthalmol Vis Sci. 2009;50:1559–65. doi: 10.1167/iovs.08-2681. [DOI] [PubMed] [Google Scholar]
- 8.Worley LA, Long MD, Onken MD, Harbour JW. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008;18:184–90. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- 9.Larsen AC, Holst L, Kaczkowski B, Andersen MT, Manfé V, Siersma VD, et al. MicroRNA expression analysis and Multiplex ligation-dependent probe amplification in metastatic and non-metastatic uveal melanoma. Acta Ophthalmol. 2014;92:541–9. doi: 10.1111/aos.12322. [DOI] [PubMed] [Google Scholar]
- 10.Triozzi PL, Singh AD. Blood biomarkers for uveal melanoma. Future Oncol. 2012;8:205–15. doi: 10.2217/fon.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 12.Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–3. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- 13.Mäkitie T, Carpén O, Vaheri A, Kivelä T. Ezrin as a prognostic indicator and its relationship to tumor characteristics in uveal malignant melanoma. Invest Ophthalmol Vis Sci. 2001;42:2442–9. [PubMed] [Google Scholar]
- 14.Chang SH, Worley LA, Onken MD, Harbour JW. Prognostic biomarkers in uveal melanoma: evidence for a stem cell-like phenotype associated with metastasis. Melanoma Res. 2008;18:191–200. doi: 10.1097/CMR.0b013e3283005270. [DOI] [PubMed] [Google Scholar]
- 15.Woodward JK, Elshaw SR, Murray AK, Nichols CE, Cross N, Laws D, Rennie IG, Sisley K. Stimulation and inhibition of uveal melanoma invasion by HGF, GRO, IL-1alpha and TGF-beta. Invest Ophthalmol Vis Sci. 2002;43:3144–52. [PubMed] [Google Scholar]
- 16.Wang ZZ, Gong BS, Wang HK, Wang HJ, Zhou M, Wang QH, et al. MicroRNA regulation constrains the organization of target genes on mammalian chromosomes. FEBS Lett. 2011;585(12):1897–904. doi: 10.1016/j.febslet.2011.04.059. [DOI] [PubMed] [Google Scholar]
- 17.Streicher KL, Zhu W, Lehmann KP, Georgantas RW, Morehouse CA, Brohawn P, et al. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene. 2012;31:1558–70. doi: 10.1038/onc.2011.345. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Liu H, Li Y, Wu J, Greenlee AR, Yang C, et al. The role of miR-506 in transformed 16HBE cells induced by anti-benzo[a]pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Toxicol Lett. 2011;205:320–6. doi: 10.1016/j.toxlet.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 19.Tan X, Zhai Y, Chang W, Hou J, He S, Lin L, et al. Global analysis of metastasis-associated gene expression in primary cultures from clinical specimens of clear-cell renal-cell carcinoma. Int J Cancer. 2008;123:1080–8. doi: 10.1002/ijc.23637. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Yang H, Lee JJ, Kim E, Lippman SM, Khuri FR, et al. MicroRNA-related genetic variations as predictors for risk of second primary tumor and/or recurrence in patients with early-stage head and neck cancer. Carcinogenesis. 2010;31:2118–23. doi: 10.1093/carcin/bgq177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Z, Wang H, Li Y, Li B, Li C, Ding C. A microRNA-related single nucleotide polymorphism of the XPO5 gene is associated with survival of small cell lung cancer patients. Biomed Rep. 2013;1:545–548. doi: 10.3892/br.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X, Wu M, Liu P, Wei F, Li L, Tang H, et al. Up-regulation and worse prognostic marker of cytoplasmic TARBP2 expression in obstinate breast cancer. Med Oncol. 2014;31:868. doi: 10.1007/s12032-014-0868-9. [DOI] [PubMed] [Google Scholar]
- 23.Grochola LF, Greither T, Taubert H, Möller P, Knippschild U, Udelnow A, et al. The stem cell-associated Hiwi gene in human adenocarcinoma of the pancreas: expression and risk of tumour-related death. Br J Cancer. 2008;99:1083–8. doi: 10.1038/sj.bjc.6604653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Z, Swede H, Cassarino D, Fleming E, Fire A, Dadras SS. Up-regulated Dicer expression in patients with cutaneous melanoma. PLoS One. 2011;6:e20494. doi: 10.1371/journal.pone.0020494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagpal N, Kulshreshtha R. miR-191: an emerging player in disease biology. Front Genet. 2014;5:99. doi: 10.3389/fgene.2014.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuang LK, Yang YT, Ma X, Han B, Wang ZS, Zhao QY, Wu LQ, Qu ZQ. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203. doi: 10.1038/cddis.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Liu R, Wang Y, Tang J, Tang S, Chen X, Xia K, Xiong W, Xu D, Wang S, He Q, Cao K. miR-199a-5p regulates the expression of metastasis-associated genes in B16F10 melanoma cells. Int J Clin Exp Pathol. 2014;7:7182–90. [PMC free article] [PubMed] [Google Scholar]
- 28.Liang H, Yan X, Pan Y, Wang Y, Wang N, Li L, Liu Y, Chen X, Zhang CY, Gu H, Zen K. MicroRNA-223 delivered by platelet-derived microvesicles promotes lung cancer cell invasion via targeting tumor suppressor EPB41L3. Mol Cancer. 2015;14:58. doi: 10.1186/s12943-015-0327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Kouchkovsky D, Esensten JH, Rosenthal WL, Morar MM, Bluestone JA, Jeker LT. microRNA-17-92 regulates IL-10 production by regulatory T cells and control of experimental autoimmune encephalomyelitis. J Immunol. 2013;191:1594–605. doi: 10.4049/jimmunol.1203567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatila WM, Criner GJ, Hancock WW, Akimova T, Moldover B, Chang JK, et al. Blunted expression of miR-199a-5p in regulatory T cells of patients with chronic obstructive pulmonary disease compared to unaffected smokers. Clin Exp Immunol. 2014;177:341–52. doi: 10.1111/cei.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Q, Zhang M, Jiang X, Zhang Z, Dai L, Min S, et al. miR-223 suppresses differentiation of tumor-induced CD11b+ Gr1+ myeloid-derived suppressor cells from bone marrow cells. Int J Cancer. 2011;129:2662–73. [DOI] [PubMed]
- 32.McKenna KC, Beatty KM, Bilonick RA, Schoenfield L, Lathrop KL, Singh AD. Activated CD11b+ CD15+ granulocytes increase in the blood of patients with uveal melanoma. Invest Ophthalmol Vis Sci. 2009;50:4295–303. doi: 10.1167/iovs.08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mougiakakos D, Johansson CC, Trocme E, All-Ericsson C, Economou MA, Larsson O, et al. Intratumoral forkhead box P3-positive regulatory T cells predict poor survival in cyclooxygenase-2-positive uveal melanoma. Cancer. 2010;116:2224–33. doi: 10.1002/cncr.24999. [DOI] [PubMed] [Google Scholar]
- 34.Torre V, Triozzi P, Eng C, Tubbs R, Schoenfiled L, Crabb JW, et al. Circulating tumor cells in uveal melanoma. Future Oncol. 2011;7:101–9. doi: 10.2217/fon.10.143. [DOI] [PubMed] [Google Scholar]
- 35.Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–18. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 38.Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010;5:e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4:e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, et al. Identification of ten serum microRNAs from a genomewide serum microRNA expression profile as novel non-invasive biomarkers for non-small cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–8. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 41.Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–9. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating MicroRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–42. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 43.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A. 2011;108:3713–8. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang QZ, Xu W, Habib N, Xu R. Potential uses of microRNA in lung cancer diagnosis, prognosis, and therapy. Curr Cancer Drug Targets. 2009;9:572–94. doi: 10.2174/156800909788486731. [DOI] [PubMed] [Google Scholar]
- 45.Davis BN, Hata A. Regulation of microRNA biogenesis: a miRiad of mechanisms. Cell Commun Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caronia-Brown G, Anderegg A, Awatramani R. Expression and functional analysis of the Wnt/beta-catenin induced mir-135a-2 locus in embryonic forebrain development. Neural Dev. 2016;11:9. doi: 10.1186/s13064-016-0065-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuidervaart W, Pavey S, van Nieuwpoort FA, Packer L, Out C, Maat W, Jager MJ, Gruis NA, Hayward NK. Expression of Wnt5a and its downstream effector beta-catenin in uveal melanoma. Melanoma Res. 2007;17:380–6. doi: 10.1097/CMR.0b013e3282f1d302. [DOI] [PubMed] [Google Scholar]
- 48.Pinheiro I, Dejager L, Libert C. X-chromosome-located microRNAs in immunity: Might they explain male/female differences?: The X chromosome-genomic context may affect X-located miRNAs and downstream signaling, thereby contributing to the enhanced immune response of females. Bioessays. 2011;33:791–802. doi: 10.1002/bies.201100047. [DOI] [PubMed] [Google Scholar]
- 49.White JS, McLean IW, Becker RL, Director-Myska AE, Nath J. Correlation of comparative genomic hybridization results of 100 archival uveal melanomas with patient survival. Cancer Genet Cytogenet. 2006;170:29–39. doi: 10.1016/j.cancergencyto.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Speicher MR, Prescher G, du Manoir S, Jauch A, Horsthemke B, Bornfeld N, et al. Chromosomal gains and losses in uveal melanomas detected by comparative genomic hybridization. Cancer Res. 1994;54:3817–23. [PubMed] [Google Scholar]
- 51.Sisley K, Parsons MA, Garnham J, Potter AM, Curtis D, Rees RC, et al. Association of specific chromosome alterations with tumour phenotype in posterior uveal melanoma. Br J Cancer. 2000;82:330–8. doi: 10.1054/bjoc.1999.0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh AD, Turell ME, Topham AK. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118:1881–5. doi: 10.1016/j.ophtha.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 53.Singh AD, Aronow ME, Sun Y, Bebek G, Saunthararajah Y, Schoenfield LR, et al. Chromosome 3 status in uveal melanoma: a comparison of fluorescence in situ hybridization and single-nucleotide polymorphism array. Invest Ophthalmol Vis Sci. 2012;53:3331–9. doi: 10.1167/iovs.11-9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de la Cruz PO, Specht CS, McLean IW. Lymphocytic infiltration in uveal malignant melanoma. Cancer. 1990;65:112–115. doi: 10.1002/1097-0142(19900101)65:1<112::AID-CNCR2820650123>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are uploaded to the Gene Expression Omnibus (GEO) under accession number GSE44297.


