Abstract
High throughput screening of a 4096 compound library of boronic acid and acridine containing branched peptides revealed compounds that have dissociation constants in the low nanomolar regime for HIV-1 RRE IIB RNA. We demonstrate that branched peptide boronic acids A5, A6, and A7 inhibit the production of p24, an HIV-1 capsid protein, in a dose-dependent manner.
The interaction of HIV-1 Rev protein with the Rev response element (RRE) RNA plays a critical role in the assembly of new viral particles. Unspliced and singly spliced mRNAs are exported to the cytoplasm by the cooperative docking of multiple Rev proteins with a highly structured intronic RRE (~350 nucleotides). Once the Rev–RRE oligomeric complex is formed and interacts with the Crm-1 pathway, mRNA nuclear export ensues.1–4 Disruption of this protein–RNA interaction presents an avenue to inhibit viral replication. The RNA component serves as a prime target because its nucleic acid sequence is highly conserved and structured. Biochemical and crystal structures studies reveal the α-helical arginine-rich motif of Rev binding in the major groove of the RNA through an extensive network of sequence specific and electrostatic contacts.5–10 Further, RRE appears to act as a scaffold in the oligomerization of Rev.11–13 In this study, we focused on targeting the primary, high-affinity binding site of Rev, RRE IIB.
As a general platform to develop compounds capable of selectively targeting folded structures of RNA, we utilized a branched peptide scaffold in order to introduce diverse structures and opportunities for multivalent interactions.14,15 Our previous studies suggest that branched peptides utilize a large surface area of RNA to achieve selective binding. The second generation library, which targeted the stem loop RRE IIB, incorporated boronic acids to capitalize on the empty p-orbital of boron. We hypothesized that the formation of a reversible covalent Lewis acid–base complex with the 2′-hydroxyl group of RNA can increase selectivity.16,17 Indeed, we showed that branched peptide boronic acids (BPBAs) demonstrated a marked increase in selectivity using identical sequence of RNA versus DNA.16 However, their binding affinities were in the low micromolar range, making them unsuitable for competing against native protein–RNA interactions where the binding affinity is in the nanomolar range.
To improve the affinity of the BPBA library towards RRE IIB, we investigated the effect of introducing an intercalator as a structural feature. We hypothesized that incorporation of an acridine18,19 moiety will facilitate intercalation whereas other functional groups on the peptide side chains will promote selective recognition of the RNA target (Fig. 1). Acridine has been conjugated to a variety of ligands in order to target RNA. For example, acridine-N-neomycin and an amphiphilic helical peptide containing two RNA acridinyl lysine residues afforded Kds of 2.4 nM and 610 pM against target RNA, respectively.20,21 In addition, a peptide-acridine conjugate has been shown to bind to a duplex RNA, where acridine is shown to interact with the RNA via a threading intercalation process.22
Fig. 1.

Possible interactions of acridine and boronic acid with target.
The BPBA library was synthesized in triplicate on Tentagel beads by split and pool synthesis, where a photocleavable linker, 3-amino-3-(2-nitrophenyl)propionic acid (ANP) was installed on the C-terminus to allow for post-screening selection and deconvolution. The library was prepared such that there were three variable amino acid positions at both the N-and C-termini (AA1–AA3 and AA4–AA6, respectively), and each variable position was composed of four side chains, generating a library containing 4096 unique sequences (Fig. 2). As the major goal of this library is to determine the effect of incorporating an intercalator on binding affinity, we chose four amino acids to represent several modes of interaction towards the RNA. For example, in each position at AA1–AA6 we used N-ε-(9-aminoacridinyl-)L-lysine (KACR) for interactions with the RNA through π stacking, Lys for electrostatic attraction and hydrogen bonding, Leu for hydrophobic interaction, and KBBA for reversible covalent bonding. Tyr was incorporated at the AA7 position for spectroscopic quantification of the peptides. The BP library was subjected to an on-bead high throughput screen against DY547-labeled HIV-1 RRE IIB RNA, after a preincubation using bovine serum albumin and tRNA to minimize non-specific binding.14,15
Fig. 2.
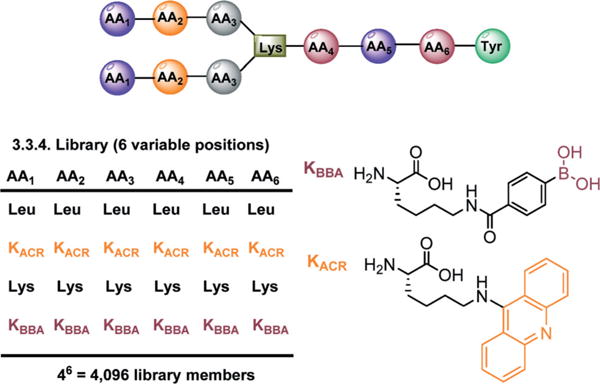
Branched peptide library and structures of KBBA and KACR.
Binding of RRE IIB to BPBAs resulted in increased bead fluorescence, which was monitored by both fluorescence and confocal microscopy. Twenty beads were selected, photocleaved via UV irradiation, and then sequenced by MALDI MS–MS analysis.23 Sequences for fourteen were obtained and the dissociation constants (Kd) were determined by electrophoretic mobility shift assays (EMSAs) (Table 1). Job plot analyses using 2-aminopurine (2-AP)-labeled RRE IIB were used to determine the stoichiometry for all acridine-containing compounds (see ESI‡). We were fortunate to find three sequences in duplicate (A1, A10, and A12), which increased confidence in the quality of the library as well as the screening process. Gratifyingly, the hits contained boronic acid, acridine or both moieties. Sequence analysis of the hits suggests that KBBA is preferred on the C-terminus, whereas KACR is present in both the N- and C-termini. In general, a substantial improvement in the dissociation constants was observed in comparison to our second generation library, where the majority of the peptides were in the low micromolar range.17 As shown in Table 1, there is a preference for lysine in positions AA1 to AA3 (entries 1–5). A preference for positively charged side chains, however, is not unexpected because the Rev protein binds RRE IIB through the polyarginine nuclear localization signal (NLS) located in its N-terminal region.9,11,24,25 The positively charged Lys side chains in our hits likely function similarly to the Arg residues of the Rev NLS and may provide the necessary electrostatic attraction with the negatively charged phosphate backbone of RRE IIB. Both acridine and boronic acid moieties were observed in these peptides, and in general, acridine-containing peptides were more potent than their boronic acid counterparts. In particular, peptide A4, which contained an acridine at position AA6, had a Kd of 20 nM. This value is significantly better than the dissociation constants obtained from the EMSA analysis of Rev17 (Suc-TRQARRNRRRRWRERQRAAAAR-am, Kd 100 nM), the minimum binding elements of the Rev protein, with wild-type RRE IIB RNA.26 The position of this group appears to be critical as moving it to a different position (i.e., A3) increased the Kd to 140 nM. However, when the lysine residues in the AA1–AA3 positions are substituted with acridine or boronic acid groups, the intercalating properties of acridine outweigh binding interactions of the boronic acid (compare entries 6 and 7 to 8 and 9). Finally, the hydrophobic nature of leucine in these positions lead to weaker binding affinities (entries 10–14). It is noteworthy that the number of Lys residues in the hit compounds did not result in increased binding affinity. For example, A11 with six Lys residues had a >7-fold higher Kd value (1430 vs. 210 nM) compared to A13 with four Lys residues. We suspect that an alternative mode of binding compensated for the loss of electrostatic interactions.
Table 1.
Sequences and Kds by EMSA
| Entry | Peptide | Sequencea | Kd (nM) |
|---|---|---|---|
| 1 | A1 | (KKK)2*LKBBAKBBAY | 280 ± 80 |
| 2 | A2 | (KKK)2*LKBBALY | 130 ± 30 |
| 3 | A3 | (KKK)2*KACRLKY | 140 ± 50 |
| 4 | A4 | (KKK)2*KLKACRY | 20 ± 4 |
| 5 | A5 | (KKK)2*KACRKACRKY | 30 ± 4 |
| 6 | A6 | (KACRKK)2*KLKBBAY | 60 ± 10 |
| 7 | A7 | (KKACRK)2*KLKBBAY | 70 ± 30 |
| 8 | A8 | (KKBBAK)2*LLKY | 660 ± 90 |
| 9 | A9 | (KKKBBA)2*KLKBBAY | 120 ± 40 |
| 10 | A10 | (KLK)2*LKKBBAY | 300 ± 40 |
| 11 | A11 | (KLK)2*KKKBBAY | 1430 ± 870 |
| 12 | A12 | (KKL)2*LKKBBAY | 330 ± 100 |
| 13 | A13 | (LLK)2*KKBBAKY | 210 ± 30 |
| 14 | A14 | (KKL)2*KKBBAKBBAY | 170 ± 50 |
* = Lysine branching unit. Each Kd value is an average at least three experiments.
With the improved binding affinities of our BPBAs, we determined whether these new generation peptides could inhibit HIV replication. First, the hit BPBAs were screened for toxicity via an MTT assay using a U87 CXCR4 human glioblastoma cell line stably transfected with CD4 and CXCR4 (Fig. 3). Almost all of the compounds were non-toxic at concentrations up to 100 μM; A4 and A6 showed slight toxicity. One compound, A7, displayed cytotoxicity in a dose-dependent manner. It is interesting that while A6 and A7 had similar Kds to RRE IIB and differed only in the sequence order of positions AA1 and AA2 (KACRK vs. KKACR), A6 was nearly non-toxic while A7 showed moderate toxicity in this cell line, suggesting that structure plays a role in toxicity. Overall, the results suggested that branched peptides containing boronic acid and acridine were non-toxic.
Fig. 3.
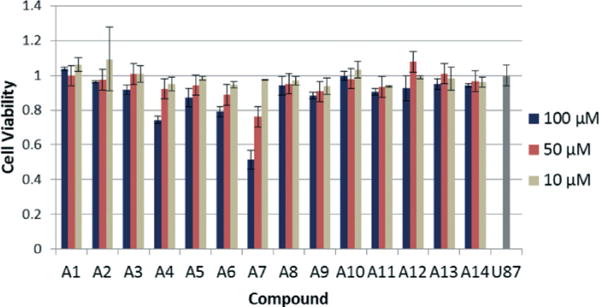
MTT assay of BPs at varying concentrations. U87 column represents the control with no compound added.
We next determined the ability of these peptides to inhibit HIV replication in cell culture by monitoring the production of the HIV-1 capsid protein, p24, by ELISA.27,28 The initial screen was performed at 10 and 100 μM (see Fig. SI.4 in the ESI‡). A significant number of compounds (A1, A3, A4, A5, A6, and A7) greatly suppressed HIV replication at 100 μM, while at a lower concentration of 10 μM, only A11 and A7 inhibited the production of p24 by 40 percent and 60 percent, respectively. Other compounds either showed slight inhibitory activities or did not demonstrate any inhibition of p24. The compounds that demonstrated dose-dependent inhibition of p24 along with complete or virtually complete inhibition of p24 at 100 μM (A1, A4, A5, A6, and A7) were subjected to another screening at 20, 40, 60, and 80 μM (Fig. 4). This assay revealed a dose-dependent inhibition of p24 for A5, A6, and A7, with approximate IC50 values of 10.2, 10.1, and 7.3 μM, respectively. For A5, A6, and A7, approximately 90% inhibition of p24 expression was achieved at 40 μM. Since the decrease in p24 concentration could be due to a loss of cell viability, an MTT assay was performed in parallel with the second screening. To our delight, the results confirmed cell viability at this concentration (Fig. SI.5 in the ESI‡). Taken together, these data support the proposal that acridine-containing BPs inhibit HIV replication in cell culture in a dose-dependent manner.
Fig. 4.

p24 inhibition assay for select compounds.
To understand the interaction of our branched peptides with RRE RNA, a footprinting experiment using ribonucleases was performed. We focused our attention on A6 rather than the strongest binders in the library (A4 or A5) due to the desire to probe peptide–RNA contacts with a more diverse sequence containing both acridine and boronic acid. Following our previous protocol,17 5′-32P-labeled RRE IIB was incubated with varying concentrations (up to 20 μM) of A6 in the presence of RNase T1, RNase A, or RNase V1 (Fig. 5). Potential binding sites were based on the ability of the peptide to protect the RNA from enzymatic cleavage. While there were no noticeable changes in band intensity with RNase T1, the most prominent cleavage bands were observed at G17 and C18 using RNase V1, which preferentially cleaves double stranded regions of RNA. The results of this assay indicate a concentration dependent protection (0.2 μM to 20 μM), which suggests that this portion of RRE IIB is a potential binding site. A6 also protected U7, C21, C29, and U36 from cleavage by RNase A, which hydrolyzes at the 3′ side of unpaired pyrimidine bases. Fortunately, this region is also the site wherein the native protein binding partner Rev binds.4,29 Taken together, the RNA footprinting assay indicated that the binding site for A6 encompasses a large surface area constituting stem B and loops A and B (for structure see Fig. SI.6 in the ESI‡). Job plots confirmed a 1: 1 binding stoichiometry between A6 and RRE IIB (see the ESI‡), and may suggest that A6 is bound in a folded groove saddled between the internal loop regions and upper stem bases of RRE IIB, as this compound is interacting with these structural elements. Although the precise nature of the RNA: peptide interaction is currently unknown, protection from enzymatic cleavage of these regions could result either from steric blockade or remodeling of its tertiary structure. The downregulation of p24 synthesis observed in vitro can, in principle, be a result of changes in RRE tertiary structure or encumbrance of the Rev binding site that impairs Rev–RRE interaction. However, since the assay shown here measures viral replication and not Rev function directly, we cannot rule out the possibility that the compounds are inhibiting other steps in the HIV life cycle.
Fig. 5.
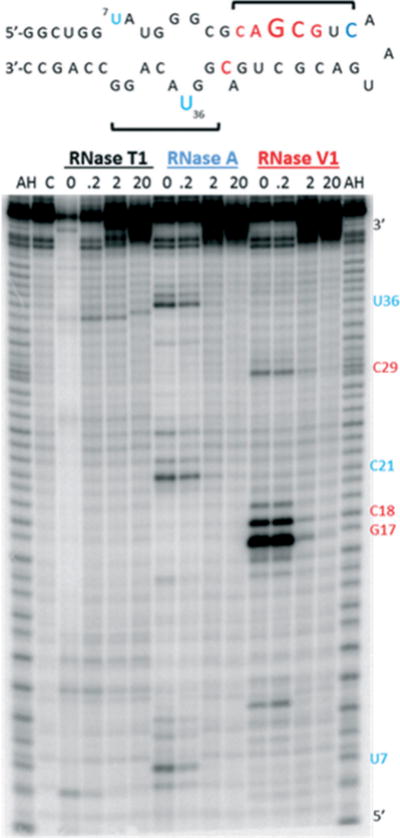
Top: Summary of RNase protection of RRE IIB with A6. Font size reflects levels of protection from RNase A (blue) and RNase VI (red), and brackets indicate regions of peptide: RNA contact; Bottom: Autoradiograph of footprinting experiment. AH is the alkaline hydrolysis ladder and C is the negative control (RRE IIB only, with no RNase or peptide).
In summary, a 3.3.4 BPBA library featuring boronic acid and acridine moieties yielded a series of peptides with improved binding affinities (double digit nM) towards HIV-1 RRE IIB RNA. Our studies support the hypothesis that incorporation of intercalating moieties in combination with boronic acids in branched peptides can have a beneficial effect in boosting binding affinity towards the target RNA. Further, we demonstrated that the majority of these compounds are non-toxic and more importantly, inhibit HIV replication in cell culture. RNase footprinting assays revealed stem B and loops A and B as the likely sites of interaction between RRE and branched peptide A6. Our studies highlight the potential of RNA-binding compounds containing acridine and boronic acid groups as a general platform to target conserved, highly structured RNA sequences that control biological pathways. Efforts towards improvement of this strategy as well as other RNA targets are currently underway.
Supplementary Material
Acknowledgments
We thank K. Ray and R. Helm at the VT mass spectrometry incubator for use of their MALDI-TOF and K. DeCourcy at the Fralin Life Science Institute for access to their microscope facility. This work was supported by NIH (RO1 GM093834 and RO1 GM110009).
Footnotes
The authors declare no competing interests.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c6md00171h
Notes and references
- 1.Mann DA, Mikaelian I, Zemmel RW, Green SM, Lowe AD, Kimura T, Singh M, Butler PJG, Gait MJ, Karn J. J Mol Biol. 1994;241:193–207. doi: 10.1006/jmbi.1994.1488. [DOI] [PubMed] [Google Scholar]
- 2.Van Ryk DI, Venkatesan S. J Biol Chem. 1999;274:17452–17463. doi: 10.1074/jbc.274.25.17452. [DOI] [PubMed] [Google Scholar]
- 3.Pollard VW, Malim MH. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 4.Kjems J, Brown M, Chang DD, Sharp PA. Proc Natl Acad Sci U S A. 1991;88:683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 6.Cook KS, Fisk GJ, Hauber J, Usman N, Daly TJ, Rusche JR. Nucleic Acids Res. 1991;19:1577–1583. doi: 10.1093/nar/19.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heaphy S, Finch JT, Gait MJ, Karn J, Singh M. Proc Natl Acad Sci U S A. 1991;88:7366–7370. doi: 10.1073/pnas.88.16.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kjems J, Brown M, Chang DD, Sharp PA. Proc Natl Acad Sci U S A. 1991;88:683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Battiste JL, Mao H, Rao NS, Tan R, Muhandiram DR, Kay LE, Frankel AD, Williamson JR. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 10.Pond SJK, Ridgeway WK, Robertson R, Wang J, Millar DP. Proc Natl Acad Sci U S A. 2009;106:1404–1408. doi: 10.1073/pnas.0807388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daugherty MD, Booth DS, Jayaraman B, Cheng Y, Frankel AD. Proc Natl Acad Sci U S A. 2010;107:12481–12486. doi: 10.1073/pnas.1007022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang X, Wang J, O’Carroll I, Mitchell M, Zuo X, Wang Y, Yu P, Liu Y, Rausch J, Dyba MA, Kjems J, Schwieters CD, Seifert S, Winans RE, Watts NR, Stahl SJ, Wingfield PT, Byrd RA, Le Grice SFJ, Rein A, Wang YX. Cell. 2013;155:594–605. doi: 10.1016/j.cell.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayaraman B, Crosby DC, Homer C, Ribeiro I, Mavor D, Frankel AD. eLife. 2014;3:e04120. doi: 10.7554/eLife.04120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryson DI, Zhang W, Ray WK, Santos WL. Mol BioSyst. 2009;5:1070–1073. doi: 10.1039/b904304g. [DOI] [PubMed] [Google Scholar]
- 15.Bryson DI, Zhang W, McLendon PM, Reineke TM, Santos WL. ACS Chem Biol. 2012;7:210–217. doi: 10.1021/cb200181v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Bryson DI, Crumpton JB, Wynn J, Santos WL. Chem Commun. 2013;49:2436–2438. doi: 10.1039/c3cc00243h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Bryson DI, Crumpton JB, Wynn J, Santos WL. Org Biomol Chem. 2013;11:6263–6271. doi: 10.1039/c3ob41053f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeJong ES, Chang CE, Gilson MK, Marino JP. Biochemistry. 2003;42:8035–8046. doi: 10.1021/bi034252z. [DOI] [PubMed] [Google Scholar]
- 19.Renner S, Ludwig V, Boden O, Scheffer U, Göbel M, Schneider G. Chem Bio Chem. 2005;6:1119–1125. doi: 10.1002/cbic.200400376. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y, Hyun S, Kim HJ, Yu J. Angew Chem, Int Ed. 2008;47:134–137. doi: 10.1002/anie.200703090. [DOI] [PubMed] [Google Scholar]
- 21.Luedtke NW, Liu Q, Tor Y. Biochemistry. 2003;42:11391–11403. doi: 10.1021/bi034766y. [DOI] [PubMed] [Google Scholar]
- 22.Gooch BD, Beal PA. J Am Chem Soc. 2004;126:10603–10610. doi: 10.1021/ja047818v. [DOI] [PubMed] [Google Scholar]
- 23.Crumpton JB, Zhang W, Santos WL. Anal Chem. 2011;83:3548–3554. doi: 10.1021/ac2002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daugherty MD, Liu B, Frankel AD. Nat Struct Mol Biol. 2010;17:1337–1342. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. Proc Natl Acad Sci U S A. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills NL, Daugherty MD, Frankel AD, Guy RK. J Am Chem Soc. 2006;128:3496–3497. doi: 10.1021/ja0582051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shuck-Lee D, Chen FF, Willard R, Raman S, Ptak R, Hammarskjold M-L, Rekosh D. Antimicrob Agents Chemother. 2008;52:3169–3179. doi: 10.1128/AAC.00274-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tebit DM, Lobritz M, Lalonde M, Immonen T, Singh K, Sarafianos S, Herchenroder O, Krausslich HG, Arts EJ. J Virol. 2010;84:9817–9830. doi: 10.1128/JVI.00991-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kjems J, Calnan BJ, Frankel AD, Sharp PA. EMBO J. 1992;11:1119–1129. doi: 10.1002/j.1460-2075.1992.tb05152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


