Abstract
Helicobacter pylori is the strongest risk factor for gastric adenocarcinoma, which develops within a hypochlorhydric environment. We sequentially isolated H. pylori (strain J99) from a patient who developed corpus-predominant gastritis and hypochlorhydia over a 6-year interval. Archival J99 survived significantly better under acidic conditions than recent J99 strains. H. pylori arsRS encodes a 2-component system critical for stress responses; recent J99 isolates harbored 2 nonsynonymous arsS mutations, and arsS inactivation abolished acid survival. In vivo, acid-resistant archival, but not recent J99, successfully colonized high-acid-secreting rodents. Thus, genetic evolution of arsS may influence progression to hypochlorhydia and gastric cancer.
Keywords: H. pylori, hypochlorhydia, acid resistance
Helicobacter pylori represents the strongest known risk factor for the development of gastric cancer; yet only 2%–3% of colonized persons ever develop this malignancy [1–3]. The most common form of gastric cancer, intestinal-type gastric adenocarcinoma, progresses from normal mucosa to antral-predominant, corpus-predominant, and atrophic gastritis, then to intestinal metaplasia, and finally to dysplasia and adenocarcinoma. During this cascade, acid secretion becomes progressively impaired, which is precipitated by the transition from antral-predominant to corpus-predominant gastritis [4].
H. pylori has evolved to survive under highly acidic conditions, and one mechanism that underpins the acid-resistant phenotype is production of urease [5–7]. Another constituent used by H. pylori within this context is the ArsRS 2-component (ArsR and ArsS) signal transduction system, which regulates genes involved in acid resistance, including urease [5, 6]. ArsS is the sensory constituent of this 2-component system, and the periplasmic domain of ArsS is required to detect external changes in pH, whereas ArsR is the response component. As acidity increases, ArsS undergoes autophosphorylation at histidine 214 within the cytoplasmic kinase domain. Phosphorylated histidine 214 subsequently transfers the phosphate residue to aspartic acid residue 52 on ArsR [5–7], liberating ArsR to function as a transcription factor regulating expression of urease complex components, amidases, and carbonic anhydrase [5–7]. These data suggest that modifications in the ability to respond to gastric acid may differ in H. pylori strains predisposed to the development of intestinal-type gastric adenocarcinoma.
To address this hypothesis, we used a unique set of paired H. pylori isolates [8]. Archival H. pylori strain J99 is a fully sequenced human clinical strain; 6 years after isolating this strain, the source patient underwent repeat endoscopy, and new (recent) J99 isolates were harvested. We now use these unique samples to define differences in acid survival, genetic evolution, topography of inflammation, and the ability of strains to colonize in vivo acidic niches, as a means to understand how a subset of H. pylori–infected individuals may be predisposed toward carcinogenesis.
MATERIALS AND METHODS
Bacterial Strains
Archival H. pylori strain J99 was isolated from a gastric biopsy specimen from a patient with a duodenal ulcer [8]. The source patient refused antimicrobial treatment and underwent repeat endoscopy 6 years after the original J99 strain was isolated, and new (recent) J99 isolates were obtained. Pooled H. pylori isolates were used for these studies. Histopathologic parameters of inflammation were determined as reported elsewhere [9]. All procedures were approved by the Vanderbilt University (IRB 000109) and Nashville Department of Veterans Affairs (IRB 5910) institutional review boards.
H. pylori strains were grown for 16–18 hours in Brucella broth with 10% fetal bovine serum [10] and then inoculated into broth adjusted to a pH of 5.0 or 7.0, incubated for 60 minutes, and plated by serial dilution. After 4 days, colonies were counted.
Construction of arsS Deletion and Reconstituted Mutants
The isogenic H. pylori J99 arsS mutant strain was constructed as follows: 3′ and 5′ recombination flanking sequences of arsS were amplified from J99 chromosomal DNA. The 3′ region was amplified using primers containing restriction enzyme sites, CTGAGCGGCCGCTTGTCGCTTTGTTTATGATAACGC (NotI) and CTGATCTAGATTTCTATTGTGGTTTCTCTAAAGCC (XbaI), and then cloned into pBSC301 vector containing a kanamycin cassette. The 5′ flanking arsS sequence was amplified using the primers CTGAGAATTCGGTGTCCAATAAAAGCAAACCTT (EcoRI) and CTGAGGTACCCCCTTCATTCCCTTCACCTC (KpnI) and cloned into the pBSC103 construct containing the 3′ flanking sequence. The resultant plasmid was used for H. pylori natural transformation with selection using 25 µg/mL kanamycin.
To generate an archival J99 mutant that harbors the recent J99 arsS, a 4286–base pair region containing the 3′ end of arsR (jhp0152), arsS, hemB, jhp0149, and the 3′ end of jhp0148 was amplified from recent J99 DNA using forward primer 5′GATTGAGAGCGAGAGCATCAACCC3″ and reverse primer 5′CATTTGGTGGCTACAACCGGTGAG3″. Amplicons were then cloned into the pGEM-T Easy vector system. A chloramphenicol resistance cassette, CAT, was excised as an EcoRV/SmaI fragment from pBSC103 and ligated into a blunt-ended BamHI site in the intergenic region between jhp0149 and jhp0148 to avoid disrupting arsS transcription. The resulting plasmid was naturally transformed into the archival J99 strain and selected using 10 µg/mL chloramphenicol. Sequencing confirmed that the 2 recent J99 arsS mutations were incorporated into the archival J99 genome.
Animal Models
All procedures were approved by the Institutional Animal Care and Use Committee of Vanderbilt University. Male C57BL/6 mice were purchased from Harlan Laboratories, and male Mongolian gerbils were purchased from Charles River Laboratories. Gerbils and mice were challenged [10] with either Brucella broth or H. pylori (109 colony-forming units per animal) and euthanized 2 weeks after the challenge.
Statistical Analysis
Analysis of variance Tukey multiple comparisons test (Figure 1A), 2-tailed χ test (Figure 1B), and 2-tailed Mann–Whitney U test (Figures 1C, 1D, and 2D) were used in GraphPad Prism software (version v5.0a). Differences were considered statistically significant at P < .05. Data are expressed as means with standard deviations.
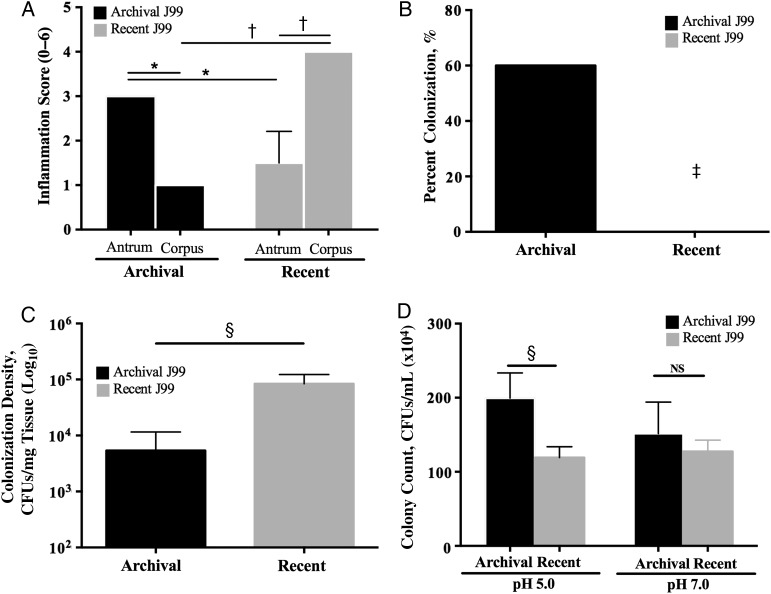
Figure 1.
Patterns of inflammation, colonization, and acid survival induced by archival versus recent Helicobacter pylori J99 isolates. A, Two antral biopsy specimens were obtained at each endoscopy from a single site 2–6 cm from the pylorus on the greater curvature of the stomach and were used for histopathologic examination. Similarly, 2 corpus biopsy specimens were obtained at each endoscopy from a single site 6–10 cm proximal to the termination of the gastric rugae on the greater curvature of the stomach. For each specimen, 10 high-powered fields were examined. Acute and chronic inflammation were each graded 0–3 in the gastric antrum and corpus, for a cumulative score ranging from 0 to 6, and data represent mean histologic scores from the 2 biopsy specimens at each site at each point in time. *P < .05; †P < .01. B, Mongolian gerbils were challenged with the H. pylori archival or recent J99 strains. Two weeks after infection, they were euthanized, and their stomachs harvested. One-fourth of the stomach was homogenized in sterile phosphate-buffered saline, as reported elsewhere [10]. After serial dilution, samples were plated on selective trypticase soy agar plates with 5% sheep blood containing vancomycin, nalidixic acid, bacitracin, and amphotericin B and were incubated at 37°C with 5% carbon dioxide for 5–7 days. Colonization efficiency was expressed as the percentage of gerbils colonized versus the percentage gerbils challenged (n = 5 per group). ‡P = .04. C, Colonization density in wild-type C57Bl/6 mice (n = 5 per group) was determined by means of quantitative culture. §P = .008. CFUs, colony-forming units (CFUs). D, Archival and recent J99 strains were cultured in Brucella broth adjusted to a pH of 5.0 or 7.0 and incubated for 60 minutes. Bacteria were then plated by serial dilution, and colonies were counted. §P = .008. Abbreviation: NS, not significant.
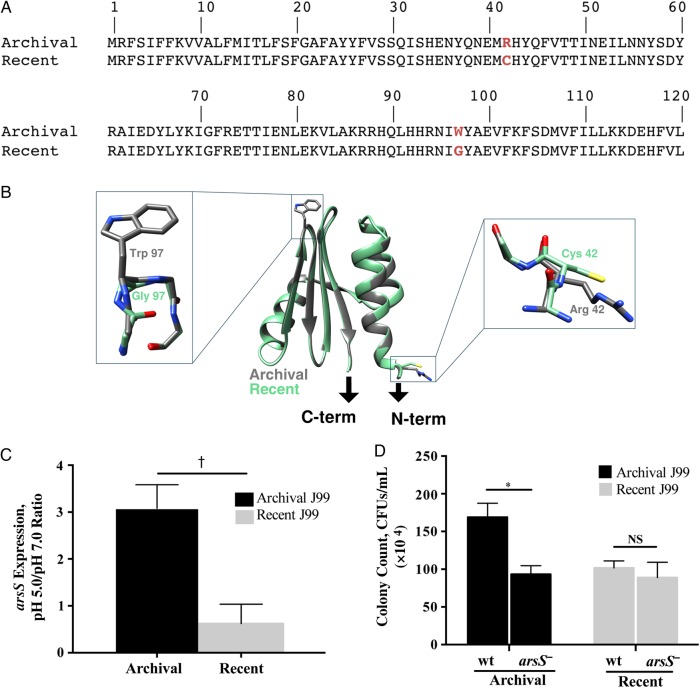
Figure 2.
The role of arsS in mediating acid survival in archival and recent Helicobacter pylori isolates. A, The arsS sequence from the archival J99 isolate was compared that from the recent J99 isolate, and 2 nonsynonymous point mutations were identified in the recent isolate. At amino acid position 42, cysteine was substituted for arginine, and at position 97, glycine was substituted for tryptophan (red). B, Ribbon diagram of a model of the ArsS sensor domain (residues 41–125). Arrows indicate connections to the putative transmembrane helices of ArsS. Side chains containing residues 42 and 97 are enlarged. C, Changes in H. pylori arsS expression in response to pH 5.0. RNA was isolated from H. pylori grown in broth adjusted to pH 5.0 or 7.0 using the RNeasy RNA purification kit (Qiagen). Reverse-transcriptase polymerase chain reaction (PCR) was performed using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems), followed by reverse-transcriptase PCR using the SYBR Green Gene Expression Assay and a 7300 real-time PCR system (Applied Biosystems), with the following primers: arsS, 5′ AAGTGAAACGCTTCGCTCAAG 3′ and 5′ ATTCGTTAGCCAAATCCCCTATT 3′; 16S, 5′ TGCGAAGTGGAGCCAATCTT 3′ and 5′ GGAACGTATTCACCGCAACA 3′. Relative gene expression levels were calculated using the 2−ΔΔCT method. Levels of arsS expression at pH 5.0 and pH 7.0 were then normalized to levels of 16S messenger RNA expression, and data are represented as the ratio of pH 5.0 to pH 7.0; 3 independent experiments were conducted. †P = .003 (archival vs recent J99). D, Archival and recent wild-type (wt) J99 strains and their arsS isogenic mutants were cultured in Brucella broth that had been adjusted to a pH of 5.0 or 7.0 and then incubated for 60 minutes. Bacteria were then plated by serial dilution, and colonies were counted. *P = .02. Abbreviations: CFUs, colony-forming units; NS, not significant. This figure is available in black and white in print and in color online.
RESULTS AND DISCUSSION
A shift in the pattern of H. pylori–induced inflammation from antral-predominant to corpus-predominant gastritis is a crucial step in the cascade to gastric carcinogenesis. Therefore, we first sought to determine whether histologic progression toward a carcinogenic phenotype occurred over time in the J99 source patient. Total inflammation scores in the gastric antrum were 3-fold higher than scores in the corpus at the initial endoscopy (P < .05); however, this pattern was reversed 6 years later, when the severity of corpus inflammation was approximately 3-fold that of antral inflammation (P < .05; Figure 1A). Importantly, the patient had not been treated with proton pump inhibitors (PPIs) during the 6-year interval, indicating that proximal migration of inflammation in the stomach was not mediated by PPI therapy. When endoscopy was repeated, the gastric pH was 5.0, further indicating that acid secretion was impaired. The transition from antral- to corpus-predominant gastritis suggested that this patient was progressing toward a hypochlorhydric phenotype and may be at higher risk for intestinal-type gastric adenocarcinoma.
We then extended these findings into 2 rodent models, Mongolian gerbils and C57BL/6 mice, which develop different inflammation and acid-related phenotypes in response to H. pylori. Mongolian gerbils are outbred and develop antral-predominant gastritis after acute H. pylori infection [11] in contrast to inbred mice, which develop corpus-predominant gastritis [12]. After challenge with the archival or recent J99 isolates for 2 weeks, only the H. pylori archival strain successfully infected gerbils (P = .04 vs recent J99; Figure 1B). However, the pattern of colonization density and efficiency differed in mice. Although all mice challenged with archival or recent H. pylori J99 isolates were successfully colonized (data not shown), colonization density was significantly increased (P = .008) in mice infected with recent J99 isolates compared with those infected with archival J99 (Figure 1C).
Given the inherent differences in inflammatory responses and the outbred versus inbred nature of these models, we recognize that other mediators could contribute to these results. For example, BabA is an outer membrane protein encoded by the babA2 gene, which binds to fucosylated Lewisb antigen on the surface of gastric epithelial cells [2, 3]. babA2 is associated with gastric cancer, but BabA expression can be lost in H. pylori isolates retrieved from human samples, as well as in mice and gerbils infected with H. pylori [2, 3]. Loss of BabA may provide a mechanism through which H. pylori can modify its outer membrane to adapt to changing conditions within an inflamed milieu. SabA is another H. pylori adhesin that binds to the sialyl-Lewisx antigen expressed on gastric epithelium and is associated with increased gastric cancer risk [2, 3]. Sialyl-Lewisx expression is induced during chronic gastric inflammation, suggesting that H. pylori modulates host cell glycosylation patterns to enhance attachment and colonization [2, 3]. Importantly, our animal results phenocopy findings from the human studies, because archival J99 isolates harvested from an acidic gastritis phenotype (antral-predominant gastritis) more robustly colonize high-acid-producing rodents (gerbils), whereas the converse is true for recent J99 isolates.
We then examined the capacity of the archival and recent J99 isolates to survive under different pH conditions in vitro as a means to understand differences observed in vivo. Archival H. pylori strain J99 survived significantly better (P = .008) than recent J99 isolates under pH 5.0 but not pH 7.0 conditions, indicating that the archival strain is more resistant to an acidic environment (Figure 1D).
Based on their known roles in acid response, the genes encoding ArsR and ArsS were completely sequenced in both isolates to identify potential mutations. There were no differences in arsR nucleotide identity between the strains (data not shown). However, when the arsS sequence of archival J99 was compared with that of the recent J99 isolate, 2 nonsynonymous point mutations were identified in the latter. At amino acid position 42, cysteine was substituted for arginine, and at amino acid position 97, glycine was substituted for tryptophan (Figure 2A). Ten additional single colony isolates from each pooled J99 strain were then sequenced, and all of the single colony isolates harbored arsS sequences that were identical to the respective pooled parental strains (data not shown).
To determine the potential impact of this 2-residue difference between the strains, we constructed homology models of the archival and recent forms of the ArsS sensor domains. The structure of the Helicobacter acinonychis ArsS sensor domain, which is 86% identical to its orthologue in H. pylori J99, was used as a template in Modeller (version 9.16) software calculations [13]. Side chains containing the amino acid changes within the archival and recent sensor domains of ArsS are both surface exposed (Figure 2B). The Trp97 to Gly mutation is located in a predicted reverse turn between 2 strands of the central β-sheet (Figure 2B), which may affect conformational responses of this domain to changes in pH, and the second mutation at position 42 replaces a charged Arg to a noncharged Cys side chain and is located at the beginning of the predicted first α-helix (Figure 2B). The location of the amino acid substitutions within the periplasmic domain of ArsS suggest that ArsS in the recent J99 isolates may be limited in its ability to respond to changes in acidity.
We also investigated potential isolate-specific differences in H. pylori arsS expression and found that archival J99 expressed arsS to levels that were significantly higher than the recent J99 isolate at pH 5.0 (Figure 2C). Thus, the ability of arsS to mediate acid survival in archival J99 may be regulated both by transcriptional up-regulation of arsS and by an enhanced capacity to sense changes in pH.
To more definitively determine the role of ArsS in the observed phenotypic changes, arsS was inactivated in the archival and recent J99 strains. Disruption of arsS in the archival strain significantly attenuated (P = .02) its ability to survive under acidic conditions; however, disruption of arsS in the recent J99 isolate had no effect on acid survival (Figure 2D). We also generated a mutant of archival J99 that contained the arsS sequence from the recent J99 strain and infected gerbils to further define the role of arsS in colonization and acid tolerance in vivo. Four of 5 gerbils challenged with the original archival J99 strain were successfully colonized, compared with none of the 5 gerbils challenged with the mutant strain (P = .01). These results reinforce our previous findings indicating that the recent J99 strain cannot successfully colonize a high-acid-producing rodent model of infection and further indicate that mutations at amino acid positions 42 and 97 are probably required for this phenotype.
In summary, these results suggest that arsS may influence the progression of gastric carcinogenesis. However, it will be critically important to validate these findings in other human populations with and without gastric cancer as well as within unique patient cohorts. These include H. pylori–infected human patients followed up longitudinally for years [14] (similarly to the J99 source patient) or patients who have been exposed to therapeutic agents such as PPIs, which alter acidity and accelerate the progression from antral-predominant to corpus-predominant gastritis, which is accompanied by gastric atrophy [15]. Collectively, our current results provide an important framework to define the roles of regulatory genes within the context of H. pylori infection and disease within the stomach.
Notes
Financial support. This work was supported by the American Gastroenterological Association (“Investing in the Future” student research fellowship) and the National Institutes of Health (grants R25DK096968, R01CA077955, R01DK058587, P30DK058404, and P01CA116087).
Potential conflict of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J et al. . GLOBOCAN 2012 v1.0. Cancer incidence and mortality worldwide: IARC CancerBase No. 11. Lyon, France: International Agency for Research on Cancer, 2013. http://globocan.iarc.fr. Accessed 12 May 2015. [Google Scholar]
- 2.Polk DB, Peek RM Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer 2010; 10:403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amieva M, Peek RM Jr. Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology 2016; 150:64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Correa P, Piazuelo MB. The gastric precancerous cascade. J Dig Dis 2012; 13:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallinger DR, Romero-Gallo J, Peek RM Jr, Forsyth MH. Polymorphisms of the acid sensing histidine kinase gene arsS in Helicobacter pylori populations from anatomically distinct gastric sites. Microb Pathog 2012; 53:227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loh JT, Gupta SS, Friedman DB, Krezel AM, Cover TL. Analysis of protein expression regulated by the Helicobacter pylori ArsRS two-component signal transduction system. J Bacteriol 2010; 192:2034–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcus EA, Sachs G, Wen Y, Scott DR. Phosphorylation-dependent and phosphorylation-independent regulation of Helicobacter pylori acid acclimation by the ArsRS two-component system. Helicobacter 2016; 21:69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Israel DA, Salama N, Krishna U et al. . Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc Natl Acad Sci USA 2001; 98:14625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tham KT, Peek RM Jr, Atherton JC et al. . Helicobacter pylori genotypes, host factors, and gastric mucosal histopathology in peptic ulcer disease. Hum Pathol 2001; 32:264–73. [DOI] [PubMed] [Google Scholar]
- 10.Romero-Gallo J, Harris EJ, Krishna U, Washington MK, Perez-Perez GI, Peek RM Jr. Effect of Helicobacter pylori eradication on gastric carcinogenesis. Lab Invest 2008; 88:328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugimoto M, Ohno T, Graham DY, Yamaoka Y. Gastric mucosal interleukin-17 and -18 mRNA expression in Helicobacter pylori-induced Mongolian gerbils. Cancer Sci 2009; 100:2152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Every AL, Ng GZ, Skene CD et al. . Localized suppression of inflammation at sites of Helicobacter pylori colonization. Infect Immun 2011; 79:4186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 2014; 47:5.6.1–5.6.32. [DOI] [PubMed] [Google Scholar]
- 14.Kennemann L, Didelot X, Aebischer T et al. . Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci USA 2011; 108:5033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuipers EJ, Lundell L, Klinkenberg-Knol EC et al. . Atrophic gastritis and Helicobacter pylori infection in patients with reflux esophagitis treated with omeprazole or fundoplication. N Engl J Med 1996; 334:1018–22. [DOI] [PubMed] [Google Scholar]