Abstract
Background and Aims:
Gut microbiota is involved in many physiological functions and its imbalance is associated with several diseases, particularly with inflammatory bowel diseases. Mucosa-associated microbiota could have a key role in induction of host immunity and in inflammatory process. Although the role of fungi has been suggested in inflammatory disease pathogenesis, the fungal microbiota has not yet been deeply explored. Here we analysed the bacterial and fungal composition of the mucosa-associated microbiota of Crohn’s disease patients and healthy subjects.
Methods:
Our prospective, observational study evaluated bacterial and fungal composition of mucosa-associated microbiota of 23 Crohn’s disease patients [16 in flare, 7 in remission] and 10 healthy subjects, using 16S [MiSeq] and ITS2 [pyrosequencing] sequencing, respectively. Global fungal load was assessed by real time quantitative polymerase chain reaction.
Results:
Bacterial microbiota in Crohn’s disease patients was characterised by a restriction in biodiversity. with an increase of Proteobacteria and Fusobacteria. Global fungus load was significantly increased in Crohn’s disease flare compared with healthy subjects [p < 0.05]. In both groups, the colonic mucosa-associated fungal microbiota was dominated by Basidiomycota and Ascomycota phyla. Cystofilobasidiaceae family and Candida glabrata species were overrepresented in Crohn’s disease. Saccharomyces cerevisiae and Filobasidium uniguttulatum species were associated with non-inflamed mucosa, whereas Xylariales order was associated with inflamed mucosa.
Conclusions:
Our study confirms the alteration of the bacterial microbiota and is the first demonstration of the existence of an altered fungal microbiota in Crohn’s disease patients, suggesting that fungi may play a role in pathogenesis.
Key Words: Inflammatory bowel disease, mucosa-associated microbiota, fungal microbiota
1. Introduction
The study of the intestinal microbiota has made dramatic progress in the past decade. However, although its bacterial fraction has been deeply investigated in both health and disease, very few data are available regarding the other microbial fractions and particularly regarding the fungal microbiota.
Bacterial population is by far the most abundant and counts about 1014 microorganisms, 10 times more than the total number of human cells in a human body.1 The gut microbiota has been shown to be involved in many physiological functions, including regulation of metabolism, detoxification of carcinogens, maturation of immune system, stimulation of epithelial barrier, and resistance to infections.2,3 An imbalance in the gut microbiota composition, named dysbiosis, is associated with several diseases and particularly with inflammatory bowel diseases [IBD]. Although the exact pathogenesis of IBD is still unknown, it is currently believed that it results from an inappropriate immune response toward the gut microbiota in genetically predisposed subjects under the influence of environmental factors.4 The most accepted features of the IBD-associated dysbiosis are: i] a reduction of the bacterial diversity and of bacteria belonging to the Firmicutes phylum; and ii] an increase in bacteria from the Proteobacteria phylum and especially of Enterobacteria.5,6,7,8 Many studies suggest that these alterations in the bacterial composition are associated with functional consequences with potential impact on disease onset, severity, or maintenance. In particular, mucosa-associated microbiota, which has been shown to differ from faecal microbiota, could have a key role in induction of host immunity and in inflammatory process.9,10,11
Although the role of fungi has been suggested for a long time in IBD pathogenesis, data on fungal microbiota composition in these patients are still scarce,12 especially at the mucosal level. One of the most comprehensive studies to date has been performed by Chehoud and colleagues who showed that paediatric IBD is associated with reduced diversity in fungal faecal microbiota.13 This aspect is also confirmed by another study performed on mucosa of distal colon in a small cohort of paediatric IBD.14 It has been notably shown many years ago that anti-Saccharomyces cerevisiae antibodies [ASCA] are associated with Crohn’s disease [CD].15,16,17 Moreover, some culture studies observed an increased load of Candida albicans in the stool of CD patients and their healthy first-degree relatives, compared with unrelated healthy subjects [HS].18 Connections between fungal microbiota and IBD are also suggested by the pro-inflammatory effects of C. albicans in colitis models in mice,19,20 whereas the probiotic yeast Saccharomyces boulardii has protective effects.21 Recently, it has been shown that mice lacking genes involved in fungi sensing, such as Dectin1 or Card9, are more susceptible to colitis and exhibit an uncontrolled fungal microbiota.22,23
Finally, many IBD-associated genetic variants are located in genes involved in fungus sensing or in response to fungus infection.12,24,25
In this study we analysed, through molecular and sequencing methods, the mucosa-associated microbiota of CD patients and HS, taking into account both bacterial and fungal fractions. We showed that, beside bacteria, a fungal dysbiosis is present in CD gut microbiota.
2. Material and Methods
2.1. Study population
Our single-centre, prospective, observational study was conducted at the S. Orsola-Malpighi’s Hospital, Bologna, Italy; 23 patients with CD [16 in flare and 7 in remission], and 10 HS were enrolled in the study [Table 1]. CD was diagnosed according to classical clinical, endoscopic, and histological parameters. Phenotype of the disease was defined following the Montreal classification. Exclusion criteria were: infectious colitis; coagulation disorders or concomitant treatment with anticoagulant therapy; and antibiotics or antifungal treatment during the 2 months before inclusion. Control group [mean age 48.3±13.43 years, 70% men] was constituted by HS without history or clinical symptoms of intestinal disorders or endoscopic or histological signs of IBD. HS underwent colonoscopy during routine surveillance screening. All the subjects gave written informed consent to participating in the study, which was approved by the local research ethical committee.
Table 1.
Demographic and clinical data of studied population.
| Active CD | CD in remission | HS | |
|---|---|---|---|
| N | 16 | 7 | 10 |
| Male n [%] | 10 [62.5] | 3 [42.9] | 7 [70] |
| Mean age [± SD, years] | 38 [± 11.9] | 47.1 [± 14.1] | 48.3[± 13.4] |
| Montreal classification n [%] | |||
| A1 | 2 [12.5] | _ | _ |
| A2 | 12 [75] | 7 [100] | _ |
| A3 | 2 [12.5] | _ | _ |
| L1 | _ | _ | _ |
| L2 | 1 [6.2] | _ | _ |
| L3 | 15 [93.8] | 7 [100] | _ |
| B1 | _ | _ | _ |
| B2 | 8 [50] | 5 [71.4] | _ |
| B3 | 8 [50] | 2 [28.6] | _ |
| Previous abdominal surgery n [%] | 5 [31.3] | 7 [100] | 1 [10] |
| Current treatment n [%] | |||
| 5-aminosalicylic acid | 8 [50] | 4 [57.1] | _ |
| Corticosteroids | 4 [25] | 1 [14.3] | _ |
| Azathioprine | 3 [18.8] | 2 [28.6] | _ |
| Infliximab | _ | _ | _ |
| Adalimumab | 1 [6.2] | _ | _ |
| Previous treatment n [%] | |||
| 5-aminosalicylic acid | 13 [81.3] | 5 [71.4] | _ |
| Corticosteroids | 11 [68.8] | 5 [71.4] | _ |
| Azathioprine | 7 [43.8] | 2 [28.6] | _ |
| Infliximab | 2 [12.5] | _ | _ |
| Adalimumab | 5 [31.3] | 1 [14.3] | _ |
CD, Crohn’s disease; HS, healthy subjects; SD, standard deviation.
2.2. Samples collection
Surgical specimens of colonic mucosa of active CD were taken during ileo-colonic resection. In particular, specimens of right colon, both in inflamed and in non-inflamed mucosa, were taken. Sampling in CD patients in remission and HS was performed during colonoscopy using standardised biopsies [Olympus forceps FB-24Q] of the right colon. There were no differences in sampling between the groups studied. All the samples were immediately put in liquid nitrogen and stored at -80°C before processing.
2.3. DNA extraction
DNA was extracted from one or two biopsies of colonic mucosa as previously described.10 For the mechanical lysis step, we used the FastPrep® [three cycles of 30s at 6.5 m/s, cooling samples in ice for 5min between each cycle] and a mix of two sizes of glass beads [0.1 and 0.5 mm] to optimise fungal DNA extraction. Nucleic acids were precipitated by isopropanol for 10min at room temperature, followed by incubation for 15min on ice, and centrifugation for 30min at 24000 g and 4°C. Pellets were suspended in 225 µl of phosphate buffer and 25 µl of potassium acetate. After the RNase treatment and DNA precipitation, nucleic acids were recovered by centrifugation at 24000 g and 4°C for 30min. The DNA pellet was finally suspended in 30 µl of TE buffer and concentration was assessed using Nanodrop® spectrophotometer.
2.4. Real-time quantitative PCR
16S or 18S ribosomal RNA gene levels were determined by real-time quantitative polymerase chain reaction [qPCR] using an ABI 7000 Sequence Detection System apparatus with 7000 system software v. 1.2.3 [Applied Biosystems, Foster City, CA]. Amplification and detection were carried out in 96-well plates and with TaqMan universal PCR 2X master mix [Applied Biosystems] to quantify total bacteria, and with SYBR-Green PCR 2X master mix to quantify total fungi. Each reaction was done in duplicate in a final volume of 25 µl with 10 µl of appropriate dilutions of DNA sample. Amplifications were performed as follows: 95°C for 10min, to denature DNA and activate Ampli-Taq Gold polymerase, followed by 40 cycles of 95°C for 15s and 60°C for 1min. To SYBR-green amplifications, a dissociation step was added and dissociation curves were analysed to confirm the identity and fidelity of amplification of SYBR-green products. The following primers were used: all bacteria: forward: CGGTGAATACGTTCCCGG; reverse: TACGGCTACCTTGTTACGACTT; probe: 6FAM-CTTGTACACACCGCCCGTC; 18S: forward: ATTGGAGGGCAAGTCTGGTG; reverse: CCGATCCCTAGTCGGCATAG.23 The threshold cycle for each sample was determined for 18S gene and normalised to the CT value of the all bacteria 16S ribosomal RNA gene. Data were calculated using the 2-{2(-Delta DeltaC[T])} method.23
2.5. 16S rRNA gene sequencing
Bacterial diversity was determined for each sample by targeting part of ribosomal genes. A 16S rRNA gene fragment comprising V3 and V4 hypervariable regions (16S [sense] TACGGRAGGCAGCAG and [antisense] CTACCNGGGTATCTAAT)26 was amplified using an optimised and standardised 16S-amplicon-library preparation protocol [Metabiote®, GenoScreen, Lille, France]. Briefly, 16S rRNA gene PCR was carried out using 5ng of genomic DNA according to Metabiote® protocol instructions, using 192 bar-coded primers [Metabiote® MiSeq Primers, GenoScreen, Lille, France] at final concentrations of 0.2 μM and an annealing temperature of 50°C for 30 cycles. PCR products were cleaned up with Agencourt AMPure XP-PCR Purification system [Beckman Coulter, Brea, CA], quantified according to the manufacturer’s protocol, and multiplexed at equal concentration. Sequencing was performed using a 300-bp paired-end sequencing protocol on the Illumina MiSeq platform [Illumina, San Diego, CA] at GenoScreen, Lille, France. Raw paired-end reads were subjected to the following process: 1] quality filtering with the PRINSEQ-lite PERL script27 by truncation of bases from the 3’end not with quality < 30 based on the Phred algorithm; and 2] paired-end read assembly using FLASH28 [Fast Length Adjustment of SHort reads to improve genome assemblies] with a minimum length overlap of 30 bases and 97% overlap identity; and 3] the search and removal of both forward and reverse primer sequences using CutAdapt, with no mismatches allowed in primers sequences. Assembled sequences for which perfect forward and reverse primers were not found were eliminated.
2.6. 16S rRNA genes sequence analysis
Assembled sequences were analysed using the Quantitative Insights Into Microbial Ecology [QIIME, version 1.8.0] software package.29 Sequences were assigned to operational taxonomic units [OTUs] using the UCLUST algorithm30 with 97% threshold of pairwise identity, and classified taxonomically using Greengenes reference database.31 Rarefaction was performed [14679–82600 sequences per sample] and used to compare abundances of OTUs across samples. Principal component analyses [PCA] of weighted Unifrac distance with each sample coloured by the disease phenotype was built and used to assess the variation between experimental groups. The number of observed species, the Shannon, the Simpson, and the Chao1 diversity indexes were calculated using rarefied data [depth = 12050 sequences/sample] and used to characterise species diversity in a community.
2.7. ITS2 sequencing
Fungal diversity was determined for each sample by 454 pyrosequencing of internal transcribed spacer 2 [ITS2]. An ITS2 fragment of about 350 bases was amplified using the primers ITS2 [sense] GTGARTCATCGAATCTTT32,33 and [antisense] GATATGCTTAAGTTCAGCGGGT34 and the optimised and standardised ITS2-amplicon-library preparation protocol [Metabiote®, GenoScreen, Lille, France]. Briefly, for each sample, 10 µl diluted genomic DNA, with a concentration of 500ng/µl, were used for a 25-µl PCR conducted under the following conditions: 94°C for 2min, 35 cycles of 15s at 94°C, 52°C for 30s, and 72°C for 45s, followed by 7min at 72°C.35 The PCR products were purified using Ampure XP Beads [Beckman Coulters, Brea, CA] and quantified using the PicoGreen staining Kit [Molecular Probes, Paris, France]. A second PCR of 9 cycles was then conducted under similar PCR conditions with purified PCR products and 10 base pair multiplex identifiers [SIM Identifiers] added to the primers at 5’ position to specifically identify each sample and avoid PCR biases. Finally, the PCR products were purified and quantified as previously described. Sequencing was then performed on a Gs-FLX Titanium Sequencing Systems [Roche Life Science, Mannheim, Germany].
2.8. ITS2 sequence analysis
The sequences were demultiplexed, quality-filtered using the Quantitative Insights Into Microbial Ecology [QIIME, version 1.8.0] software package.29 Sequences were trimmed for barcodes and PCR primers, and binned for a minimal sequence length of 150 pb, a minimal base quality threshold of 25, a maximum homopolymers length of 7. Sequences were then assigned to OTUs using the UCLUST algorithm30 with 97% threshold of pairwise identity, and classified taxonomically using UNITE ITS database [alpha version 12_11].36 Rarefaction was performed [2013–8714 sequences per sample] and used to compare abundances of OTUs across samples. PCA of the Bray-Curtis distance with each sample coloured by the disease phenotype was built and used to assess the variation between experimental groups. The number of observed species, the Shannon, the Simpson, and the Chao1 diversity indexes were calculated using rarefied data [depth = 1552 sequences/sample] and used to characterise species diversity in a community.
2.9. Statistical analysis
GraphPad Prism version 6.0 [San Diego, CA] was used for all analyses and graph preparation. For all data in graphs, results were expressed as mean ± standard error of the mean [SEM] and statistical analyses were performed using the two-tailed Student t test for unpaired data [if normal distribution according to D’Agostino and Pearson omnibus test] or by the non-parametric Mann–Whitney test. Differences with a p-value less than 0.05 were considered significant. Differential abundance analysis were performed using Linear Discriminant Analysis Effect Size [LEfSe].37
3. Results
3.1. Bacterial microbiota diversity
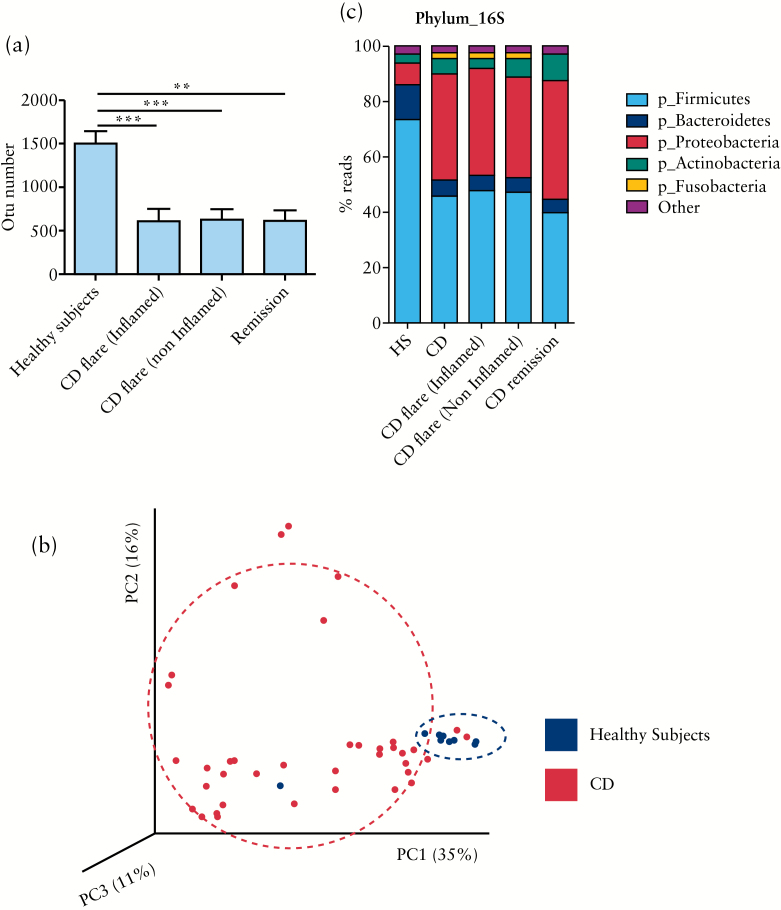
Based on four different indicators [number of OTU observed, Chao1, Shannon, and Simpson indexes] we observed a decline in biodiversity [alpha diversity] of CD patients samples compared with HS. This result was observed in samples from both active and quiescent CD patients and in inflamed and non-inflamed mucosa [Figure 1A and Supplementary Figure 1A [available as Supplementary data at ECCO-JCC online]]. Confirming previous studies, no statistical difference between samples from CD patients in flare or in remission, in inflamed or non-inflamed mucosa was observed [Figure 1A and Supplementar7 Figure 1A].38,39
Figure 1.
Bacterial microbiota biodiversity and composition. [A] Operational taxonomic units [OTUs] number describing the diversity of the bacterial microbiota in the different groups studied [t test, ** = p < 0.01; *** = p < 0.001]. [B] Beta diversity. Principal coordinate analysis of weighted Unifrac distance with each sample coloured by the disease phenotype. PC1, PC2, and PC3 represent the top three principal coordinates that captured most of the diversity, with the fraction of diversity captured by that coordinate shown percent. [C] Global composition of bacteria at phylum level. Healthy subjects [HS] and patient sub-groups are labelled on the x-axis and expressed as relative OTUs abundance per each group.
Between-samples diversity [beta diversity] represented by a principal coordinate analysis clearly showed a clustering of healthy subjects samples versus CD samples [Figure 1B].
3.2. Bacterial microbiota composition
As expected, the large majority of the bacteria from both CD and HS samples belonged to the phyla Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria [Figure 1C]. Distribution of the major phyla in HS and CD patients was in accordance with published data. Firmicutes and Bacteroidetes were the most abundant in HS, followed by Proteobacteria and Actinobacteria. In CD patients, a decrease of Firmicutes and Bacteroidetes was observed with an increase of Proteobacteria.
Interestingly, Fusobacteria, identified as a minor bacterial phylum in HS, was more represented in CD flare, on both inflamed and non-inflamed mucosa [Figure 1C]. Although they represent a minor fraction of the bacterial microbiota, five phyla were detected only in CD flare patients: Acidobacteria, Chloroflexi, Tenericutes, Chlorobi, and Elusimicrobia. Notably, Chlorobi was present only on inflamed mucosa of CD flare [Supplementary Table 1, available as Supplementary data at ECCO-JCC online].
3.2.1. Differential bacterial composition
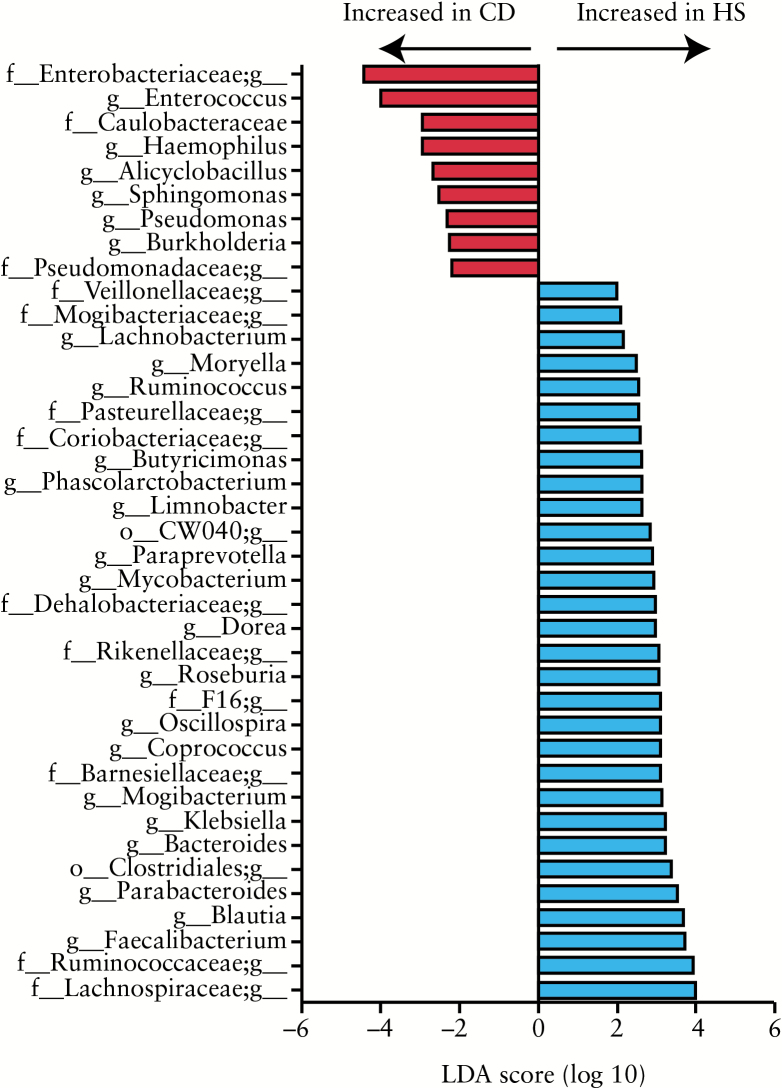
We then directly compared the bacterial composition between studied groups using LEfSe [Linear discriminant analysis Effect Size].40 The major differences observed between CD and HS were in Proteobacteria and Firmicutes phyla [Figure 2 and Supplementary Table 2, available as Supplementary data at ECCO-JCC online]. Gammaproteobacteria and particularly Enterobacteriaceae family were overrepresented in CD. Enterococcus, Alicyclobacillus and Lactobacillus were the only three Firmicutes genera significantly overrepresented in CD patients compared with HS. On the other hand, Firmicutes belonging to the Clostridiales order and particularly to the Lachnospiraceae and Ruminococcaceae families were overrepresented in HS. Among the Firmicutes, Faecalibacterium, Blautia, Dorea, Coprococcus, and Roseburia genera were associated with HS status. Within Bacteroidetes, the Porphyromonadaceae family, and particularly the genus Parabacteroides, were overrepresented in HS [Supplementary Figure 1B, available as Supplementary data at ECCO-JCC online and Supplementary Table 3, available as Supplementary data at ECCO-JCC online].
Figure 2.
Bacterial genus differentially abundant in Crohn’s disease [CD] and healthy subject [HS] mucosa. HS [n = 10] versus CD [n = 23] patients. The histogram shows the Linear Discriminant Analysis [LDA] score computed for genus differentially abundant between the CD and HS mucosa and identified using linear discriminant analysis effect size [LEfSe].
Similar results were obtained when the comparison was restricted to CD flare and HS [SupplementalryTable 4 and Supplementary Figure 2, available as Supplementary data at ECCO-JCC online]. Bacteria belonging to the Actinobacteria and Proteobacteria phyla were more represented in CD in remission compared with the other groups.
Strikingly, no statistical difference between inflamed and non-inflamed mucosa was detected in CD flare patients.
3.3. Fungal microbiota quantification and diversity
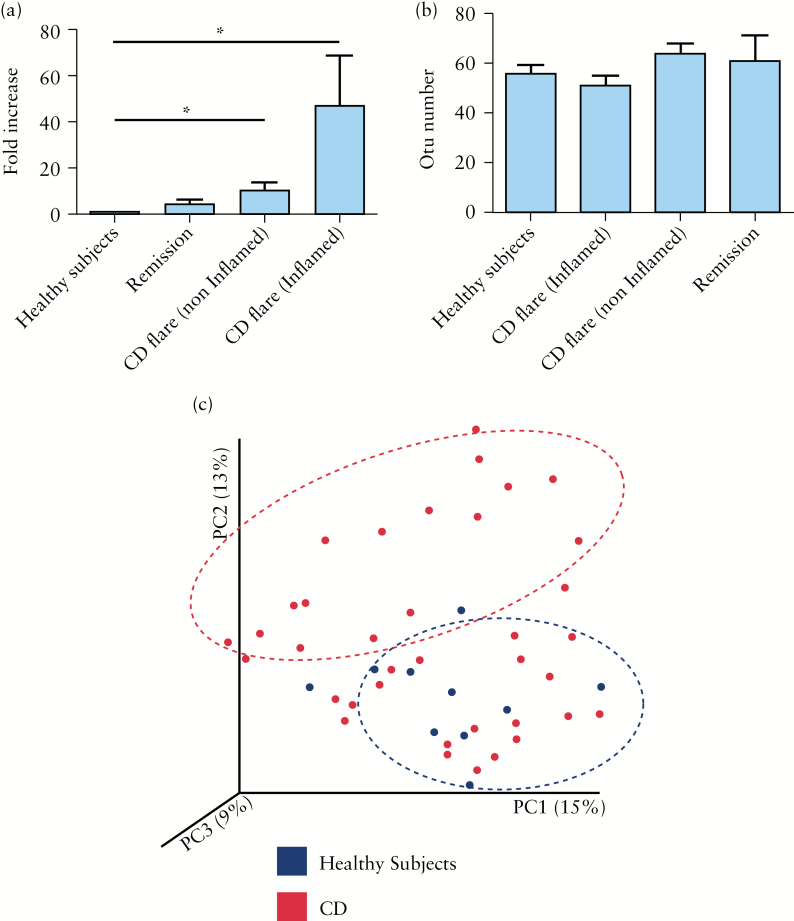
Using qPCR, we observed that global fungus load was significantly increased in CD flare, in both inflamed and non-inflamed mucosa compared with HS [p < 0.05; Figure 3A].
Figure 3.
Fungal microbiota quantification and biodiversity. [A] Real-time quantitative polymerase chain reaction [qPCR] results of fungi normalised to total bacterial 16S rRNA gene copies. Global fungus level was significantly increased in Crohn’s disease [CD] flare, both in inflamed [n = 16] and non-inflamed [n = 16] mucosa compared with healthy subjects [HS] [n = 10] [t test, * = p < 0.05]. [B] Operational taxonomic units [OTUs] number describing the biodiversity of fungal microbiota in the different groups studied. [C] Beta diversity. Principal coordinate analysis of the BrayCurtis distance [for internal transcribed spacer 2 ITS2] with each sample coloured by the disease phenotype. PC1, PC2, and PC3 represent the top three principal coordinates that captured most of the diversity, with the fraction of diversity captured by that coordinate shown percent.
Contrary to bacterial microbiota data, we did not observe any significant difference in fungus biodiversity [alpha diversity] between the studied groups [Figure 3B and Supplementary Figure 3A, available as Supplementary data at ECCO-JCC online]. Between-sample diversity [beta diversity] analysis showed that one subgroup of CD samples segregated from HS whereas another subgroup overlapped with HS, suggesting a difference with respect to HS but also a non-homogeneous diversity within CD samples [Figure 3C].
3.4. Fungal microbiota composition
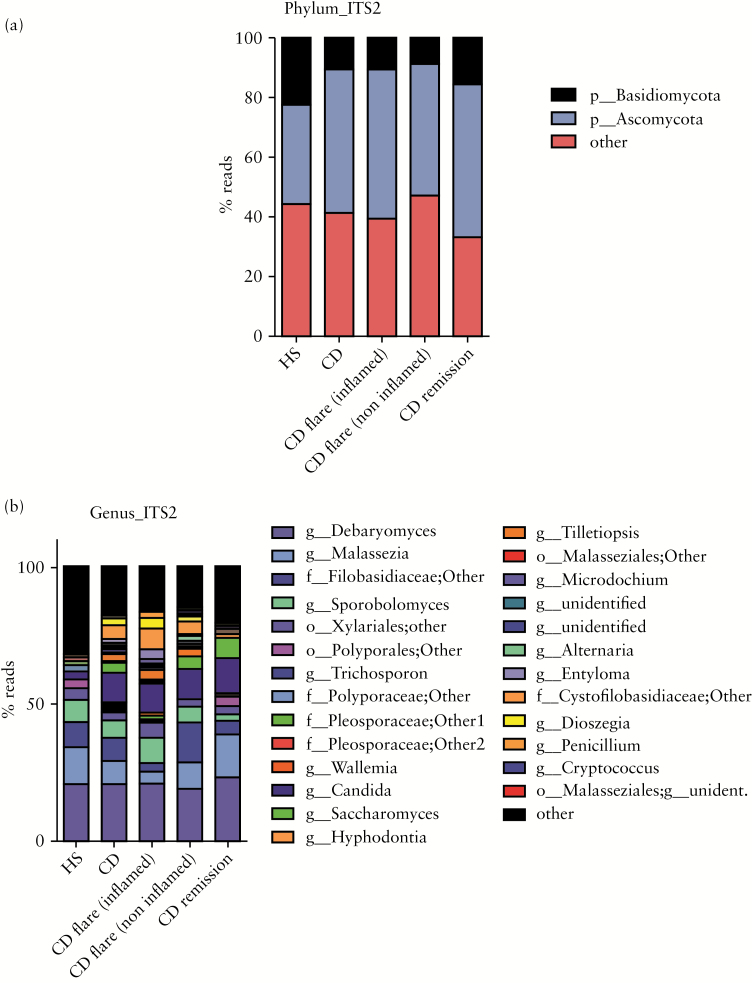
In both HS and CD, the colonic mucosa-associated fungal microbiota was dominated by two phyla, Basidiomycota [44% ± 20% in HS and 41% ± 22% in CD] and Ascomycota [34% ± 14% in HS and 48% ± 23% in CD]. Importantly, a significant proportion of the sequences were assigned to unidentified fungi [22% ± 19% in HS and 10% ± 10% in CD], reflecting the poor annotation of the current fungi database [Figure 4A]. The Tremellomycetes and Saccharomycetes classes were the dominant in Basidiomycota and Ascomycota phyla, respectively. Eurotiomycetes were present only in CD. The most represented genera were Candida, Debaryomyces, Saccharomyces, Malassezia, Sporobolomyces, Trichosporon, Wallemia, unidentified Filobasidiaceae, and unidentified Xylariales [Figure 4B and Supplementary Table 5, available as Supplementary data at ECCO-JCC online].
Figure 4.
Global composition of fungi. [A] Fungal composition at phylum level. [B] Fungal composition at genus level. Healthy subjects [HS] and patients sub-groups are labelled on the x-axis and expressed as relative perational taxonomic units [OTUs] abundance per each group.
3.4.1. Differential fungal composition
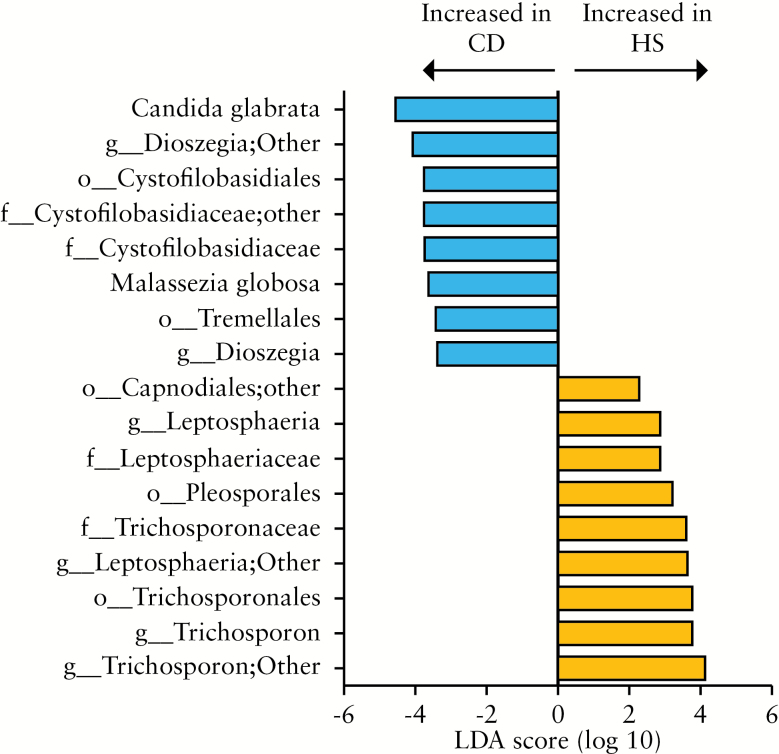
Using LEfSe, we compared samples from HS and CD subjects and observed several differences [Figure 5]. Saccharomycetes, Exobasidiomycetes, and Sordariomycetes were overrepresented in CD compared with HS. Notably, Cystofilobasidiaceae family, Dioszegia genera, and Candida glabrata species were overrepresented in CD whereas Leptosphaeria and Trichosporon genera were decreased compared with HS. Interestingly, Cystofilobasidiaceae family was also more abundant in CD samples in flare than in remission [Figure 6A and Supplementary Figure 3B].
Figure 5.
Fungal taxa differentially abundant in Crohn’s disease [CD] and healthy subject [HS] mucosa. HS [n = 10] versus CD [n = 23] patients. The histogram shows the Linear Discriminant Analysis [LDA] score computed for taxa differentially abundant between the CD and HS mucosa and identified using linear discriminant analysis effect size [LEfSe].
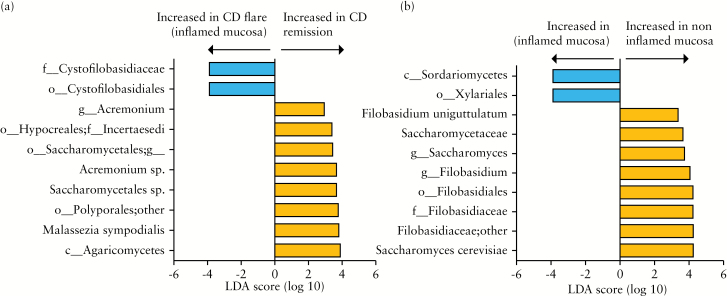
Figure 6.
Fungal taxa differentially abundant in Crohn’s disease [CD] flare, remission, inflamed, and non-inflamed mucosa: CD remission [n = 7], CD flare [n = 16]: inflamed mucosa [n = 16], and non-inflamed [n = 16]. The histogram shows the Linear Discriminant Analysis [LDA] score computed for taxa differentially abundant between the different groups: [A] CD flare inflamed mucosa [n = 16] versus CD in remission [n = 7]. [B] CD flare inflamed mucosa [n = 16] versus CD flare non-inflamed mucosa [n = 16].
In contrast to what was observed with bacterial microbiota, we detected several differences between inflamed and non-inflamed mucosa from CD patients in flare. Saccharomyces cerevisiae and Filobasidium uniguttulatum species were associated with non-inflamed mucosa, whereas Xylariales order was associated with inflamed mucosa [Figure 6B].
4. Discussion
IBD pathogenesis is still unknown but it is now accepted that it involves a microbial factor. Many different microorganisms have been investigated as potential culprit,41,42,43,44 but the hypothesis of an intestinal dysbiosis affecting the immune response and perpetuating inflammation gained ground during the past years.8,45 In the current study, we described for the first time the fungal mucosa-associated microbiota in HS and CD patients and showed that a dysbiosis is also present at the fungal level in CD, suggesting a potential role of fungi in IBD pathogenesis.
We first analysed the bacterial microbiota and confirmed published data showing that bacterial biodiversity was significantly reduced in CD patients and that composition was modified with reduction of Firmicutes and Bacteroidetes and increase of Proteobacteria. Proteobacteria are Gram-negative bacteria that include pathogenic species exhibiting pro-inflammatory effects and potentially responsible for diarrhoea in humans.46,47 In this study, we have identified several members of this phylum and particularly of the Gammaproteobacteria associated with mucosal inflammation such as Haemophilus and Pseudomonas. Beside Proteobacteria, other genera with potential pathogenic effects, such as Enterococcus and Fusobacterium, were also associated with mucosal inflammation. CD patients have a reduced bactericidal capability, which leads to overexposure of microbial factors to the immune system, resulting in activation of immune cells.48 In particular, several studies have shown that CD patients with ileitis exhibit a decreased production of alpha-defensins which are antimicrobial peptides with bactericidal properties notably against Enterobacteriaceae.49,50 Moreover, Fusobacterium nucleatum stimulates the expression of tumour necrosis factor [TNF]-alpha in colonic epithelial cells.51
On the other hand, we observed that several genera within the Ruminococcaceae and Lachnospiraceae families, such as Faecalibacterium, Roseburia, Blautia, Ruminococcus, Coprococcus, and Dorea, were less represented in CD compared with HS. Faecalibacterium prausnitzii has been showed to have anti-inflammatory properties, reducing the severity of colitis and improving the intestinal barrier function.52,53,54 Additionally, its lower concentration in CD could be predictive of endoscopic relapse 6 months after ileocecal resection and of clinical relapse following anti-TNF treatment withdrawal.55,56
Data regarding the non-bacterial components of the intestinal microbiota are still scarce. A recent study on human faeces showed that there is an interesting relationship between bacterial, fungal, and archeal fractions, creating a cooperative microbial consortium able to be modified by diet. 57
There are several clues suggesting a role for fungi in IBD pathogenesis.58,59 The current study is the first one describing the mucosa-associated fungal microbiota in CD patients compared with HS, through sequencing methods. The first important result is the significant increase of fungus load in CD flare, on both inflamed and non-inflamed mucosa. Despite the increased level, no significant difference in fungal diversity between the studied groups was observed. This result is in contradiction with previous results published by the only two studies describing the mucosa-associated fungal microbiota in the context of IBD,58,59 where authors showed a slight increase in diversity in inflamed patients. However, in both publications, the diversity indexes were obtained using the denaturing gradient gel electrophoresis [DGGE] technique, which has a lower power of detection and discrimination than the pyrosequencing. Moreover, Ott and collaborators used, in addition to DGGE, sequenced clones for OTU identification and in this dataset the OTU numbers were equal between HS, CD, and UC patients.59 In opposition to these data, a recent study on faecal fungal microbiota showed that paediatric IBD is associated with reduced diversity in fungal faecal microbiota.13 The two dominant phyla in mucosa-associated fungal microbiota, in both HS and CD, were Basidiomycota and Ascomycota; however, the composition in CD differed from that in HS. Among the numerous differences, Candida glabrata in the Saccharomycetes class was overrepresented in CD and particularly in flare. C. glabrata is one of the most important fungal pathogens and it has been shown to promote inflammatory response,60,61 suggesting a possible role in gut inflammation.
Interestingly, some fungi were detected only in CD patients and notably in flare. This was the case for Eurotiomycetes class. The Penicillium genus belongs to Eurotiomycetes and some species are well known for their antibacterial and/or antifungal activities, like the penicillin producer A. chrysogenum or new Penicillium sp. isolated from plants inhibiting Salmonella sp., Staphyloccocus aureus, Listeria monocytogenes, and Candida albicans.62 Other Penicillium species are considered plant pathogens producing metabolites dangerous for humans. This is the case with patulin, a neurotoxic carcinogenic substance with genotoxic effects, produced by P. expansum.63
We also observed that members of the Cystofilobasidiaceae family were more present in CD patients and particularly during flare. Interestingly, species belonging to this family have antagonist effects against fungal plant pathogens including Penicillium, in citrus and apple diseases, and against fungal human pathogens like Candida species.64,65 The increase of Cystofilobasidiaceae in CD might thus reflect a reaction to the overgrowth of potentially harmful fungi such as C. glabrata and more globally to the CD-associated fungal dysbiosis.
The increase of Eurotiomycetes might reveal how the fungal microbiota could modulate the bacterial microbiota equilibrium through the overgrowth of fungal species with antibacterial activities.
One of the most striking results was that, unlike bacterial microbiota, the fungal microbiota was different between inflamed and non-inflamed mucosa in CD flare patients. Fungi from the Xylariales order were more abundant on inflamed mucosa, whereas Saccharomyces cerevisiae and Filobasidium uniguttulatum were more abundant in non-inflamed mucosa.
Many species belonging to Xylariales produce compounds with antibacterial properties against human pathogens such as Mycobacterium tubercolosis, Yersinia, Listeria and Salmonella, and metabolites with cytotoxic and antioxidant activities.66,67,68 Thus, Xylariales increase could reflect its adaptation to inflammatory environment.
Saccharomyces boulardii, a probiotic strain very close to S. cerevisiae, has been shown to interact with transmembrane receptors promoting intestinal cell restitution,69,70 to prevent C. albicans colonisation during intestinal inflammation19 and to inhibit adherence of Citrobacter rodentium, a bacterial mice pathogen, to intestinal epithelial cells, improving colitis.71 Filobasidium uniguttulatum is a human pathogen firstly isolated from an infected human nail but more recently implicated in cryptococcal meningitis: globally very little information is available on its biology and pathogenicity.72 Interestingly, F. capsuligenum, a related species, was shown to produce mycotoxin active against several fungal species [Cryptoccocus neoformans, Fillibasidium sp., Tremella sp.]. Obviously, the poverty of the information available on most of the fungal strains recently identified hampers greatly our power of analysis of the fungal microbiota.
These differences between inflamed and non-inflamed mucosa could be interpreted as a direct effect of host alterations on fungal populations or as an argument for the role of fungi in onset or perpetuation of lesions. Mechanistic studies should be performed in a near future to address this question.
In conclusion, this study confirms an alteration of bacterial mucosal microbiota in CD and demonstrates the presence of an associated fungal dysbiosis characterised by an increased load of fungi, the presence of particular species with potential pro-inflammatory effects, and a decrease in fungi with potential beneficial effects. New studies should be performed to assess whether the fungal microbiota plays a role in IBD pathogenesis and if fungal microbiota could be a therapeutic target.
Funding
This work was supported by Xeda International S.a.
Conflict of Interest
The authors disclose no conflicts.
Author Contributions
GL, HS, GB, PL, and MC designed the study. GL, HS, BL, GD, and TWH performed acquisition of data. HS and GL performed analysis and interpretation of data. GL and HS wrote the manuscript. HS and MLR revised the manuscript for important intellectual content. HS performed the statistical analysis. GL, GP, CC, and MPDS gave administrative, technical and material support. HS, MC, and PL supervised the study.
Supplementary Data
Supplementary data are available at ECCO-JCC online.
References
- 1. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 1977;31:107–33. [DOI] [PubMed] [Google Scholar]
- 2. Reiners JJ, Clift R, Mathieu P. Suppression of cell cycle progression by flavonoids: dependence on the aryl hydrocarbon receptor. Carcinogenesis 1999;20:1561–6. [DOI] [PubMed] [Google Scholar]
- 3. Bernet MF, Brassart D, Neeser JR, Servin AL. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut 1994;35:483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007;448:427–34. [DOI] [PubMed] [Google Scholar]
- 5. Frank DN, Amand ALS, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baumgart M, Dogan B, Rishniw M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J 2007;1:403–18. [DOI] [PubMed] [Google Scholar]
- 7. Sokol H, Seksik P. The intestinal microbiota in inflammatory bowel diseases: time to connect with the host. Curr Opin Gastroenterol 2010;26:327–31. [DOI] [PubMed] [Google Scholar]
- 8. Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol 2012; 9:599–608. [DOI] [PubMed] [Google Scholar]
- 9. Zoetendal EG, Von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 2002;68:3401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lepage P, Seksik P, Sutren M, et al. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm Bowel Dis 2005;11:473–80. [DOI] [PubMed] [Google Scholar]
- 11. Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science 2005;308:1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Richard ML, Lamas B, Liguori G, Hoffmann TW, Sokol H. Gut Fungal Microbiota: The Yin and Yang of Inflammatory Bowel Disease. Inflamm Bowel Dis 2015;21:656 65. [DOI] [PubMed] [Google Scholar]
- 13. Chehoud C, Albenberg LG, Judge C, et al. Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflamm Bowel Dis 2015;21:1948–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mukhopadhya I, Hansen R, Meharg C, et al. The fungal microbiota of de-novo paediatric inflammatory bowel disease. Microbes Infect 2015;17:304–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKenzie H, Main J, Pennington C, Parratt D. Antibody to selected strains of Saccharomyces cerevisiae [baker’s and brewer’s yeast] and Candida albicans in Crohn’s disease. Gut 1990;31:536–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vermeire S, Peeters M, Vlietinck R, et al. Anti-saccharomyces cerevisiae antibodies [ASCA], phenotypes of IBD, and intestinal permeability: a study in IBD families. Inflamm Bowel Dis 2001;7:8–15. [DOI] [PubMed] [Google Scholar]
- 17. Quinton JF, Sendid B, Reumaux D, et al. Anti-saccharomyces cerevisiae mannan antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: prevalence and diagnostic role. Gut 1998;42:788–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Standaert-Vitse A, Sendid B, Joossens M, et al. Candida albicans colonization and ASCA in familial Crohn’s disease. Am J Gastroenterol 2009;104:1745–53. [DOI] [PubMed] [Google Scholar]
- 19. Jawhara S, Thuru X, Standaert-Vitse A, et al. Colonization of mice by Candida albicans is promoted by chemically induced colitis and augments inflammatory responses through galectin-3. J Infect Dis 2008;197:972–80. [DOI] [PubMed] [Google Scholar]
- 20. Zwolinska-Wcislo M, Brzozowski T, Budak A, et al. Effect of Candida colonization on human ulcerative colitis and the healing of inflammatory changes of the colon in the experimental model of colitis ulcerosa. J Physiol Pharmacol 2009;60:107–18. [PubMed] [Google Scholar]
- 21. Jawhara S, Poulain D. Decreases inflammation and intestinal colonization by Candida albicans in a mouse model of chemically-induced colitis. Med Mycol 2007;45:691–700. [DOI] [PubMed] [Google Scholar]
- 22. Iliev ID, Funari VA, Taylor KD, et al. Interaction between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012; 336:1314–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sokol H, Conway KL, Zhang M, et al. Card9 mediates intestinal epithelial cell restitution, T-helper 17 responses, and control of bacterial infection in mice. Gastroenterology 2013;145:591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Franke A, Balschun T, Sina C, et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 [IL17REL]. Nat Genet 2010;42:292–4. [DOI] [PubMed] [Google Scholar]
- 25. McGovern DP, Gardet A, Törkvist L, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet 2010;42:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013;22:971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011;27:863–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- 31. McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ihrmark K, Bödeker IT, Cruz-Martinez K, et al. New primers to amplify the fungal ITS2 region-evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol 2012;82:666–77. [DOI] [PubMed] [Google Scholar]
- 33. Op De Beeck M, Lievens B, Busschaert P, Declerck S, Vangronsveld J, Colpaert JV. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One 2014;16:e97629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lohtander K, Myllys L, Sundin R, Källersjö M, Tehler A. The species pair concept in the lichen Dendrographa leucophaea [Arthoniales]: Analyses based on ITS sequences. Bryologist 1998;101:404–11. [Google Scholar]
- 35. Reinhard A, Bressenot A, Dassonneville R, et al. Photodynamic therapy relieves colitis and prevents colitis-associated carcinogenesis in mice. Inflamm Bowel Dis 2015;21:985–95. [DOI] [PubMed] [Google Scholar]
- 36. Kõljalg U, Nilsson RH, Abarenkov K, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 2013;22:5271–7. [DOI] [PubMed] [Google Scholar]
- 37. Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bibiloni R, Mangold M, Madsen KL, Fedorak RN, Tannock GW. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn’s disease and ulcerative colitis patients. J Med Microbiol 2006;55:1141–9. [DOI] [PubMed] [Google Scholar]
- 39. Walker AW, Sanderson JD, Churcher C, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol 2011;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morgan XC, Huttenhower C. Meta’omic analytic techniques for studying the intestinal microbiome. Gastroenterology 2014;146:1437–48. [DOI] [PubMed] [Google Scholar]
- 41. Munro J, Mayberry JF, Matthews N, Rhodes J. Chlamydia and Crohn’s disease. Lancet 1979;2:45–6. [DOI] [PubMed] [Google Scholar]
- 42. Liu Y, Van Kruiningen HJ, West AB, Cartun RW, Cortot A, Colombel JF. Immunocytochemical evidence of Listeria, Escherichia coli, and Streptococcus antigens in Crohn’s disease. Gastroenterology 1995;108:1396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martin HM, Campbell BJ, Hart CA, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 2004;127:80–93. [DOI] [PubMed] [Google Scholar]
- 44. Subramanian S, Rhodes JM, Hart CA, et al. Characterization of epithelial IL-8 response to inflammatory bowel disease mucosal E. coli and its inhibition by mesalamine. Inflamm Bowel Dis 2008;14:162–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012;489:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol 2012;9:219–30. [DOI] [PubMed] [Google Scholar]
- 47. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009;9:313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Korzenik JR. Is Crohn’s disease due to defective immunity? Gut 2007;56:2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nuding S, Fellermann K, Wehkamp J, Stange EF. Reduced mucosal antimicrobial activity in Crohn’s disease of the colon. Gut 2007;56:1240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wehkamp J, Harder J, Weichenthal M, et al. NOD2 [CARD15] mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut 2004;53:1658–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun 2011;79:2597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Qiu X, Zhang M, Yang X, Hong N, Yu C. Faecalibacterium prausnitzii upregulates regulatory T cells and anti-inflammatory cytokines in treating TNBS-induced colitis. J Crohns Colitis 2013;7:558–68. [DOI] [PubMed] [Google Scholar]
- 54. Carlsson AH, Yakymenko O, Olivier I, et al. Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scand J Gastroenterol 2013;48:1136–44. [DOI] [PubMed] [Google Scholar]
- 55. Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 2009;15:1183–9. [DOI] [PubMed] [Google Scholar]
- 56. Rajca S, Grondin V, Louis E, et al. Alterations in the intestinal microbiome [dysbiosis] as a predictor of relapse after infliximab withdrawal in Crohn’s disease. Inflamm Bowel Dis 2014;20:978–86. [DOI] [PubMed] [Google Scholar]
- 57. Hoffmann C, Dollive S, Grunberg S, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 2013;8:e66019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Q, Wang C, Tang C, He Q, Li N, Li J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J Clin Gastroenterol 2014;48:513–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ott SJ, Kuhbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol 2008;43:831–41. [DOI] [PubMed] [Google Scholar]
- 60. Seider K, Brunke S, Schild L, et al. The facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J Immunol 2011;187:3072–86. [DOI] [PubMed] [Google Scholar]
- 61. Seider K, Gerwien F, Kasper L, et al. Immune evasion, stress resistance, and efficient nutrient acquisition are crucial for intracellular survival of Candida glabrata within macrophages. Eukaryot Cell 2014;13:170–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shen XY, Cheng YL, Cai CJ, Fan L, Gao J, Hou CL. Diversity and antimicrobial activity of culturable endophytic fungi isolated from moso bamboo seeds. PLoS One 2014;9:e95838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morales H, Marín S, Rovira A, Ramos AJ, Sanchis V. Patulin accumulation in apples by Penicillium expansum during postharvest stages. Lett Appl Microbiol 2007;44: 30–5. [DOI] [PubMed] [Google Scholar]
- 64. Liu J, Wisniewski M, Droby S, Vero S, Tian S, Hershkovitz V. Glycine betaine improves oxidative stress tolerance and biocontrol efficacy of the antagonistic yeast Cystofilobasidium infirmominiatum. Int J Food Microbiol 2011;146:76–83. [DOI] [PubMed] [Google Scholar]
- 65. Vadkertiová R, Sláviková E. Killer activity of yeasts isolated from natural environments against some medically important Candida species. Pol J Microbiol 2007;56:39–43. [PubMed] [Google Scholar]
- 66. Mitchell AM, Strobel GA, Moore E, Robison R, Sears J. Volatile antimicrobials from Muscodor crispans, a novel endophytic fungus. Microbiology 2010;156[Pt 1]:270–7. [DOI] [PubMed] [Google Scholar]
- 67. Chang CW, Chang HS, Cheng MJ, et al. Inhibitory effects of constituents of an endophytic fungus Hypoxylon investiens on nitric oxide and interleukin-6 production in RAW264.7 macrophages. Chem Biodivers 2014;11:949–61. [DOI] [PubMed] [Google Scholar]
- 68. Chen C, Hu SY, Luo DQ, Zhu SY, Zhou CQ. Potential antitumor agent from the endophytic fungus Pestalotiopsis photiniae induces apoptosis via the mitochondrial pathway in HeLa cells. Oncol Rep 2013;30:1773–81. [DOI] [PubMed] [Google Scholar]
- 69. Canonici A, Pellegrino E, Siret C, et al. Saccharomyces boulardii improves intestinal epithelial cell restitution by inhibiting αvβ5 integrin activation state. PLoS One 2012;7:e45047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Canonici A, Siret C, Pellegrino E, et al. Saccharomyces boulardii improves intestinal cell restitution through activation of the α2β1 integrin collagen receptor. PLoS One 2011;6:e18427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu X, Vallance BA, Boyer L, et al. Saccharomyces boulardii ameliorates Citrobacter rodentium-induced colitis through actions on bacterial virulence factors. Am J Physiol Gastrointest Liver Physiol 2008;294:G295–G306. [DOI] [PubMed] [Google Scholar]
- 72. Pan W, Liao W, Hagen F, et al. Meningitis caused by Filobasidium uniguttulatum: case report and overview of the literature. Mycoses 2012;55:105–9. [DOI] [PubMed] [Google Scholar]