Abstract
Background. Women with bacterial vaginosis (BV) have complex communities of anaerobic bacteria. There are no cultivated isolates of several bacteria identified using molecular methods and associated with BV. It is unclear whether this is due to the inability to adequately propagate these bacteria or to correctly identify them in culture.
Methods. Vaginal fluid from 15 women was plated on 6 different media using classical cultivation approaches. Individual isolates were identified by 16S ribosomal RNA (rRNA) gene sequencing and compared with validly described species. Bacterial community profiles in vaginal samples were determined using broad-range 16S rRNA gene polymerase chain reaction and pyrosequencing.
Results. We isolated and identified 101 distinct bacterial strains spanning 6 phyla including (1) novel strains with <98% 16S rRNA sequence identity to validly described species, (2) closely related species within a genus, (3) bacteria previously isolated from body sites other than the vagina, and (4) known bacteria formerly isolated from the vagina. Pyrosequencing showed that novel strains Peptoniphilaceae DNF01163 and Prevotellaceae DNF00733 were prevalent in women with BV.
Conclusions. We isolated a diverse set of novel and clinically significant anaerobes from the human vagina using conventional approaches with systematic molecular identification. Several previously “uncultivated” bacteria are amenable to conventional cultivation.
Keywords: bacterial vaginosis, anaerobes, cultivation, vagina
Bacterial vaginosis (BV) is characterized by a dramatic increase in the abundance of fastidious anaerobes and facultative bacteria, with a concomitant reduction in select Lactobacillus species that are typically markers of health [1–5]. The microbiota in BV is heterogeneous; subgroups of women have vaginal bacterial communities dominated by different bacteria such as Prevotella spp., Leptotrichia/Sneathia spp., BV-associated bacterium (BVAB)1, or Gardnerella vaginalis, but in others there is no dominant bacterium [4]. Classical cultivation methods have routinely isolated numerous bacteria representing several different phyla from the human vagina in women with BV [6–10], but many bacteria detected in the vagina using molecular methods have not been successfully cultivated.
The lack of cultivated isolates that are clearly identified as BVAB hinders our ability to explore the pathogenesis of BV. Cultivation provides critical information on bacterial phenotypes and allows us to explore bacterial interactions among themselves, and with the human host. Cultivation also enables experimental manipulation of the isolates in the laboratory for testing hypotheses about pathogenesis and the role of virulence factors [11–15]. Isolates are important for generation of whole-genome sequencing (WGS) data and interpretation of metagenomic data from vaginal samples, which inform the functional potential of individual species and their community.
Molecular surveys of the vaginal microbiota have shown that there are numerous bacteria associated with BV that are yet to be easily cultivated, such as Eggerthella, Megasphaera, Dialister, Leptotrichia, and Sneathia species, and members of the order Clostridiales, including BVAB1 and BVAB2 [4, 16]. Cultivation studies of vaginal bacteria are typically not paired with identification of bacteria using advanced molecular methods, potentially leading to misclassification or underrepresentation of isolates obtained using these conventional approaches. In this study, we created a collection of >100 different vaginal bacterial isolates obtained using classical cultivation methods coupled with systematic 16S ribosomal RNA (rRNA) sequencing for isolate identification. We submitted DNA from bacteria that were underrepresented in available culture collections for WGS by the Human Microbiome Project (HMP) initiative to contribute to the bank of reference genomes [17].
METHODS
Sample Collection
Baseline vaginal samples were collected from 15 women attending the Public Health–Seattle & King County Sexually Transmitted Diseases Clinic between April 2011 and July 2012 (Supplementary Table 1). We collected additional vaginal samples from 6 of 15 women at follow-up visits that were at least a month apart; 1 woman returned for 1 follow-up visit, 4 returned for 2 visits and 1 returned for 3 visits, for a total of 27 samples that were used for cultivation experiments and deep sequencing analysis (Supplementary Table 1). BV was diagnosed by Amsel criteria [18], and all 27 samples were also subjected to Gram stain interpretation using the Nugent method [19]. Of the 27 samples, 19 were collected when women had BV determined by both Amsel and Nugent criteria. Eight samples were collected when women did not have BV by Amsel criteria, but 3 of the 8 were positive for BV by Nugent criteria (Supplementary Table 1). The study was approved by the institutional review board at the Fred Hutchinson Cancer Research Center, and all women provided written informed consent.
A pelvic examination with a speculum was performed for the collection of samples. Polyurethane foam swabs (Epicenter Biotechnologies) were brushed against the lateral vaginal wall to collect vaginal bacteria. Swabs for cultivation experiments were transported in anaerobic Port-A-Cul tubes (Becton Dickinson) from the clinic to the laboratory within 2 hours of collection and processed in the laboratory within 4 hours of sample collection to maximize recovery of bacterial isolates. Swab samples for molecular studies were stored at −80°C until DNA extraction. Vaginal fluid was also collected for Gram staining, pH determination, saline preparation with microscopy, and potassium hydroxide preparation with microscopy.
Direct Plating for Isolation of Bacteria
The format for plating has been described in detail [7]. Briefly, vaginal swab samples were vortexed in 1.5 mL of reduced Hank's solution [20, 21], and the resulting vaginal fluid was serially diluted in reduced saline. Aliquots (100 µL) from a range of dilutions were plated on Brucella blood agar with hemin and vitamin K (Hardy Diagnostics); Rogosa agar, selective for lactobacilli [22]; human blood Tween (HBT) bilayer, selective for G. vaginalis; and Brucella laked blood agar with kanamycin, hemin, and vitamin K, selective for Prevotella spp. (both from Becton Dickinson); they were then incubated under anaerobic conditions at 37°C for 5–7 days. Another set of plates including HBT bilayer, Columbia blood agar with 5% sheep blood and chocolate agar (both from Hardy Diagnostics) were incubated for up to 72 hours at 37°C in 5%–10% carbon dioxide. Bacterial isolates from the plates were selected based on morphologic characteristics of the bacterial colonies using stereomicroscopy, Gram staining, and acridine orange staining, followed by epifluorescence microscopy.
Identification and Phylogeny of Cultivated Isolates
DNA from bacterial pellets was extracted using the Biostic Bacteremia DNA Isolation Kit (MoBio Laboratories) and eluted in 75–150 µL of buffer. Bacterial DNA was subjected to broad-range polymerase chain reaction (PCR), using primers targeting a conserved sequence of the 16S rRNA gene [23, 24]. Amplification was confirmed by gel electrophoresis, and amplicons were cleaned with the DNA Clean & Concentrator-5 Kit (Zymo Research) and submitted for Sanger sequencing (Shared Resources, Fred Hutchinson Cancer Research Center). Sequence alignments were performed using BioEdit software (version 7.1.11) [25]. The 16S rRNA sequences were compared with sequences in GenBank [26] and EzTaxon [27] for taxonomic identification of each cultivated isolate. Phylogenetic trees were constructed using MEGA software (version 6) [28]. Select strains were submitted to the Biodefense and Emerging Infections Research Resources Repository, and 16S rRNA sequences have been submitted to the National Center for Biotechnology Information GenBank database (see Supplementary Table 1 for accession numbers).
Molecular Profiling of the Vaginal Microbiota
DNA from vaginal swabs was extracted using the Biostic Bacteremia DNA Isolation Kit, eluted in 150 µL of buffer. Sham swabs without human contact were processed as controls to assess contamination from reaction buffers or sample collection swabs. We performed community 16S rRNA gene PCR with pyrosequencing using 454 Life Sciences Titanium technology (Roche), targeting the V3–V4 region of the 16S rRNA gene [4, 5] to determine the bacterial community in each vaginal swab sample. Sequence reads were classified using a phylogenetic placement tool pplacer [29] and a curated reference set of vaginal bacteria [4]. The prevalence of each bacterial isolate was determined by comparing isolate sequences to pyrosequencing reads with vsearch software (version 1.9.10; https://github.com/torognes/vsearch) with an alignment threshold of 99% identity. Pyrosequencing reads have been submitted to the National Center for Biotechnology Information Short Read Archive (SRP071678, SRX950298-SRX950303, SRX950306, SRX950317, SRX950348, and SRX950350).
Preparation of DNA for WGS
Genomic DNA (gDNA) from bacterial pellets was extracted using the ZR Fungal/Bacterial DNA MidiPrep Kit (Zymo Research) and eluted in 150 µL of buffer. A NanoDrop 1000 spectrophotometer was used to determine gDNA concentrations, and gDNA quality was assessed using A260/280 and A260/230 ratios. Before submission for WGS, isolate gDNA was sequence confirmed by broad-range 16S rRNA gene PCR and Sanger sequencing. DNA from 31 strains (Supplementary Table 1) was submitted to 1 of 2 HMP sequencing centers: Human Genomic Medicine, J. Craig Venter Institute, Rockville, Maryland, or The Genome Center, Washington University School of Medicine, St Louis, Missouri. A subset of these isolates were deposited in the Biodefense and Emerging Infections Research Resources Repository.
RESULTS
Bacterial Isolates
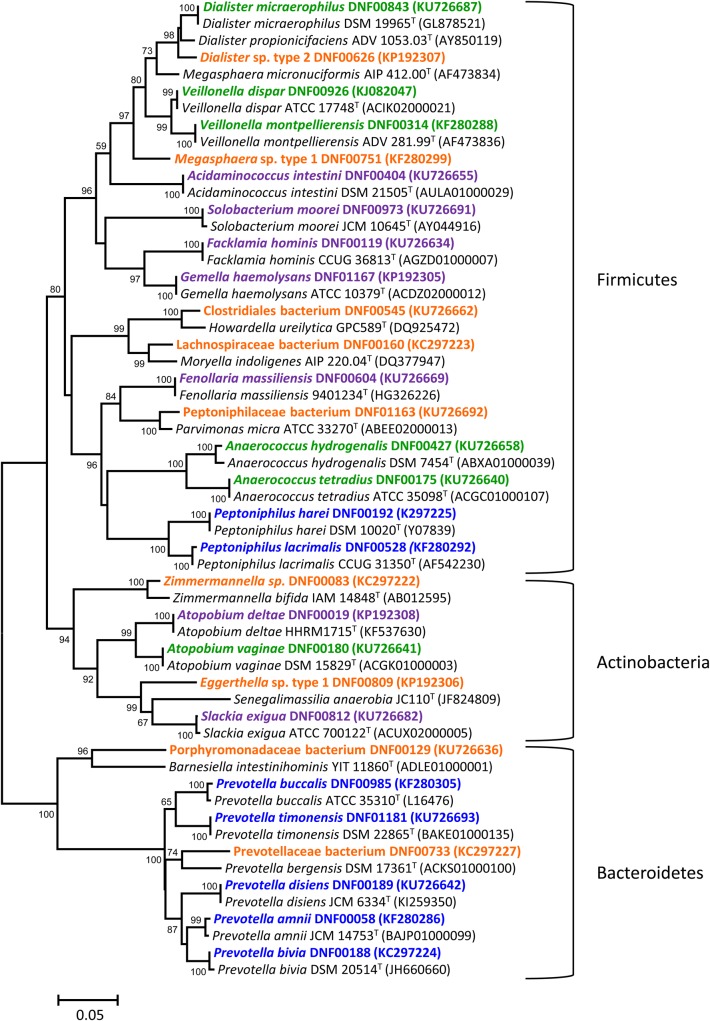
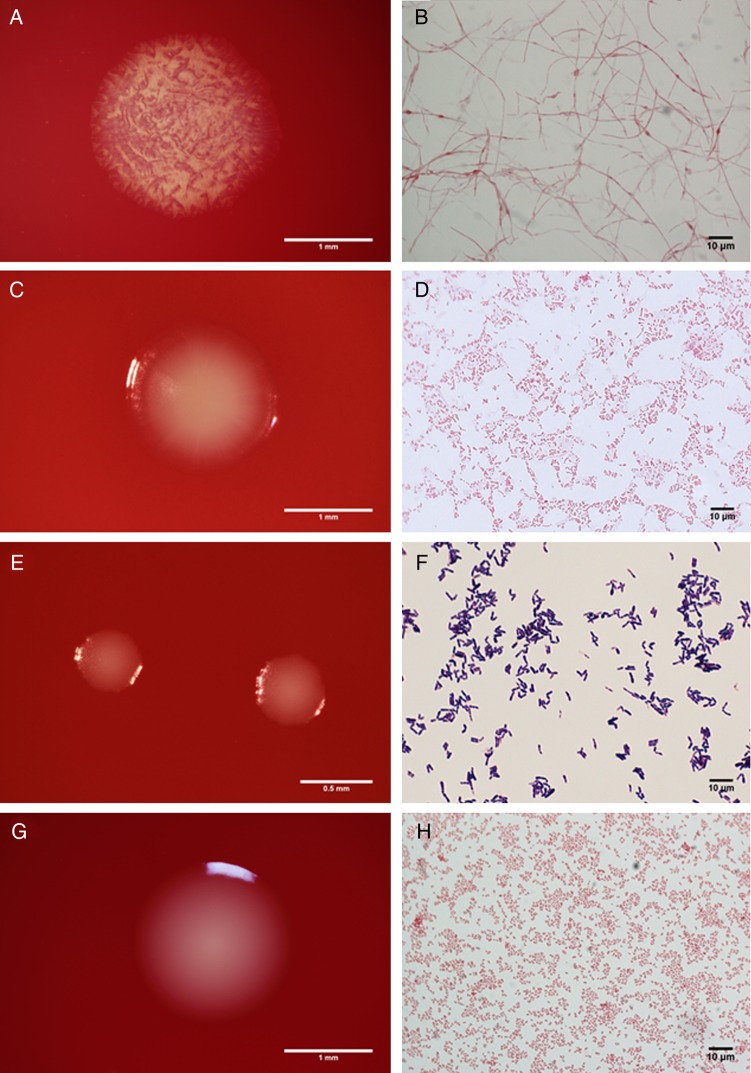
We isolated and identified 101 different bacterial strains spanning 6 phyla from a screen of 704 isolates obtained from 27 vaginal samples (Supplementary Table 1). As expected, the vast majority of isolates were from the Firmicutes (n = 43) and Actinobacteria (n = 36). We also isolated bacteria belonging to the Bacteroidetes (n = 14), Proteobacteria (n = 6), Fusobacteria (Fusobacterium nucleatum), and Tenericutes (Mycoplasma hominis). Bacterial isolates were categorized into 4 groups described below. Select strains are depicted in the phylogenetic tree (Figure 1). Morphologic characteristics of the bacterial colonies and Gram stains of 1 isolate from each of the 4 groups below are displayed in Figure 2.
Figure 1.
Diversity of bacterial isolates obtained with classical cultivation methods. Minimum evolution phylogenetic dendrogram shows relationships of representative 16S ribosomal RNA (rRNA) gene sequences of vaginal bacterial isolates among closely related validly described species. Numbers at branch points depict bootstrap support based on analysis of 1000 replicates. GenBank accession numbers for the 16S rRNA sequences are provided in parentheses. Strains are color coded based on categories developed in this study. Orange indicates bacteria with <98% sequence identity to validly published species; blue, closely related species within a genus; purple: bacteria previously isolated from other human body sites but not the vagina; and green, known bacteria previously isolated from the vagina. Bar represents 5% sequence divergence.
Figure 2.
Colony morphology (left) and Gram stains (right) of vaginal bacteria. Representative isolates from the 4 groups described in the study are shown, including Lachnospiraceae bacterium DNF00160 (A, B), Prevotella massiliensis DNF00663 (C, D), Corynebacterium kroppenstedtii DNF00591 (E, F), and Peptoniphilus lacrimalis DNF00528 (G, H).
Novel Strains
Of the 101 strains, 11 (10.9%) had <98% sequence identity to validly described species (Supplementary Table 1 and Figure 1) including Dialister sp. type 2 DNF00626 (95.7% sequence identity) and Eggerthella sp. type 1 DNF00809 (90.3% sequence identity). Both are fastidious bacteria of clinical significance to BV [4, 30]. All novel isolates belonged to the phyla Actinobacteria, Firmicutes, or Bacteroidetes.
Closely Related Species Within a Genus
We obtained ≥3 phylogenetically distinct species from some genera, including Prevotella (n = 9), Peptoniphilus (n = 3), and Lactobacillus (n = 7) (Supplementary Table 1). Among the Prevotella species, we obtained both Prevotella amnii DNF00058 and Prevotella timonensis DNF01181 (Figure 1), designated as Prevotella genogroups 1 and 2 in earlier molecular studies of the vaginal microbiota [16, 30]. For the lactobacilli, we isolated the following Lactobacillus species: L. crispatus, L. jensenii, L. coleohominis, and L. vaginalis, mostly from women without BV, and L. iners and L. gasseri, from both women with and women without BV. L. iners isolates were obtained only using media containing blood, such as HBT, Columbia blood agar or Brucella blood agar, but not from Rogosa agar, which is consistent with previous observations [31].
Identical Species Found in Other Body Sites but not the Vagina
Additional searches of databases were conducted for bacteria that are not typically noted in molecular screens of the vaginal microbiota, and with >99% sequence identity with validly described species, to determine whether they had been previously isolated from the vagina. Searches were conducted in National Center for Biotechnology Information PubMed (http://www.ncbi.nlm.nih.gov/pubmed) and GenBank (http://www.ncbi.nlm.nih.gov/genbank/) databases with the search string “name of bacterium, vagina.” Bacteria were scored as “not previously isolated from the vagina” if there were no publications reporting these bacteria as vaginal isolates or if no sequence data were available from cultured vaginal isolates. Seventeen bacteria (16.8%) not previously isolated from the vagina were identified in the study (Supplementary Table 1). Examples include Atopobium deltae DNF00019, previously isolated from human blood [32], and Fenollaria massiliensis DNF00604, formerly isolated from human osteoarticular samples [33] (Supplementary Table 1, Figure 1).
Previously Cultivated Members of the Vaginal Microbiota
Nineteen strains obtained (18.8%) have been previously isolated from the vagina, including Dialister micraerophilus, Veillonella spp., Anaerococcus spp., and Peptostreptococcus anaerobius (Supplementary Table 1 and Figure 1). Several isolates of Atopobium vaginae and G. vaginalis were cultivated; A. vaginae was isolated from HBT and Brucella blood agar plates that were incubated under anaerobic conditions, whereas G. vaginalis was isolated from all plates tested except Rogosa agar, under either aerobic or anaerobic conditions (data not shown).
Comparative Analysis of 16S rRNA Sequences Obtained From Isolates and Those Found by Community PCR With High-Throughput Sequencing
The prevalence of each bacterial isolate was determined by comparing isolate sequences to pyrosequencing reads from the set of 25 samples. Two samples had <500 sequence reads and were excluded from this analysis. The 16S rRNA sequences of 60 isolates (59.4%) were detected in the vaginal community (Supplementary Table 2). Seven of the 11 sequences (63.6%) from novel bacteria were present, and Dialister sp. type 2 DNF00626, Eggerthella sp. type 1 DNF00809, Peptoniphilaceae DNF01163, and Megasphaera sp. type 1 DNF00751 sequences were detected in >50% of samples. Prevotellaceae DNF00733 (designated Prevotella genogroup 7 in molecular screens) was detected in 40% of samples.
Among the genera with multiple species isolated, we noted that the number of species detected with deep sequencing was consistently lower than the number isolated. For example, our molecular survey did not detect certain bacteria cultivated in this study, such as L. vaginalis DNF00112, several Actinomyces, and Bifidobacterium spp., or any Corynebacterium or Propionibacterium sequences. Likewise, only 4 of 18 sequences (22.2%) from isolates classified as previously isolated from other body sites were detected with deep sequencing, including A. deltae DNF00019, Facklamia hominis DNF00119, F. massiliensis DNF00604, and Gemella haemolysans DNF01167. In contrast, most isolates (73.4%) classified as previously isolated from the vagina were detected by means of their 16S rRNA sequences in the community.
To identify bacteria that were both cultivated and numerically abundant in this study (>0.1% of all reads), we compared bacterial sequence reads with isolates obtained (Table 1). We isolated 23 of 36 abundant bacteria (63.9%) using conventional plating methods. The analysis also highlighted notable exceptions including Leptotrichia amnionii and BVAB1 (Table 1).
Table 1.
Abundant Bacteria Isolated by Conventional Cultivation
| Bacteria Detected With Broad-Range PCR and Pyrosequencinga | Isolates Obtained |
|---|---|
| Anaerococcus prevotii | Yes |
| Atopobium vaginae | Yes |
| Dialister micraerophilus | Yes |
| Dialister sp. type 2 | Yes |
| Eggerthella sp. type 1 | Yes |
| Finegoldia magna | Yes |
| Fusobacterium nucleatum | Yes |
| Gardnerella vaginalis | Yes |
| Lactobacillus crispatus | Yes |
| Lactobacillus gasseri | Yes |
| Lactobacillus iners | Yes |
| Lactobacillus jensenii | Yes |
| Megasphaera sp. type 1 | Yes |
| Mycoplasma hominis | Yes |
| Peptoniphilus harei | Yes |
| Prevotella amnii | Yes |
| Prevotella bivia | Yes |
| Prevotella disiens | Yes |
| Prevotella melaninogenica | Yes |
| Prevotella timonensis | Yes |
| Streptococcus agalactiae | Yes |
| Streptococcus anginosus | Yes |
| Streptococcus mitis/oralis | Yes |
| Aerococcus christensenii | No |
| BV-associated bacterium 1 | No |
| BV-associated bacterium 2 | No |
| Gemella asaccharolytica | No |
| Leptotrichia amnionii | No |
| Mageeibacillus indolicus | No |
| Megasphaera sp. type 2 | No |
| Parvimonas micra | No |
| Porphyromonas asaccharolytica | No |
| Prevotella genogroup 3 | No |
| Prevotella genogroup 4 | No |
| Sneathia sanguinegens | No |
| Streptococcus gallolyticus | No |
Abbreviations: BV, bacterial vaginosis; PCR, polymerase chain reaction.
a Bacteria with >0.1% abundance across 25 samples in the study.
DISCUSSION
Meta-omics technologies can provide great insight into the composition and function of bacterial communities but do not fully substitute for the information gleaned from observing the properties of isolated microbial cells. Metagenomic and metatranscriptomic methods are useful for obtaining important clues regarding the genomic potential and gene expression profiles of bacteria in a community [34, 35], but so far they have not provided a complete picture of microbial properties and interactions. Many microbes remain resistant to cultivation in the laboratory [36, 37], and this is a serious impediment to better understanding of microbial interactions and pathogenesis. In the current study, we sought to expand the bank of cultivated bacterial isolates from the human vagina, based on the assumption that many bacteria will grow in the laboratory with conventional cultivation methods but are poorly identified with conventional techniques. We predicted that using molecular identification tools would help close this gap.
We used plating on conventional media to isolate vaginal bacteria and paired this with molecular identification of isolates using 16S rRNA gene sequencing. A set of 101 strains belonging to 6 phyla were obtained in the study, illustrating that a wide range of bacteria can be cultivated from the vagina using classic approaches. We also examined the composition of the vaginal microbiota in the same set of samples using broad-range PCR and pyrosequencing. We determined the prevalence of the bacterial isolate sequences in these samples and identified bacteria that were not isolated using conventional cultivation in this study. A subset of 31 isolates has been submitted for WGS based on the guidelines set by the HMP for the selection of reference genomes (http://hmpdacc.org/reference_genomes/reference_genomes.php). Briefly, bacterial isolates for WGS were selected for ≥1 of the following criteria: novelty or uniqueness of species, established clinical significance, abundance in the vagina, identical species found in a different body site, or the opportunity to explore pangenomes.
By using systematic 16S rRNA-based identification, we obtained 11 novel bacteria whose 16S rRNA sequences were <98% identical to validly described species. Of the 11 isolates, 3 bacteria are highly associated with BV (Dialister sp. type 2, Eggerthella sp. type 1, and Megasphaera sp. type 1) [4, 16, 30]. Although not dominant members of the vaginal bacterial community, these strains are prevalent in women with BV. Eggerthella sp. type 1 16S rRNA sequence was detected in 85.5% of women with BV versus 9.5% of women without BV; Dialister sp. type 2 in 72.6% versus 4.8%, respectively; and Megasphaera sp. type 1 in 67.5% versus 12.4%, respectively [4]. Interestingly, in an analysis that examined association of bacterial taxa with individual Amsel clinical characteristics, Dialister sp. type 2 and Megasphaera sp. type 1 were correlated with clue cells, whereas Eggerthella sp. type 1 was one of only 2 bacteria associated with all 4 clinical criteria [4]. In another study that examined vaginal bacteria in 264 women from Seattle, Megasphaera sp. type 1, combined with BVAB2, was found to be highly sensitive and specific for the diagnosis of BV [30]. This is the first report of cultivated isolates of Dialister sp. type 2 and Eggerthella sp. type 1, and their DNA has been submitted for WGS by HMP to generate reference genomes. The Hillier laboratory at Magee Women's Research Institute, Pittsburgh, has previously cultivated Megasphaera sp. type 1 (strain 28L), whose genome has been sequenced (ADGP00000000; http://hmpdacc.org/catalog/).
We cultivated several bacterial isolates that were minority members of the bacterial community but were highly prevalent. Minority status does not necessarily imply that these isolates are insignificant in the pathogenesis of BV. Low abundance organisms in the human body can function as pathogens under particular conditions. For example, Porphyromonas gingivalis, a low-abundance but prevalent bacterium in the oral niche, can disrupt host-microbe homeostasis, leading to inflammatory periodontal disease [38]. Having cultivated isolates of minority vaginal bacteria provides opportunities for future hypothesis development and in vitro experimentation. Potentially important novel isolates include Peptoniphilaceae DNF01163 and Prevotellaceae DNF00733, which were present in 68% and 40% of women in this study, but at low abundance.
Similarly, little is known about the role of rare members of the bacterial community in the human microbiome that are neither prevalent, nor abundant. Cultivation strategies have the potential to be more sensitive than deep sequencing methods in detecting these rare members if favorable growth conditions are provided. Theoretically, cultivating a bacterium requires just one bacterial cell to divide and form a colony under optimal growth conditions. Examples of rare bacteria isolated in this study include novel bacteria, such as Corynebacterium sp. DNF00584 and Olsenella sp. DNF959, and many members of Actinomyces, Bifidobacterium, and Corynebacterium. It is conceivable that some rare bacteria isolated in the vagina are not long-term colonizers of the vaginal microbiota but are transient members of the community. For example, the Actinomyces are present in oral bacterial communities [39], Bifidobacterium spp. are gut colonizers [40], and Corynebacterium spp. are key members of the skin microbiota [41], and these bacteria may have been inoculated into the vagina through sexual or other practices. It is also plausible that duplicate species may have differing metabolic capabilities depending on the body site they are present in, and studies examining such questions are emerging as more isolates and genomes are available for comparison [42]. With this in mind, the HMP has recommended that duplicate species from different body sites be selected for WGS to help unravel the role of similar bacterial species in different body sites (http://hmpdacc.org/reference_genomes/reference_genomes.php); several bacteria from our study have been submitted for WGS (Supplementary Table 1).
Human microbial communities frequently have multiple species within a bacterial genus, but there is limited understanding of the function of closely related species in each ecological niche. Several species of Prevotella and Lactobacillus were isolated in this study. There is emerging evidence that not all Lactobacillus species are functionally equivalent in the vaginal niche. For example, L. crispatus and L. jensenii have often been associated with health in many cultivation and molecular studies. Recent molecular investigations have shown that L. iners is present in high abundance both in women with and women without BV [2, 4]. We have previously noted that L. iners often dramatically increases in concentrations after antibiotic treatment for BV [43, 44], and this bacterium has metabolic profiles that are intermediate between those of the lactobacilli often associated with health, L. crispatus and L. jensenii, and the BVAB [5]. These observations highlight that closely related members of a bacterial community can have different metabolic capabilities in the same ecological niche. Similarly, several Prevotella species found in the vagina are associated with BV, but the role of each species is poorly understood. P. amnii and P. timonensis are both associated with BV [4, 30, 45] and have been isolated in this study. P. buccalis, P. disiens, and P. bivia sequences were correlated with a positive amine odor, whereas P. bivia sequences were associated with elevated pH [4]. Having multiple isolates of these closely related Prevotella species will enable us to explore pangenomes, the full complement of genes in a phylogenetic clade, and facilitate investigations about the genetic variation and functional redundancy of closely related species.
An important question we addressed was what fraction of bacterial species identified in the vagina using PCR methods are ultimately cultivated using the conventional plating method. We successfully cultivated a large fraction of the bacteria known to be present in the vagina, though several species remained recalcitrant to cultivation. Prominent bacteria that we did not isolate include Leptotrichia/Sneathia spp., Megasphaera sp. type 2, BVAB1, BVAB2, and Mageeibacillus indolicus (BVAB3), among others. Recently, M. indolicus was isolated on Brucella blood agar from an endometrial biopsy specimen and has the ability to produce indole, hence its name [46]. In another study, Sneathia amnii was isolated on chocolate agar plates from a vaginal swab sample [47]. A genome of Megasphaera sp. type 2 isolated by the Hillier laboratory is available from the HMP collection of reference genomes (www.hmpdacc.org/catalog). Nonetheless, there are still uncultivated species from the human vagina, and novel approaches may be required to routinely culture these crucial members of the vaginal microbiota.
In conclusion, classical cultivation paired with methodical 16S rRNA sequencing for identification yielded a diverse and varied set of bacterial isolates from the human vagina, including 2 key members of the BV microbiota, Dialister sp. type 2 and Eggerthella sp. type 1, that were known only by their molecular signatures. Identification of isolates using 16S rRNA sequencing and phylogeny was valuable and robust in differentiating between bacterial strains. However, the approach was labor intensive, and neither high throughput nor real time. Bacteria isolated from this study are being subjected to WGS and used to study microbial interactions in vitro with the goal of further illuminating the pathogenesis of BV.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Supplement sponsorship. This article appears as part of the supplement “Proceedings of the 2015 NIH/NIAID Bacterial Vaginosis Expert Consultation,” sponsored by the Division of Microbiology and Infectious Diseases of the National Institute of Allergy and Infectious Diseases in partnership with the University of Alabama at Birmingham Sexually Transmitted Infections Clinical Trials Group; contract HHSN272201300012I.
Acknowledgments. We thank Dwyn Dithmer and Kathy Ringwood for their clinical contributions. We are grateful to Kevin Depner for his assistance in DNA extractions, broad-range polymerase chain reaction, and sequencing.
Financial support. The work was supported by the National Institute of Health (grant R01 HG005816-01 to D. N. F.), as part of the Human Microbiome Project initiative focused on technology development.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Gajer P, Brotman RM, Bai G et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med 2012; 4:132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hummelen R, Fernandes AD, Macklaim JM et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One 2010; 5:e12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ravel J, Gajer P, Abdo Z et al. Microbes and health Sackler Colloquium: vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA 2010; 108(suppl 1):4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srinivasan S, Hoffman NG, Morgan MT et al. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One 2012; 7:e37818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srinivasan S, Morgan MT, Fiedler TL et al. Metabolic signatures of bacterial vaginosis. mBio 2015; 6:e00204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillier S, Marrazzo JM, Holmes KK. Bacterial vaginosis. In: Holmes KK, Sparling P-A. Sexually transmitted diseases. New York, NY: McGraw-Hill, 2008:737–68. [Google Scholar]
- 7.Hillier SL, Krohn MA, Rabe LK, Klebanoff SJ, Eschenbach DA. The normal vaginal flora, H2O2-producing lactobacilli, and bacterial vaginosis in pregnant-women. Clin Infect Dis 1993; 16:S273–81. [DOI] [PubMed] [Google Scholar]
- 8.Paavonen J, Miettinen A, Stevens CE, Chen KC, Holmes KK. Mycoplasma hominis in nonspecific vaginitis. Sex Transm Dis 1983; 10(4 suppl):271–5. [PubMed] [Google Scholar]
- 9.Spiegel CA, Roberts M. Mobiluncus gen-nov, Mobiluncus curtisii subsp curtisii sp nov, Mobiluncus curtisii subsp holmesii subsp nov, and Mobiluncus mulieris sp nov, curved rods from the human vagina. Intl J Sys Bacteriol 1984; 34:177–84. [Google Scholar]
- 10.Totten PA, Amsel R, Hale J, Piot P, Holmes KK. Selective differential human blood bilayer media for isolation of Gardnerella (Haemophilus) vaginalis. J Clin Microbiol 1982; 15:141–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hymes SR, Randis TM, Sun TY, Ratner AJ. DNase inhibits Gardnerella vaginalis biofilms in vitro and in vivo. J Infect Dis 2013; 207:1491–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis WG, Robinson LS, Gilbert NM, Perry JC, Lewis AL. Degradation, foraging, and depletion of mucus sialoglycans by the vagina-adapted Actinobacterium Gardnerella vaginalis. J Biol Chem 2013; 288:12067–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson JL, Stull-Lane A, Girerd PH, Jefferson KK. Analysis of adherence, biofilm formation and cytotoxicity suggests a greater virulence potential of Gardnerella vaginalis relative to other bacterial-vaginosis-associated anaerobes. Microbiology 2010; 156(pt 2):392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rampersaud R, Planet PJ, Randis TM et al. Inerolysin, a cholesterol-dependent cytolysin produced by Lactobacillus iners. J Bacteriol 2011; 193:1034–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santiago GL, Deschaght P, El Aila N et al. Gardnerella vaginalis comprises three distinct genotypes of which only two produce sialidase. Am J Obstetr Gynecol 2011; 204:450 e1–7. [DOI] [PubMed] [Google Scholar]
- 16.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med 2005; 353:1899–11. [DOI] [PubMed] [Google Scholar]
- 17.Human Microbiome Jumpstart Reference Strains Consortium, Nelson KE, Weinstock GM, Highlander SK et al. A catalog of reference genomes from the human microbiome. Science 2010; 328:994–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amsel R, Totten PA, Spiegel CA, Chen KC, Eschenbach D, Holmes KK. Nonspecific vaginitis: diagnostic criteria and microbial and epidemiologic associations. Am J Med 1983; 74:14–22. [DOI] [PubMed] [Google Scholar]
- 19.Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J Clin Microbiol 1991; 29:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanks JH, Wallace RE. Relation of oxygen and temperature in the preservation of tissues by refrigeration. Pro Soc Exp Biol Med 1949; 71:196–200. [DOI] [PubMed] [Google Scholar]
- 21.Martius J, Krohn MA, Hillier SL, Stamm WE, Holmes KK, Eschenbach DA. Relationships of vaginal Lactobacillus species, cervical Chlamydia trachomatis, and bacterial vaginosis to preterm birth. Obstet Gynecol 1988; 71:89–95. [PubMed] [Google Scholar]
- 22.Rogosa M, Mitchell JA, Wiseman RF. A selective medium for the isolation and enumeration of oral and fecal lactobacilli. J Bacteriol 1951; 62:132–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 2008; 74:2461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loy A, Maixner F, Wagner M, Horn M. probeBase—an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res 2007; 35(database issue):D800–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999; 41:95–8. [Google Scholar]
- 26.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res 2014; 42(database issue):D32–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim OS, Cho YJ, Lee K et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Sys Evol Microbiol 2012; 62(pt 3):716–21. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsen FA, Kodner RB, Armbrust EV. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinform 2010; 11:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fredricks DN, Fiedler TL, Thomas KK, Oakley BB, Marrazzo JM. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J Clin Microbiol 2007; 45:3270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falsen E, Pascual C, Sjoden B, Ohlen M, Collins MD. Phenotypic and phylogenetic characterization of a novel Lactobacillus species from human sources: description of Lactobacillus iners sp. nov. Int J Syst Bacteriol 1999; 49(pt 1):217–21. [DOI] [PubMed] [Google Scholar]
- 32.Cools P, Oyaert M, Vaneechoutte M, De Laere E, Vervaeke S. Atopobium deltae sp. nov., isolated from the blood of a patient with Fournier's gangrene. Int J Sys Evol Microbiol 2014; 64(pt 9):3140–5. [DOI] [PubMed] [Google Scholar]
- 33.Pagnier I, Croce O, Robert C, Raoult D, La Scola B. Non-contiguous finished genome sequence and description of Fenollaria massiliensis gen. nov., sp. nov., a new genus of anaerobic bacterium. Stand Genomic Sci 2014; 9:704–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 2004; 68:669–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y, Xiong X, Danska J, Parkinson J. Metatranscriptomic analysis of diverse microbial communities reveals core metabolic pathways and microbiome-specific functionality. Microbiome 2016; 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rappe MS, Giovannoni SJ. The uncultured microbial majority. Ann Rev Microbiol 2003; 57:369–94. [DOI] [PubMed] [Google Scholar]
- 37.Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Ann Rev Microbiol 1985; 39:321–46. [DOI] [PubMed] [Google Scholar]
- 38.Hajishengallis G, Liang S, Payne MA et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 2011; 10:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aruni AW, Dou Y, Mishra A, Fletcher HM. The biofilm community-rebels with a cause. Curr Oral Health Rep 2015; 2:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jost T, Lacroix C, Braegger C, Chassard C. Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutr Rev 2015; 73:426–37. [DOI] [PubMed] [Google Scholar]
- 41.Kong HH, Oh J, Deming C et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 2012; 22:850–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendes-Soares H, Suzuki H, Hickey RJ, Forney LJ. Comparative functional genomics of Lactobacillus spp. reveals possible mechanisms for specialization of vaginal lactobacilli to their environment. J Bacteriol 2014; 196:1458–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayer BT, Srinivasan S, Fiedler TL, Marrazzo JM, Fredricks DN, Schiffer JT. Rapid and profound shifts in the vaginal microbiota following antibiotic treatment for bacterial vaginosis. J Infect Dis 2015; 212:793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivasan S, Liu C, Mitchell CM et al. Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PLoS One 2010; 5:e10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fredricks DN, Fiedler TL, Thomas KK, Mitchell CM, Marrazzo JM. Changes in vaginal bacterial concentrations with intravaginal metronidazole therapy for bacterial vaginosis as assessed by quantitative PCR. J Clin Microbiol 2009; 47:721–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Austin MN, Rabe LK, Srinivasan S, Fredricks DN, Wiesenfeld HC, Hillier SL. Mageeibacillus indolicus gen. nov., sp. nov.: a novel bacterium isolated from the female genital tract. Anaerobe 2015; 32:37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harwich MD Jr, Serrano MG, Fettweis JM et al. Genomic sequence analysis and characterization of Sneathia amnii sp. nov. BMC Genomics 2012; 13(suppl 8):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.