Abstract
CD8+ T cells use contact-dependent cytolysis of target cells to protect the host against intracellular pathogens. We have previously shown that CD8+ T cells and perforin are required to protect against the extracellular pathogen Yersinia pseudotuberculosis. Here we establish an experimental system where CD8+ T cells specific to a single model antigen are the only memory response present at time of challenge. Using mice immunized with a vaccine strain of Listeria monocytogenes that expresses secreted ovalbumin (Lm-OVA), we show that OVA-specific CD8+ T cells are generated and provide limited protection against challenge with virulent OVA+ Y. pseudotuberculosis. Perforin expression by OVA-specific CD8+ T cells was required, as Lm-OVA-immunized perforin-deficient mice showed higher bacterial burden as compared to Lm-OVA-immunized perforin-sufficient mice. Surprisingly, antigen-specific T cell protection waned over time, as Lm-OVA-immune mice eventually succumbed to Yersinia infection. Kinetic analysis of infection in mice with and without OVA-specific CD8+ T cells revealed that bacterial numbers increased sharply in OVA-naïve mice until death, while OVA-immune mice held bacterial burden to a lower level throughout the duration of illness until death. Clonal analysis of bacterial populations in OVA-naïve and OVA-immune mice at distinct time points revealed equivalent and severe bottle-neck effects for bacteria in both sets of mice immediately after intravenous challenge, demonstrating a dominant role for other aspects of the immune system regardless of CD8+ T cell status. These studies indicate that CD8+ T cells against a single antigen can restrict Y. pseudotuberculosis colonization in a perforin-dependent manner, but ultimately are insufficient in their ability to provide sterilizing immunity and protect against death.
Keywords: Yersinia pseudotuberculosis, CD8+ T cells, YopE, perforin, bottle-neck, ovalbumin
1. INTRODUCTION
Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis are the three human-pathogenic yersiniae that cause different diseases, but have in common the ability to disseminate from initial colonization sites and cause systemic disease. Y. pestis can enter the skin through infected flea bite and disseminate to draining lymph nodes, eventually reaching the bloodstream and lungs to cause septicemic and pneumonic plague, respectively. In contrast, enteric Y. enterocolitica and pseudotuberculosis, acquired via contaminated food stuffs, are generally restricted to the intestine, intestinal lymphoid tissue, and draining mesenteric lymph nodes, but in unique circumstances, such as during hemochromatosis (an iron overload disease), can disseminate to cause systemic disease (Logsdon and Mecsas, 2003; Moalem et al., 2004; Vadillo et al., 1994). Yersinia fatalities typically result from failure of the host immune system to eliminate the bacterium and subsequent sepsis and multi-organ failure. To prevent its clearance and facilitate its survival, this microorganism uses multiple virulence factors that disarm phagocytic cells. Yersinia injects Type 3 secretion system (T3SS) effector proteins called Yops (Yersinia outer proteins) directly into host cells (Plano and Schesser, 2013), where they misregulate proteins that function in cell signaling pathways critical for defense. For example, YopE, a GTPase-Activating Protein (GAP), inactivates Rho family GTPases to inhibit activities associated with actin cytoskeleton rearrangement, MAP kinase signal generation, and reactive oxygen species (ROS) production (Black and Bliska, 2000; Songsungthong et al., 2010; Viboud et al., 2006; Von Pawel-Rammingen et al., 2000). YopH is a tyrosine phosphatase interferes with calcium flux, cytokine production and phagocytosis (Andersson et al., 1999; Grosdent et al., 2002; Rolan et al., 2013).
Proof that innate immune cells are a target for Yersinia-mediated immune-disruption comes from in vivo studies showing that Yersinia injects Yops into neutrophils and macrophages (Durand et al., 2010; Koberle et al., 2009; Marketon et al., 2005) and that yop mutants are rescued for avirulence when neutrophils are depleted (Rolan et al., 2013; Westermark et al., 2014; Ye et al., 2009). It has been previously shown that Yersinia-specific CD8+ T cells are generated in response to Yops and this contributes towards vaccine-driven protective immunity against Y. pseudotuberculosis (Bergman et al., 2009; Wiedig et al., 2005; Zhang et al., 2012). Along such lines, Bergsbaken and Bevan showed that after oral infection with Y. pseudotuberculosis the pro-inflammatory microenvironment within the intestine regulated the differentiation of tissue resident CD8+ T cells to a CD103(−) subset. The latter was required for controlling the infection and pathogen clearance (Bergsbaken and Bevan, 2015). A recent report showed that Y. pestis antigen YopE69–77 was recognized by CD8 T cells in vivo and conferred protection against Y. pestis in a TNF-α and IFN-γ dependent manner (Szaba et al., 2014). Herein, we sought to determine if this adaptive immune response was alone sufficient to confer protection in the absence of other Yersinia-specific memory responses. To do this, we established an immunization-challenge system to study Yersinia-responsive CD8+ T cells in isolation. The results of these studies indicate that CD8+ T cell are critical for limiting the extent of early disease and suggests that Yersinia-specific vaccine efforts must engage multiple immune responses to ensure sterilizing immunity.
2. MATERIALS AND METHODS
2.1 Mouse Strains and Procedures
All animal use procedures were performed in strict accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at Tufts University (Protocol #51-07) and at the University of Texas Health Sciences Center San Antonio (Protocol #12030X). C57BL/6-Prf1tm1Sdz/J (perforin-deficient) and aged-matched C57BL/6 mice were obtained from The Jackson Laboratory (Bar Harbor, ME), all other C57BL/6 mice were obtained from Charles River (Wilmington, MA). Female 8–10 week-old mice were used for all experiments and were allowed to acclimatize for 5–7 days prior to use. Mice were housed in specific-pathogen-free conditions. For depletion studies, mice were injected intraperitoneally with 200 μg monoclonal antibody against murine CD8 in 200 μL phosphate-buffered saline (PBS), at days 3 and 1 prior to infection, then days 1, 4, 7, and 10 post-infection; control mice received PBS only (Coligan et al., 2005). Antibodies used for CD8 depletion were purified from hybridoma supernatants (clone 2.43, American Type Culture Collection by Dr. Douglas Jefferson, Tufts University Medical Center and determined to be endotoxin-free (data not shown). The CD8 deficiency status of the knockout or depleted mice was confirmed by fluorescence-activated cell sorting (FACS) analysis of spleen cells stained with mouse antibodies against CD8 (see Flow Cytometry below); depleted mice routinely displayed a 97–98% reduction in splenic CD8+ T cells.
2.2 Bacterial Strains
Y. pseudotuberculosis serotype III strain YPIII pIB1 was used to cause virulent disease in animals (Bolin et al., 1982), while the attenuated ksgA− mutant strain on the same background was used to immunize mice against Y. pseudotuberculosis (Mecsas et al., 2001). Fusions of yopE and ovalbumin DNA were constructed exactly as previously described by Wiedig et al., where either the first 18 or 138 codons of yopE were fused to an ovalbumin gene fragment encoding amino acids 247–355 (Wiedig et al., 2005). The cloned yopE allele included DNA encoding the gene upstream of yopE, sycE (chaperone for YopE, transcribed in opposite direction), and the intergenic DNA (containing native promoters). DNA encoding the fusions was inserted between fragments containing ~500 bp that flank the lacZ gene, cloned in a suicide vector (gift of Dr. Joan Mecsas, Tuft Medical School), to allow for simultaneous disruption of the lacZ gene and knock-in of the yopE::ovalbumin fusion. Allelic exchange was accomplished using the suicide vector pCVD442 (λpir-dependent replicon, AmpR, sacB gene conferring sucrose sensitivity): pCVD442 carrying the ΔlacZ::yopE::ovalbumin allele was transferred to Y. pseudotuberculosis strains YPIII pIB1 and the ksgA− mutant by conjugation, and the resulting AmpR LacZ− Y. pseudotuberculosis (irgasan-resistant) integrants were then grown in the absence of ampicillin selection and plated on nutrient agar containing sucrose and X-gal to identify those clones that had lost sacB gene (and by inference, the linked plasmid DNA) and lacZ genes. Individual AmpS, sucroseR, LacZ− colonies were screened by Western to confirm expression of ovalbumin (see below). The control YPIII pIB1 ΔlacZ and ksgA− ΔlacZ strains were constructed similarly, except the yopE::OVA fusions were not inserted. The attenuated L. monocytogenes ΔactA ΔplcB strain in the 10403S background (Angelakopoulos et al., 2002), carrying an actA::ovalbumin gene fusion, which contains the first 100 codons of actA fused to ovalbumin and integrated at the tRNAArg gene on the L. monocytogenes genome (Lauer et al., 2002) was a kind gift from Dr. Darren Higgins of Harvard Medical School.
2.3 YopE::OVA Expression and Secretion Assays
To generate cultures induced for Yop expression and secretion, overnight cultures of Y. pseudotuberculosis were back-diluted into low calcium medium (2X YT broth with 20 mM sodium oxalate and 20 mM MgCl2) to give an estimated optical density at 600 nm of 0.2, then incubated at 26° C with rolling for 1.5 hours, then at 37°C for an additional 1.5 hours. Cultures were centrifuged at 12,000 × g for 1 min. Pellets were washed with 1× PBS and re-suspended in 20 μL 2× SDS sample buffer. Then, 900 μL of culture supernatant was added to 100 μL of chilled 100% TCA and incubated for 30 min on ice. Samples were then centrifuged at 12,000 × g for 20 min at 4° C. Pellets were rinsed with ice cold acetone and centrifuge at 12,000 × g for 20 min, twice. Pellets were air dried and resuspended in 20 μL 2X SDS sample buffer. Yersinia protein preparations were separated by gradient SDS-PAGE, transferred to nitrocellulose, probed with polyclonal sera specific to ovalbumin, followed by goat anti-rabbit IgG conjugated to horseradish peroxidase (HRP). Reactive HRP was detected by enhanced chemiluminescence (Amersham Biosciences, Piscataway, NJ).
2.4 In Vivo Inoculations, Survival and Organ CFU Assays
Y. pseudotuberculosis strains were grown overnight in LB (Lennox) broth at 26°C with aeration, diluted in PBS to the desired concentration, and 200 μL of the indicated colony-forming units (CFU) intravenously inoculated into the lateral tail vein of mice (30½ gauge needle, Becton Dickinson & Co., Franklin Lakes NJ). L. monocytogenes strains were grown overnight in brain-heart infusion broth at 30°C without aeration, washed and diluted in PBS to the desired concentration, and 200 μL of 3.0 × 107 CFU delivered intravenously. For survival experiments, animals were sacrificed upon displaying unresponsiveness to touch and signs of morbidity (hunched, scruffy fur, unresponsiveness to touch) and scored as non-surviving. To determine Y. pseudotuberculosis CFU levels in organs, mice were sacrificed at the indicated time points and tissues harvested, placed in pre-weighed tubes containing sterile PBS, and weighed to determine tissue weight. Tissues were mechanically homogenized using a tissue homogenizer (Omni, Marietta GA) and 100 μL of dilutions of tissue homogenate were plated on LB agar plates. After 48 hours incubation at room temperature, CFU were enumerated and normalized to the gram weight of each tissue.
2.5 Flow Cytometry
To prepare hepatic lymphocytes we first perfused each mouse with 15 mL of PBS + heparin 75 U/mL (H3149-100KU sigma). Perfused livers were dissected, cut into small pieces and fully separated with a syringe plunger on Falcon® 70 μm Cell Strainer (Falcon, BD Biosciences, San Jose CA). Collections were centrifuged at 400 × g for 10 min at 25° C. Supernatant was removed and pellets were re-suspended in 15 mL of 35% Percoll (GE 250 mL 17-0891-02). Collections were centrifuged at 400 × g for 15 min at 25° C, and washed with HBSS. Single cell suspensions of spleen were prepared by gently dissociating the tissue through Falcon® 70 μm Cell Strainer (BD Biosciences, San Jose CA). After removal of red blood cells by PharmLyse (BD Biosciences, San Jose CA), 1 × 106 cells were blocked with FcBlock (clone 2.4G2) for 15 min on ice. Then cells were stained with antibodies to murine CD4 (L3T4, clone RM4-4, FITC conjugated), CD8 (Ly-2, clone 53-6.7, PE or FITC conjugated), CD19 (IgG2a, clone 6D5, PE/Cy5 conjugated) and CD69 (Very Early Activation Antigen, clone H1.2F3, PECy7 conjugated) for 30 min on ice in the dark. Stained cells were fixed with 2% paraformaldehyde (Electron Microscopy Services, Hatfield PA), and data acquired on a FACSCalibur (BD Biosciences Immunocytometry Systems, San Diego CA). All flow cytometry reagents and antibodies were purchased from BD Biosciences Pharmingen (San Jose CA).
2.6 Yptb-OVA clones – generation, inoculation, sequencing, analysis
The Yptb-OVA clone library was generated similarly as previously described (29). A mutated pSC189 vector containing the Himar1 transposon with an MmeI restriction site was introduced to the Yptb-OVA strain via mating. Briefly, 25 mL of Yptb-OVA was grown overnight in 2XYT broth at 26°C, and 75 mL of Escherichia coli SM10λpir/pSC189-Himar1(MmeI) was grown O/N at 37°C in LB containing 30 μg/mL kanamycin and 100 μg/mL ampicillin The SM10λpir bacteria were washed 3X with PBS, pelleted, resuspended in the Yersinia culture, and the suspension filtered onto nitrocellulose, which was placed gently placed on minimal media agar. Mating was allowed to proceed for 16–24 hours at 26°C. Bacteria were then scraped off the filter paper, resuspended in 5 mL 2XYT, and spread on 10 LB plates containing 30 μg/ml kanamycin and 2 μg/ml irgasan. 200 colonies were picked, inoculated into 2XYT broth containing 30 μg/ml kanamycin and 2 μg/ml irgasan in multiwall plates. After overnight growth at 26°C, glycerol was added to 25% final concentration and the stocks stored at −80°C. After screening clones individually to confirm each was an independent clone (Southern blot analysis, data not shown), 150 clones were chosen to comprise the final population.
For mouse inoculations, clones in glycerol stocks were individually inoculated into wells of deep-well multiwall plates containing 2XYT broth with 30 μg/ml kanamycin and 2 μg/ml irgasan in multiwall plates. After overnight growth, the clones were combined (normalizing for OD600) and diluted in PBS to give an estimated concentration of 150 colony-forming units (CFU)/200 μL. Animals were injected with 200 μL (estimated ISO clones per animal) via tail vein. The remaining portion of the inoculum stock was plated onto LB plates containing 30 μg/ml kanamycin and 2 μg/ml irgasan for overnight growth at 26°C, both to determine the actual concentration of the inoculum and to provide source material to assess the number of clones in inoculum – all colonies were scraped for genomic DNA isolation (input sample). At various times post infection, animals were sacrificed, organs homogenized and the entire homogenate plated onto LB medium containing 30 μg/ml kanamycin and 2 μg/ml irgasan. Colonies were scraped off plates (output samples) and genomic DNA from all samples isolated using Qiagen DNeasy kit. Samples were prepared for Illumina sequencing, as described (29). After Illumina sequencing, the number of reads for each clone in a given sample was normalized for amount of DNA added to sequencing run (total number of reads) and the number of clones present in input and output samples enumerated.
2.7 Data and Statistical Analysis
FlowJo (Tree Star, Ashland OR) was used to analyze flow cytometry data. Prism (GraphPad Software, La Jolla CA) was used for graphing and statistical analysis. Survival curves were estimated using the Kaplan Meier method and significance calculated using the log-rank test. The nonparametric Mann-Whitney U test was used to determine statistical differences between groups of animals.
3. RESULTS
3.1 An immunization-challenge system to examine memory CD8+ T cell responses to Y. pseudotuberculosis
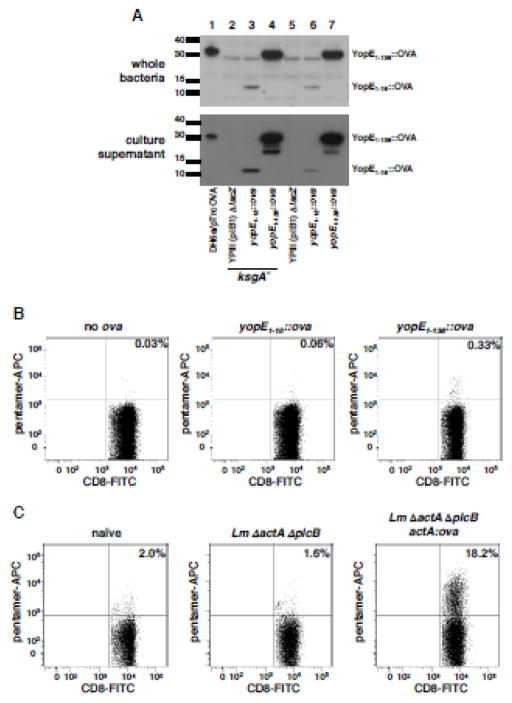
We first designed an immunization-challenge system whereby CD8+ T cells were the only Yersinia-specific memory response at the time of challenge. As the natural Yersinia antigens recognized by CD8+ T cells were unknown at the start of our studies, we instead expressed the model antigen ovalbumin in Y. pseudotuberculosis. To do this, we used a previously constructed fusion containing the amino terminal 138 residues of YopE tagged with residues 247–355 of ovalbumin at its C-terminal end (Wiedig et al., 2005). This construct, which includes the YopE immunodominant CD8 epitope (aa 69–77), the ovalbumin dominant epitope SIINFEKL, translocation signals, and the YopE chaperone SycE, was inserted in the Y. pseudotuberculosis chromosome at the lacZ gene (henceforth referred to as Yptb YopE1-138::OVA). We confirmed that disruption of lacZ did not alter the ability of either virulent Y. pseudotuberculosis (strain YPIII pIB1, hereafter called virulent or wildtype, WT Yptb) to cause disease or attenuated Y. pseudotuberculosis (YPIII pIB1 ksgA−, hereafter called attenuated Yptb) to colonize target organs in inoculated animals (Supplementary Figure 1). Both wildtype and the attenuated Yptb were capable of expressing the translocation-competent YopE1-138::OVA fusion and the control fusion protein, secretion-competent but translocation-defective YopE1-18::OVA (Figure 1A). In the case of the secretion competent fusion, robust accumulation of the fusion was observed in culture supernatant, indicating that the protein could be recognized and secreted by the TTSS (Figure 1A).
Figure 1. YopE-OVA fusion protein is secreted by Yersinia, and is processed and presented in vivo to stimulate OVA-specific CD8+ T cell responses.
(A) The indicated Y. pseudotuberculosis strains were grown in Yop secretion conditions (Materials and Methods) and processed for western blotting of whole cell lysates and supernatant proteins, with anti-ovalbumin polyclonal antisera. (B) ksgA− strains expressing YopE18::OVA or YopE138::OVA were delivered to animals via intravenous inoculation and 10 days later, spleens removed, single cell suspensions generated and cells incubated with MHC Class I H2-Kb pentamers specific to OVA residues 257–264 and antibodies against CD8 and CD19. Shown are representative histograms showing pentamer and CD8 labeling of CD8+ CD19− cells. (C) Immunization with L. monocytogeneses ΔactA ΔplcB ActA100::OVA generates OVA-specific CD8+ T cells. 3 × 107 CFU of noted L. monocytogeneses strains were delivered to animals via intravenous inoculation. 7 days later spleens were removed, single cell suspensions generated and cells were incubated with H2-Kb OVA257-264 pentamers and antibodies against CD8 and CD19. Shown are representative scatter plots of 2 separated experiments showing pentamer and CD8 labeling of CD8+ CD19− cells (n=3 mice per experiment).
We subsequently immunized animals with the OVA+ ksgA− strains and 10 days later evaluated the frequency and number of CD8+ T cells specific to the H-2Kb-restricted SIINFEKL epitope in spleens from immunized animals. Animals exposed to attenuated OVA+ Y. pseudotuberculosis only generated detectable OVA-specific CD8+ T cells when the immunizing bacteria expressed the translocation-competent YopE1-138::OVA (Figure 1B). To generate OVA257-264-specific CD8+ memory T cell responses in animals that were naïve to Yersinia, we employed an attenuated Listeria monocytogenes strain lacking the virulence genes actA and plcB (Angelakopoulos et al., 2002) and encoding an ActA-ovalbumin fusion protein (Lauer et al., 2002), as used previously to generate OVA-specific CD8+ T cells (Wiedig et al., 2005). As expected, immunization of C57BL/6 mice with the ΔactA ΔplcB actA::ova strain (hereafter called Lm-OVA) generated large numbers of OVA257-264-specific CD8+ T cells (Figure 1C); 18.2% of CD8+ cells, compared to 2% in naïve mice). As such mice challenged with Lm OVA could be used to test the impact of specific CD8+ T cells in isolation on Yersinia infection when using Y. pseudotuberculosis expressing YopE1-138::OVA fusion (hereafter called Yptb-OVA).
3.2 OVA-specific CD8+ T cells are sufficient to protect hosts against Y. pseudotuberculosis in a perforin-dependent manner
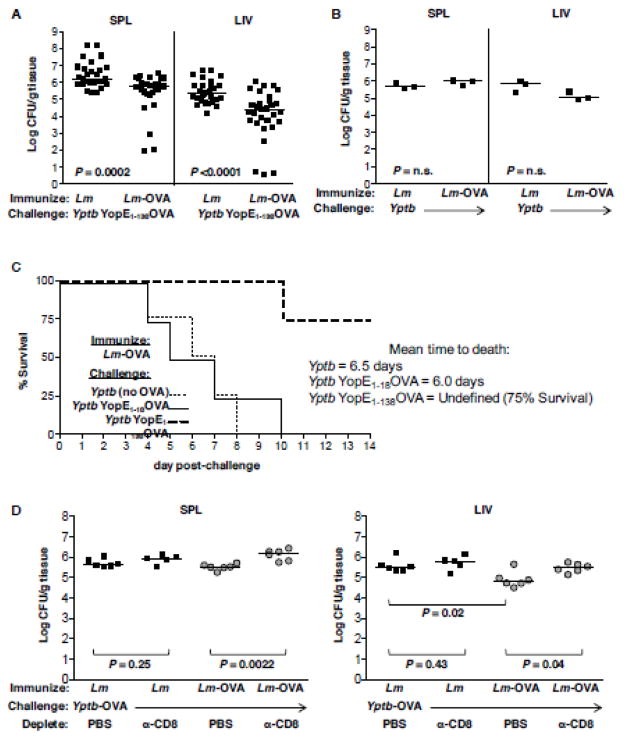
We first tested if OVA-specific CD8+ T cells were alone sufficient to protect animals from lethal challenge with Y. pseudotuberculosis expressing translocated OVA 60 days after immunization. Yptb-OVA bacterial numbers were significantly reduced in both the spleen and liver of Lm-OVA-immune mice as compared to Lm-immune mice at 5 days post-challenge (Figure 2A). Immunization with Lm-OVA per se did not confer nonspecific immunity to mice, as Lm-OVA- and Lm-immune mice were equally susceptible to challenge with OVA-negative Y. pseudotuberculosis (Figure 2B). Protection required translocation of ovalbumin by the challenge Yersinia bacteria, as virulent Y. pseudotuberculosis expressing the non-translocated YopE1-18::OVA fusion protein were equivalently virulent for Lm-OVA-immune mice as OVA-negative Y. pseudotuberculosisis in a survival assay. Mice challenged with YopE1-138::OVA fusion protein experienced only a 25 percent decrease in survival 14 days post-challenge (Figure 2C). Depletion of CD8+ T cells during challenge of the Lm-OVA-immune mice increased the number of Yptb-OVA CFU in the spleen (Figure 2D) and liver (Figure 2E) by almost ten-fold as compared to mock-depleted animals, supporting the notion that OVA-specific protective immunity was CD8+ T cell-dependent. Changes between the Lm controls versus the Lm-OVA vaccine upon challenge with Yptb-OVA were only seen in the liver (Figure 2E).
Figure 2. OVA-specific CD8+ T cells are sufficient to protect against Y. pseudotuberculosis expressing a YopE-OVA fusion protein and required for Lm-OVA immune mice to reduce Yptb-OVA burden.
Mice were immunized with 3 ×107 CFU L. monocytogenes ΔactA ΔplcB (Lm, A, B) or the same strain carrying the integrated ActA::OVA fusion protein (Lm-OVA, A, B, C), then 60 days later, challenged with 100 CFU virulent Y. pseudotuberculosis expressing YopE1-138::OVA (A, C), YopE1-18::OVA (C) or the parental strain YPIII(pIB1) (Yptb, B, C). (A, B) Mice were either sacrificed at day 5 and bacterial burden per gram tissue of spleen and liver determined or (C) observed until day to death. Mice were immunized with either Lm or Lm-OVA and at days 60 post-immunization challenged with Yptb-OVA. At days −3, -1, +1, +3 and +5 relative to challenge, mice were intraperitoneally injected with 200μg anti-CD8 monoclonal antibody or PBS. At day 6 post-challenge, mice were sacrificed and bacteria burden per gram (D) spleen or (E) liver determined. Bars indicate median values.
3.3 CD8+ T cells use perforin to protect hosts against Y. pseudotuberculosis in a tissue-specific manner
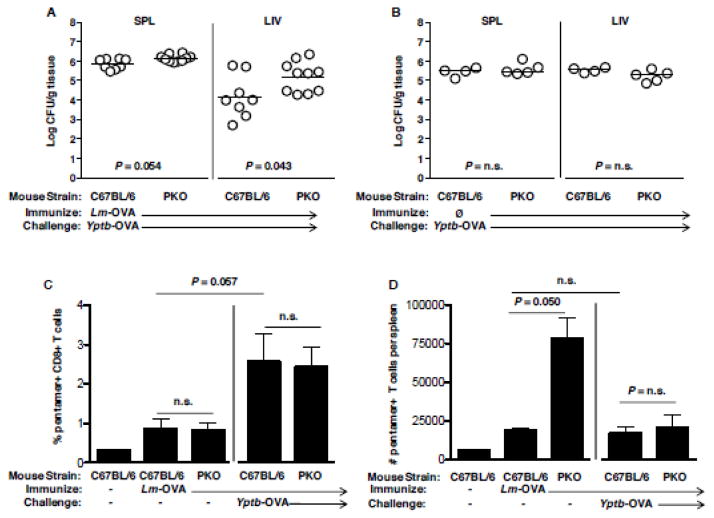
We previously observed that perforin-deficient animals were more susceptible to Y. pseudotuberculosis (Bergman et al., 2009). To reevaluate this requirement in context of only a CD8+ T cell response to Yersinia, we immunized perforin knockout (PKO) and C57BL/6 mice with Lm-OVA and challenged mice with Yptb-OVA. Bacterial burden in the liver of PKO mice was significantly greater than WT C57BL/6 mice at day 5 post-infection, although no differences were observed in the spleen (Figure 3A). Importantly, PKO mice were not inherently more susceptible to Y. pseudotuberculosis, as PKO and C57BL/6 mice displayed equivalent levels of Yptb-OVA burden in the absence of prior Lm-OVA immunization (Figure 3B). Lm-OVA-immunized PKO and C57BL/6 mice displayed equivalent frequencies of splenic OVA-specific CD8+ T cells at the time of Yptb-OVA challenge, and also at day 6 post-challenge (Figure 3C), although total numbers were larger at day 0 in PKO mice than C57BL/6 (Figure 3D), as has been previously observed in PKO exposed to Lm (Humphreys et al., 2008). In addition, challenge with Yptb-OVA increased the percentage of OVA+ CD8 T cells but not their number. These results indicate that the increased susceptibility of OVA-immune PKO mice was not due to reduced numbers of antigen-specific CD8+ T cells, but rather due to the lack of perforin in those T cells. Therefore, CD8-mediated protection against Y. pseudotuberculosis requires both perforin-independent and -dependent mechanisms, with the latter supporting the clearance of bacteria from liver tissues.
Figure 3. Perforin is required for OVA-specific CD8+ T cells to protect against Y. pseudotuberculosis and OVA-specific CD8+ T cells are present in Lm-OVA-immunized PKO animals and increase after exposure to Yptb-OVA.
C57BL/6 and PKO mice were immunized with Lm-OVA, then 60 days later, challenged with Yptb YopE1-138::OVA (hereafter called Yptb-OVA). Mice were sacrificed on day 5 and the bacterial burden per gram tissue of (A) spleen and (B) liver determined. C57BL/6 and PKO mice were immunized with 3 × 107 CFU Lm-OVA, then 60 days later a subset of the animals challenged with Yptb-OVA. 7 days post-challenge all animals were sacrificed, spleens harvested and single cell suspensions stained with pentamer specific to H2-Kb OVA257-264 and antibodies against CD8 and CD19. Shown are the calculated percent (C) and number (D) of splenic pentamer+ CD8+ cells.
3.4 Antigen-specific CD8+ T cells fail to contain Y. pseudotuberculosis over time
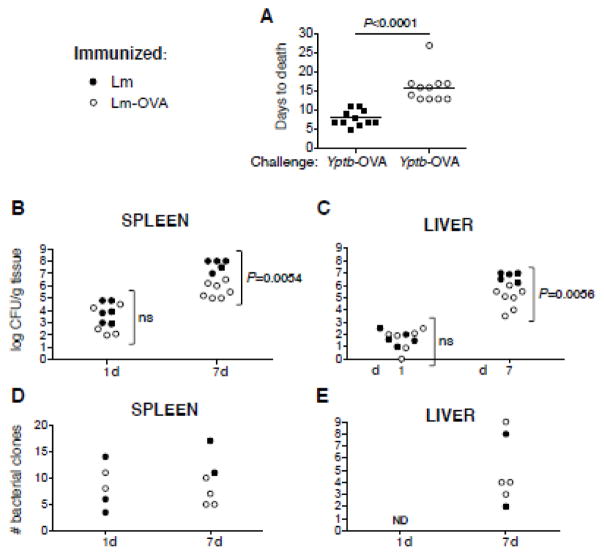
We subsequently tested whether OVA-specific CD8+ T cells were sufficient to provide sterilizing immunity against Yptb-OVA challenge bacteria. Lm-OVA-immune mice succumbed to challenge with Yptb-OVA after 15 days, whereas those immunized with Lm alone succumbed on day 7 (Fig 4A). Seven days is a time frame comparable to when naïve mice succumb to virulent Yptb infection (Barnes et al., 2006). At day 7 Lm-OVA-immune mice had no outward signs of infection, however, mice became ill and moribund identical to the control mice immediately prior to death (data not shown). Thus, CD8+ T cells significantly delayed but did not prevent death.
Figure 4. Lm-OVA-immune mice restrict Yptb-OVA burden early but eventually succumb to infection.
C57BL/6 mice were immunized with 3 ×107 CFU Lm or Lm-OVA, then 60 days later challenged with 100 CFU Yptb-OVA. Mice were sacrificed upon observation of morbidity and (A) day of death noted. Results shown are pooled from 3 separate experiments. Lm or Lm-OVA-immune mice were challenged with 150 clones of a Lm-OVA clone library, sacrificed at days 1, 7 and output bacterial burden (B, C) and clone numbers (D, E) in the spleen (B, D) and liver (D, E) determined. ND, not detected. Results shown are from two separate experiments.
To examine why CD8+ T cell immunity failed to contain infection, we evaluated the kinetics of bacterial burden in Ytpb-OVA-challenged mice. Although bacterial yields were similar at early timepoints, at day 7 Yptb-OVA titers in the spleen were 10 to 100-fold greater for mice that had received Lm versus those that had received Lm-OVA (Figure 4B; P = 0.0054). In the liver, titers were 10-fold greater in the Lm-alone immunized animals (Figure 4C; P = 0.0056). Together these results suggest that that pathogen surveillance by CD8+ T cells is most evident to the liver.
To investigate in greater detail the impact of CD8+ T cells on Yptb, we challenged immunized mice with mariner transposon-tagged bacterial clones and examined population dynamics (Barnes et al., 2006; Crimmins et al., 2012). A pooled library of 150 Yptb-OVA clones were used to challenge Lm and Lm-OVA-immunized mice, and deep-sequenced genomic DNA from input and output pools of splenic and hepatic-derived bacterial CFU to determine clone numbers. This analysis revealed that in each organ, only a fraction of the input clones could be detected at each of the time points analyzed, while there was no appreciable difference in the number of clones present in the spleens or livers of immunized relative to naïve mice (Fig 4D, E). Thus, the initial reduction in bacterial burden was not due to CD8+ T cells but to other host-imposed restrictions such as a “bottleneck” effect after intravenous challenge.
4. DISCUSSION
This report shows that antigen-specific CD8+ T cells can recognize and reduce Y. pseudotuberculosis burden in the mouse and in a perforin- and tissue-dependent manner. While CD8+ T cells conferred early protection against challenge, they were insufficient to provide sterilizing immunity, such that bacterial levels quickly climbed thereafter and animals succumbed to infection. Our observations indicate that Yersinia can either escape or undermine one arm of adaptive immune responses to persist in host tissues.
Overall, CD8+ T cells conferred greater protection against bacterial colonization in the liver than the spleen. This conclusion is based on two observations. First, at 1-week post-Yptb-OVA challenge, OVA-specific CD8+ T cells reduced burden more dramatically in the liver than in the spleen as compared to control mice. Secondly, the requirement for perforin in CD8+ T cell-mediated protection was only observed in the liver. T cells may protect the liver better than the spleen because liver has a larger number of antigen-specific CD8+ T cells. Pope et. al. found that the Listeria-specific CD8 response was significantly greater in the murine liver as compared to the spleen following primary and secondary exposure to L. monocytogenes (Pope et al., 2001). Other studies have found that influenza-specific CD8+ T cells accumulate in the liver to greater levels as compared to the spleen (Kedzierska et al., 2007; Polakos et al., 2006). It is surprising, however, that OVA-specific CD8+ T cells did not reduce levels of splenic OVA+ Yptb more significantly than what we observed, given that long-lived potent effector CD8+ T cells and Yersinia both localize to the red pulp in the spleen (Carter, 1975; Olson et al., 2013). Apparently, some aspect of the splenic environment is unfavorable for maximal CD8+ T cell activity against Yersinia. It is possible that microcolony formation of the bacterium in this organ site forms a niche that is protected from immune attack. Yet why this would be specific only to the spleen and not liver is unclear. One possibility is that bacteria in the spleen are to a greater extent surrounded by poorly functioning live neutrophils that have been rendered ineffective for antibacterial function by the translocated Yops (Davis et al., 2015). The ineffectiveness of our model to produce protective immunity might be also the result of the lack of an immunological boost during the vaccination process, as Lm-OVA only expressed a single antigen.
The inability to efficiently block disease after immunization with a single antigen, such as Lm-OVA, has precedence in the literature. Mice intranasally immunized with YopE, which contain a dominant CD8 epitope, YopE69-77, (Lin et al., 2011) were only partially protected from oral Y. pseudotuberculosis challenge, with 36% of the mice unable to survive the challenge out to 28 days (Lin et al., 2011; Zhang et al., 2015). Taken together, with our data demonstrating tissue-specific immunity, these data suggest that CD8+ T cell driven immunity, as previously reported (Bergman et al., 2009), requires a polyclonal response.
Our clonal analysis experiments are consistent with microbial escape, with or without antigen-specific T cell recognition, by a small number of clones that became established in tissues. This notion is based on the observation that the absolute clonal numbers of Yptb-OVA in the spleen were substantially reduced to less than 20 clones. No evidence was observed that this initial bottleneck was the result of T cell recognition, although subsequent protection was still observed in immunized mice before the infection eventually began to overwhelm the animal. One possible explanation for this early clearance are natural killer cells, which have been recently shown to mediate clearance of Y. pseudotuberculosis in the mesenteric lymph nodes (Rosenheinrich et al., 2015). Beyond early clearance, the possibility exists that Yersinia could escape CD8+ T cell recognition by down-regulating antigen expression to avoid antigen-specific T cell recognition, or inducing T cell exhaustion (Durward-Diioia et al., 2015). The latter is due to the semi-chronic nature of Yersinia infection in the Lm-OVA-immune mice. Regarding the latter hypothesis, we did not find any evidence that exhaustion markers were upregulated on OVA-specific CD8+ T cells, nor did blockage of exhaustion markers rescue Lm-OVA-immune mice from eventually succumbing to Yptb-OVA challenge (data not shown). Of note, we have previously demonstrated the presence of an intestinal bottleneck upon Yersinia dissemination in naïve mice (Barnes et al., 2006), which has also been described for enteric Salmonella (Maier et al., 2014). More recently a kidney-to-blood bottleneck was found for uropathogenic Escherichia coli (Walters et al., 2012). Thus our observations are not unprecedented.
5. CONCLUSIONS
In summary this study demonstrates that adaptive immunity can play an important role in restricting initial Y. pseudotuberculosis replication, most dramatically in the liver, during an infection. Yet this is insufficient for sterilization. At time of infection there is a dramatic reduction in the clonal diversity of Yersinia, indicating that other immune mechanisms clear most bacteria from the host, yet those that remain recover and can overcome a monoclonal CD8+ T cell defense. Future studies to dissect the additional host factors that restrict bacterial population dynamics will illuminate the importance of different immune responses during infection.
Supplementary Material
Highlights.
Perforin and CD8+ T-cells are required to protect against Yersinia pseuotuberculosis
CD8+ T-cells provide preferential protection in the liver vs. the spleen
Immunity to a single antigen is insufficient to generate sterilizing immunity
Acknowledgments
We acknowledge Rose Seoanes for technical assistance. This work was supported by National Institutes of Health, Grant AI085116 from the NIAID to MAB, HHMI support of RI, grant AI114800 to CJO, and AI067716, AI070412 and the Cancer Prevention Research Institute of Texas grant RP160512 to PHD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson K, Magnusson KE, Majeed M, Stendahl O, Fallman M. Yersinia pseudotuberculosis-induced calcium signaling in neutrophils is blocked by the virulence effector YopH. Infection and immunity. 1999;67:2567–2574. doi: 10.1128/iai.67.5.2567-2574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelakopoulos H, Loock K, Sisul DM, Jensen ER, Miller JF, Hohmann EL. Safety and shedding of an attenuated strain of Listeria monocytogenes with a deletion of actA/plcB in adult volunteers: a dose escalation study of oral inoculation. Infection and immunity. 2002;70:3592–3601. doi: 10.1128/IAI.70.7.3592-3601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PD, Bergman MA, Mecsas J, Isberg RR. Yersinia pseudotuberculosis disseminates directly from a replicating bacterial pool in the intestine. J Exp Med. 2006;203:1591–1601. doi: 10.1084/jem.20060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman MA, Loomis WP, Mecsas J, Starnbach MN, Isberg RR. CD8(+) T cells restrict Yersinia pseudotuberculosis infection: bypass of anti-phagocytosis by targeting antigen-presenting cells. PLoS Pathog. 2009;5:e1000573. doi: 10.1371/journal.ppat.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken T, Bevan MJ. Proinflammatory microenvironments within the intestine regulate the differentiation of tissue-resident CD8(+) T cells responding to infection. Nature immunology. 2015;16:406–414. doi: 10.1038/ni.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DS, Bliska JB. The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol Microbiol. 2000;37:515–527. doi: 10.1046/j.1365-2958.2000.02021.x. [DOI] [PubMed] [Google Scholar]
- Bolin I, Norlander L, Wolf-Watz H. Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with the virulence plasmid. Infection and immunity. 1982;37:506–512. doi: 10.1128/iai.37.2.506-512.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter PB. Pathogenecity of Yersinia enterocolitica for mice. Infection and immunity. 1975;11:164–170. doi: 10.1128/iai.11.1.164-170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coligan JE, Bierer BE, Margulies DH, Shevach EM, Strober W. Current Protocols in Immunology. John Wiley and Sons, Inc; New York, NY: 2005. [Google Scholar]
- Crimmins GT, Mohammadi S, Green ER, Bergman MA, Isberg RR, Mecsas J. Identification of MrtAB, an ABC transporter specifically required for Yersinia pseudotuberculosis to colonize the mesenteric lymph nodes. PLoS pathogens. 2012;8:e1002828. doi: 10.1371/journal.ppat.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KM, Mohammadi S, Isberg RR. Community behavior and spatial regulation within a bacterial microcolony in deep tissue sites serves to protect against host attack. Cell host & microbe. 2015;17:21–31. doi: 10.1016/j.chom.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EA, Maldonado-Arocho FJ, Castillo C, Walsh RL, Mecsas J. The presence of professional phagocytes dictates the number of host cells targeted for Yop translocation during infection. Cell Microbiol. 2010;12:1064–1082. doi: 10.1111/j.1462-5822.2010.01451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durward-Diioia M, Harms J, Khan M, Hall C, Smith JA, Splitter GA. CD8+ T Cell Exhaustion, Suppressed Gamma Interferon Production, and Delayed Memory Response Induced by Chronic Brucella melitensis Infection. Infection and immunity. 2015;83:4759–4771. doi: 10.1128/IAI.01184-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdent N, Maridonneau-Parini I, Sory MP, Cornelis GR. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infection and immunity. 2002;70:4165–4176. doi: 10.1128/IAI.70.8.4165-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys TD, Khanolkar A, Badovinac VP, Harty JT. Generation and maintenance of Listeria-specific CD8+ T cell responses in perforin-deficient mice chronically infected with LCMV. Virology. 2008;370:310–322. doi: 10.1016/j.virol.2007.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierska K, Stambas J, Jenkins MR, Keating R, Turner SJ, Doherty PC. Location rather than CD62L phenotype is critical in the early establishment of influenza-specific CD8+ T cell memory. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9782–9787. doi: 10.1073/pnas.0703699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberle M, Klein-Gunther A, Schutz M, Fritz M, Berchtold S, Tolosa E, Autenrieth IB, Bohn E. Yersinia enterocolitica targets cells of the innate and adaptive immune system by injection of Yops in a mouse infection model. PLoS pathogens. 2009;5:e1000551. doi: 10.1371/journal.ppat.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. Journal of bacteriology. 2002;184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JS, Szaba FM, Kummer LW, Chromy BA, Smiley ST. Yersinia pestis YopE contains a dominant CD8 T cell epitope that confers protection in a mouse model of pneumonic plague. Journal of immunology. 2011;187:897–904. doi: 10.4049/jimmunol.1100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon LK, Mecsas J. Requirement of the Yersinia pseudotuberculosis effectors YopH and YopE in colonization and persistence in intestinal and lymph tissues. Infection and immunity. 2003;71:4595–4607. doi: 10.1128/IAI.71.8.4595-4607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L, Diard M, Sellin ME, Chouffane ES, Trautwein-Weidner K, Periaswamy B, Slack E, Dolowschiak T, Stecher B, Loverdo C, Regoes RR, Hardt WD. Granulocytes impose a tight bottleneck upon the gut luminal pathogen population during Salmonella typhimurium colitis. PLoS pathogens. 2014;10:e1004557. doi: 10.1371/journal.ppat.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marketon MM, DePaolo RW, DeBord KL, Jabri B, Schneewind O. Plague bacteria target immune cells during infection. Science. 2005;309:1739–1741. doi: 10.1126/science.1114580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecsas J, Bilis I, Falkow S. Identification of attenuated Yersinia pseudotuberculosis strains and characterization of an orogastric infection in BALB/c mice on day 5 postinfection by signature-tagged mutagenesis. Infect Immun. 2001;69:2779–2787. doi: 10.1128/IAI.67.5.2779-2787.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moalem S, Weinberg ED, Percy ME. Hemochromatosis and the enigma of misplaced iron: implications for infectious disease and survival. Biometals. 2004;17:135–139. doi: 10.1023/b:biom.0000018375.20026.b3. [DOI] [PubMed] [Google Scholar]
- Olson JA, McDonald-Hyman C, Jameson SC, Hamilton SE. Effector-like CD8(+) T cells in the memory population mediate potent protective immunity. Immunity. 2013;38:1250–1260. doi: 10.1016/j.immuni.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plano GV, Schesser K. The Yersinia pestis type III secretion system: expression, assembly and role in the evasion of host defenses. Immunol Res. 2013;57:237–245. doi: 10.1007/s12026-013-8454-3. [DOI] [PubMed] [Google Scholar]
- Polakos NK, Cornejo JC, Murray DA, Wright KO, Treanor JJ, Crispe IN, Topham DJ, Pierce RH. Kupffer cell-dependent hepatitis occurs during influenza infection. The American journal of pathology. 2006;168:1169–1178. doi: 10.2353/ajpath.2006.050875. quiz 1404–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope C, Kim SK, Marzo A, Masopust D, Williams K, Jiang J, Shen H, Lefrancois L. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. Journal of immunology. 2001;166:3402–3409. doi: 10.4049/jimmunol.166.5.3402. [DOI] [PubMed] [Google Scholar]
- Rolan HG, Durand EA, Mecsas J. Identifying Yersinia YopH-targeted signal transduction pathways that impair neutrophil responses during in vivo murine infection. Cell host & microbe. 2013;14:306–317. doi: 10.1016/j.chom.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenheinrich M, Heine W, Schmuhl CM, Pisano F, Dersch P. Natural Killer Cells Mediate Protection against Yersinia pseudotuberculosis in the Mesenteric Lymph Nodes. PLoS One. 2015;10:e0136290. doi: 10.1371/journal.pone.0136290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songsungthong W, Higgins MC, Rolan HG, Murphy JL, Mecsas J. ROS-inhibitory activity of YopE is required for full virulence of Yersinia in mice. Cell Microbiol. 2010;12:988–1001. doi: 10.1111/j.1462-5822.2010.01448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaba FM, Kummer LW, Duso DK, Koroleva EP, Tumanov AV, Cooper AM, Bliska JB, Smiley ST, Lin JS. TNFalpha and IFNgamma but not perforin are critical for CD8 T cell-mediated protection against pulmonary Yersinia pestis infection. PLoS pathogens. 2014;10:e1004142. doi: 10.1371/journal.ppat.1004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadillo M, Corbella X, Pac V, Fernandez-Viladrich P, Pujol R. Multiple liver abscesses due to Yersinia enterocolitica discloses primary hemochromatosis: three cases reports and review. Clin Infect Dis. 1994;18:938–941. doi: 10.1093/clinids/18.6.938. [DOI] [PubMed] [Google Scholar]
- Viboud GI, Mejia E, Bliska JB. Comparison of YopE and YopT activities in counteracting host signalling responses to Yersinia pseudotuberculosis infection. Cell Microbiol. 2006;8:1504–1515. doi: 10.1111/j.1462-5822.2006.00729.x. [DOI] [PubMed] [Google Scholar]
- Von Pawel-Rammingen U, Telepnev MV, Schmidt G, Aktories K, Wolf-Watz H, Rosqvist R. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol Microbiol. 2000;36:737–748. doi: 10.1046/j.1365-2958.2000.01898.x. [DOI] [PubMed] [Google Scholar]
- Walters MS, Lane MC, Vigil PD, Smith SN, Walk ST, Mobley HL. Kinetics of uropathogenic Escherichia coli metapopulation movement during urinary tract infection. mBio. 2012:3. doi: 10.1128/mBio.00303-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark L, Fahlgren A, Fallman M. Yersinia pseudotuberculosis efficiently escapes polymorphonuclear neutrophils during early infection. Infection and immunity. 2014;82:1181–1191. doi: 10.1128/IAI.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedig CA, Kramer U, Garbom S, Wolf-Watz H, Autenrieth IB. Induction of CD8+ T cell responses by Yersinia vaccine carrier strains. Vaccine. 2005;23:4984–4998. doi: 10.1016/j.vaccine.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Ye Z, Kerschen EJ, Cohen DA, Kaplan AM, van Rooijen N, Straley SC. Gr1+ cells control growth of YopM-negative yersinia pestis during systemic plague. Infection and immunity. 2009;77:3791–3806. doi: 10.1128/IAI.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mena P, Romanov G, Lin JS, Smiley ST, Bliska JB. A protective epitope in type III effector YopE is a major CD8 T cell antigen during primary infection with Yersinia pseudotuberculosis. Infect Immun. 2012;80:206–214. doi: 10.1128/IAI.05971-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tam JW, Mena P, van der Velden AW, Bliska JB. CCR2+ Inflammatory Dendritic Cells and Translocation of Antigen by Type III Secretion Are Required for the Exceptionally Large CD8+ T Cell Response to the Protective YopE69-77 Epitope during Yersinia Infection. PLoS pathogens. 2015;11:e1005167. doi: 10.1371/journal.ppat.1005167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.