Abstract
Compared with controlled terminologies (e.g., MedDRA, CTCAE, and WHO-ART), the community-based Ontology of AEs (OAE) has many advantages in adverse event (AE) classifications. The OAE-derived Ontology of Vaccine AEs (OVAE) and Ontology of Drug Neuropathy AEs (ODNAE) serve as AE knowledge bases and support data integration and analysis. The Immune Response Gene Network Theory explains molecular mechanisms of vaccine-related AEs. The OneNet Theory of Life treats the whole process of a life of an organism as a single complex and dynamic network (i.e., OneNet). A new “OneNet effectiveness” tenet is proposed here to expand the OneNet theory. Derived from the OneNet theory, the author hypothesizes that one human uses one single genotype-rooted mechanism to respond to different vaccinations and drug treatments, and experimentally identified mechanisms are manifestations of the OneNet blueprint mechanism under specific conditions. The theories and ontologies interact together as semantic frameworks to support integrative pharmacovigilance research.
Keywords: Pharmacovigilance, bioinformatics, vaccine, drug, adverse event, Ontology of Adverse Events (OAE), Ontology of Vaccine Adverse Events (OVAE), Ontology of Drug Neuropathy Adverse Events (ODNAE), the Immune Response Gene Network Theory, the OneNet Theory of Life
Introduction
Although various types of drugs have dramatically improved public health, many drugs are associated with various types of adverse events (AEs, or called adverse reactions) including severe AEs. While most vaccine AEs are mild, many vaccines are occasionally associated with severe AEs and even death [1–4]. Every year there are over two million severe adverse drug reactions (ADRs) [5]. Many countries spend 15–20% of hospital budgets to treat drug complications [6]. ADRs cause 100,000 human deaths yearly, making ADRs the fourth leading cause of human mortality in the United States [5, 7]. To prevent ADRs, it is critical to accurately monitor and assess ADRs, identify the drug-ADR associations, and understand the fundamental ADR causal mechanisms. These become the important topics of pharmacovigilance.
Different AE case reporting systems have been developed. USA has two federal level systems to monitor AEs. The USA Food and Drug Administration (FDA) AE Reporting System (FAERS) is a database that contains information on AEs and medication error reports and is designed to support the FDA post-marketing safety surveillance program for drug and therapeutic biologic products [8]. The Vaccine AE Reporting System (VAERS) is a web-based vaccine safety surveillance program co-sponsored by the USA FDA and Centers for Disease Control and Prevention (CDC) [9, 10]. Broadly speaking, a vaccine is a special type of drug [11]. However, vaccines and chemical drugs also differ in many aspects. For example, vaccine administration is usually given to humans under healthy conditions for a preventive purpose, but chemical drugs are typically given to a sick patient for therapeutic purpose. In general, the dose, time, route, and frequency of vaccine administration are well-defined; however, for chemical drug administration, the dose, time, and frequency are often difficult to establish for patients with various conditions [11]. As a result of these differences, it is more challenging to identify the causality of ADRs since ADRs are associated with more confounding factors than vaccine AEs (VAEs). On the other hand, vaccine AEs can be spread to more of the population quickly due to its often wide use in a short period of time. To improve public health, it is important to monitor both ADRs and VAEs closely.
To support data comparison and sharing, it is important to establish and use a controlled AE terminology system. The Medical Dictionary for Regulatory Activities (MedDRA) [12, 13] is the standard terminology system used in FAERS and VAERS AE classifications in the USA. MedDRA has also been used in European Union and Japan. The Common Terminology Criteria for Adverse Events (CTCAE) is a product of the US National Cancer Institute (NCI) that provides the criteria for the standardized classification of adverse effects of drugs used in cancer therapy [14]. The World Health Organization (WHO) Adverse Reactions Terminology (WHO-ART) is a dictionary that serves as a basis for rational coding of adverse reaction terms [15]. As introduced later in this article, although MedDRA, CTCAE, and WHO-ART have been widely used, these classical AE terminology systems have many drawbacks to be addressed.
A biomedical ontology is a human- and computer-interpretable set of terms and relations (sometimes called ‘classes’ and ‘object-properties’) that represent entities in a specific biomedical domain and how they relate to each other. Ontologies have played a critical role in biomedical data and knowledge representation, exchange, integration, as well as inferring new knowledge. For example, the Gene Ontology (GO) provides controlled and standardized terms for naming different types of biological processes, cellular components, and molecular functions [16]. Creating such ontology-based annotations is highly valuable for both querying databases and analyzing high throughput data. Since its first publication in 2000 [16], GO has been cited by over 9,000 peer-reviewed publications in PubMed and over 110,000 hits in Google Scholar. The Ontology for Biomedical Investigations (OBI) [17] is a biomedical ontology co-developed by over 20 biomedical communities with the collaborative aim of representing life-science and clinical investigations. OBI has been used as the basis for developing more than 10 ontologies and for representing a wide range of investigations [18–21]. The community-based Vaccine Ontology (VO) [22–24] has been used in different applications, such as vaccine data integration [25–28] and literature mining [23, 29]. The Drug Ontology (DrON) has recently been developed to support drug classification [30]. A community-based Ontology of Adverse Events (OAE) has also been developed to support AE classification and different applications [31].
In addition to clinical AE surveillance and analysis, another important research effort is to identify the molecular mechanisms of causal AEs. Extensive research has been conducted in pharmacovigilance mechanism studies, resulting in a large amount of knowledge and data. However, it is still very difficult to integratively represent and analyze the results using bioinformatics approaches. Scientific theories may provide a better framework to support such studies. For the mechanism studies in pharmacovigilance, there have been two recently proposed theories, i.e., the “Immune Response Gene Network” theory [32, 33] and the OneNet Theory of Life (abbreviated as “the OneNet theory”) [34]. Ontology and ontology-based Semantic Web technologies may provide a robust platform for reproducible data representation, exchange, sharing, and analysis [35, 36]. It is possible to use the theories to guide ontology development, and use theory-guided ontologies to build up an integrative foundation and framework to support the representation of reported knowledge and prediction of unknown knowledge and adverse events.
The overall goal of this article is to review pharmacovigilance-related terminologies, ontologies, and theories, and propose the combination of ontologies and theories to support integrative research on clinical AE phenotypes and fundamental AE mechanisms. Specific objectives include: (i) a review of existing controlled terminologies and ontologies related to pharmacovigilance, (ii) a review of two theories applicable to understand the causal mechanisms of vaccine and drug safety, (iii) extension and applications of the OneNet theory, and (iv) proposing that the ontologies and theories can interact with each other and be applied to support integrative pharmacovigilance research.
Conventional controlled AE vocabulary systems
Major controlled AE vocabulary systems that are widely used in different settings are first introduced below.
The Medical Dictionary for Regulatory Activities (MedDRA)
MedDRA is organized with a five level hierarchy, with Lowest Level Terms (LLT) at the bottom, followed by Preferred Terms (PT), High Level Term (HLT), High Level Group Terms (HLGT), and with System Organ Class (SOC) at the top [12] (Figure 1A). The LLT level has more than 70,000 terms. Each LLT term is linked to only one PT term. Each PT has at least one LLT (itself) and may include many synonyms and lexical variants. Related PTs are grouped into HLTs and higher level HLGTs based on anatomy, pathology, etiology or function. Finally, HLGTs are grouped into SOCs based on etiology, manifestation site, purpose, or social circumstances.
Figure 1. MedDRA features.
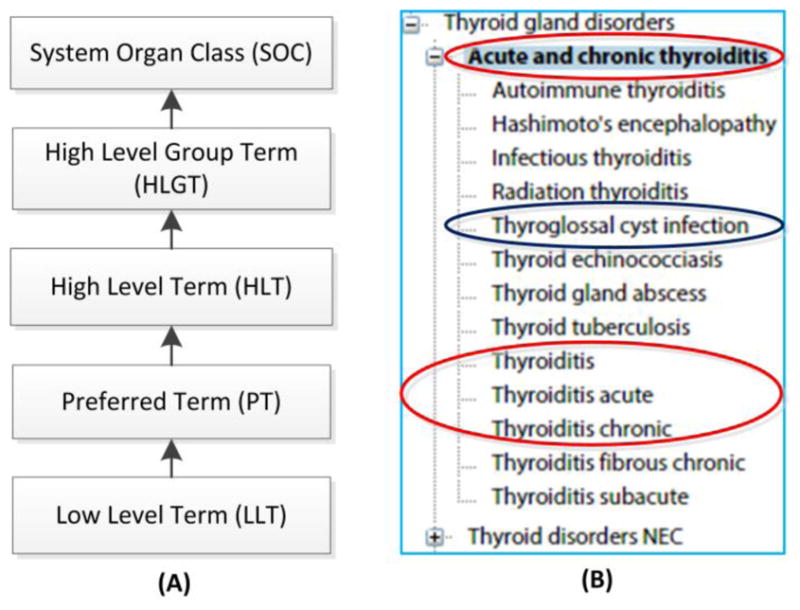
(A) The five layers of MedDRA. (B) A subset of the MedDRA hierarchy. It is a screenshot made from the MedDRA visualization in NCBO BioPortal (http://bioportal.bioontology.org/ontologies/MEDDRA). It is problematic to list a term ‘Acute and chronic thyroiditis’ as the parent term of other thyroiditis terms including ‘thyroiditis’ itself. Instead of being a subtype of ‘Acute and chronic thyroiditis’, ‘Thyroglossal cyst infection’ is an infection process that may cause thyroiditis.
MedDRA [12] has several limitations. First, the terms in MedDRA do not include term definitions, which may cause confusion and misunderstanding, especially when terms may have different meanings. Second, synonyms are often incorrectly assigned. For example, ‘swollen eyelid’ and ‘swollen eyes’ are defined incorrectly as synonyms in MedDRA. Third, hierarchical relations among MedDRA terms are often invalid. As seen in Figure 1B, the MedDRA class ‘Acute and chronic thyroiditis’ has many subclasses including ‘Thyroiditis’, ‘Thyroiditis acute’, ‘Thyroiditis chronic’, and ‘Thyroglossal cyst infection’. The class ‘Acute and chronic thyroiditis’ is problematic because it is likely difficult to find a type of thyroiditis which is both acute and chronic at the same time. Based on the structure, it would be ideal to replace the ‘Acute and chronic thyroiditis’ with ‘Thyroiditis’; and the classes ‘Thyroiditis acute’, ‘Thyroiditis chronic’ and other specific thyroiditis should be subclasses of ‘Thyroiditis’. In addition, ‘Thyroglossal cyst infection’ should not be considered as a subclass of ‘Acute and chronic thyroiditis’ or ‘thyroiditis’. Instead, ‘Thyroglossal cyst infection’ is an infection process that may be a possible cause of thyroiditis. The poorly defined hierarchy limits its ability to support valid VAE classification [3]. These issues described here often resulted in inconsistency and failure in identifying AEs [37]. In addition, the MedDRA licensing requirement prevents wide public software development and dissemination.
The Common Terminology Criteria for Adverse Events (CTCAE)
As a product of the US National Cancer Institute (NCI), CTCAE (http://evs.nci.nih.gov/ftp1/CTCAE) is a set of criteria for the standardized classification of AEs of drugs used in cancer therapy. Most US and UK drug trials encode their observations based on CTCAE. The CTCAE version 4.0 released in 2009 includes a relatively small number (790) of AE terms. This version of CTCAE is in general in harmony with the MedDRA’s terminology. Different from MedDRA, CTCAE terms are often well-defined with text definition. An AE in CTCAE may be assigned to Grades 0 through 5 that indicate different levels of clinical severity. The general guideline of assigning CTCAE severity is: Grade 0 – sign and symptom within normal limits; Grade 1 - Mild AE; Grade 2 - Moderate AE; Grade 3 - Severe AE; Grade 4: life-threatening or disabling AE; and Grade 5 - Death related to AE [14].
The WHO Adverse Reaction Terminology (WHO-ART)
WHO-ART has been developed and maintained by the Uppsala Monitoring Centre, the WHO Collaborating Centre for International Drug Monitoring [38]. The WHO-ART hierarchical structure has four levels: Included Term (IT), Preferred Terms (PT), High Level Term (HLT), and System Organ Class (SOC). The 2015Q1 version of WHO-ART includes 6,410 terms with 2,123 terms being the PTs - principal terms for describing adverse reactions [38]. Like MedDRA, WHO-ART does not provide formal definitions of terms.
In addition to MedDRA, CTCAE, and WHO-ART, there exist other controlled terminology systems for AE case reporting and analysis, including the Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT) [39], and the International Classification of Disease (ICD) [40]. There have also been efforts to compare, map, and combinatorically use different terminology systems [41–46].
Ontology-based AE representation and analysis
Compared to the conventional AE terminologies as described above, the formal ontology representation of AEs happened more recently and has offered more advantages [47–50]. A terminology is a structured collection of concepts and terms in a certain language in a specific subject field. An ontology is a formal naming and definition of the classes (or called types), properties, and the relations among the entities that really or fundamentally exist for a particular domain of discourse. An ontology provides formal and explicit models of entities in a human- and machine-readable representation, and relations among entities are logically represented as “axioms”. The conventional AE terminologies (e.g., MedDRA) are roughly equivalent to class hierarchies in ontologies. This means that a MedDRA concept “10037844” defines the class of all the individual AE instances that match the criteria for “rash”. The is_a relation relates classes between a superclass and its subclasses (e.g., members). In addition to the is_a relation, a formal ontology typically includes more sophisticated relations to present various necessary and/or sufficient conditions [29]. The logical relations between entities are more rigorously and explicitly defined in an ontology. Ontologies are developed by using ontology specific languages, including the OBO format [51] and the Web Ontology Language (OWL) [52]. The OWL description logics (DL) format supports asserted hierarchy and inferred hierarchy [29]. In addition, many ontology-based Semantic Web technologies [36] are available to support advanced data linkage, sharing, and analysis.
A formal ontology is developed by following specific principles and guidelines. The Open Biological and Biomedical Ontologies (OBO) Foundry is a collaborative initiative aimed at building up consensus-based ontology development principles and establishing ontologies following the set of principles in an evolving non-redundant suite [53]. Example OBO Foundry principles include: (1) ontologies should be open; (2) ontologies are developed in a collaborative effort; and (3) ontologies use common unambiguously defined relations [53]. These principles are widely accepted as the guidance for new ontology development.
In this section, we will introduce three ontologies that represent the general AEs and specific AEs induced by vaccines and chemical drugs. These ontologies are all developed by following OBO Foundry principles.
The Ontology of Adverse Events (OAE)
OAE represents various AEs as pathological bodily processes that occur after a medical intervention [31]. The OAE-defined adverse event does not have to be causally induced by a medical intervention, which is consistent with its definition in commonly used clinical scenarios, including their uses in VAERS and FAERS. OAE also defines the term ‘causal adverse event’ to present an adverse event is known to be causally induced by a medical intervention [31].
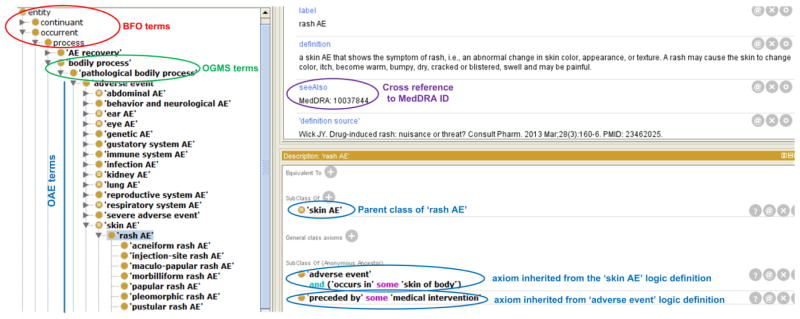
OAE is developed by importing existing ontology terms and generating new OAE-specific terms. OAE is aligned with the Basic Formal Ontology (BFO) [53, 54] (Figure 2). BFO contains two branches, continuant and occurrent [53, 54]. The continuant branch represents time-independent entities such as material entity and spatial region, and the occurrent branch represents time-related entities such as process and time interval. Using BFO as the upper level ontology makes our ontologies integrated seamlessly with >100 other ontologies that also align with BFO. To support ontology reuse and interoperability, OAE also imports terms (e.g., ‘pathological bodily process’) from over 20 existing ontologies (e.g., the Ontology of General Medical Science or OGMS) [55] (Figure 2). In addition to imported ontology terms, OAE has included over 2,600 OAE-specific terms whose ontology IDs start with “OAE_” followed with seven digital numbers. Most of these OAE-specific terms are various types of AE terms (Figure 2). Each term is well-defined, referenced, and linked to possible MedDRA ID using the annotation property seeAlso. The linkage supports cross-referencing and term mapping. As of February 16, 2016, OAE has 4,666 terms. The statistics of OAE can be found on the Ontobee [56] website: http://www.ontobee.org/ontostat/OAE.
Figure 2. OAE hierarchical structure.
This screenshot displays part of the OAE hierarchy using the Protégé-OWL editor. For each OAE term such as ‘rash AE’, clear label and definition are provided. Its cross-references to other resources (e.g., MedDRA) are cited if available. OAE is aligned with the upper level BFO and OGMS.
OAE was first demonstrated to offer better classification of AEs than MedDRA in a bioinformatics analysis of VAERS VAE case reports [3]. This study used the VAERS VAEs associated with four trivalent (killed) inactivated influenza vaccines (TIV) and FluMist (the only trivalent live attenuated influenza vaccine or LAIV). After classical statistical analysis, 48 TIV-enriched and 68 LAIV-enriched AEs were identified. The MedDRA terms of these AEs were first mapped to OAE terms. The analysis of the OAE-based hierarchies of TIV and LAIV-enriched AE terms allowed better classification of these AE terms. Different patterns associated with these VAE subsets were identified. Specifically, TIV-enriched AEs include neurological and muscular processing such as paralysis, movement disorders, and muscular weakness. In contrast, LAIV-enriched AEs include inflammatory response and respiratory system disorders. Furthermore, LAIV was found to have lower chance of inducing two severe AEs, Guillain-Barre Syndrome (GBS) and paralysis, than TIV [3]. In this study, a side-by-side comparison on how OAE, MedDRA [12], and SNOMED [39] classified TIV- and LAIV-associated VAEs provided strong empirical evidence on the clear advantages of OAE over MedDRA and SNOMED in AE classification [3].
As demonstrated below, OAE has also been used in developing domain-specific ontologies that act as vaccine or drug AE knowledge bases.
The Ontology of Vaccine Adverse Events (OVAE) as a knowledge base of vaccine AEs
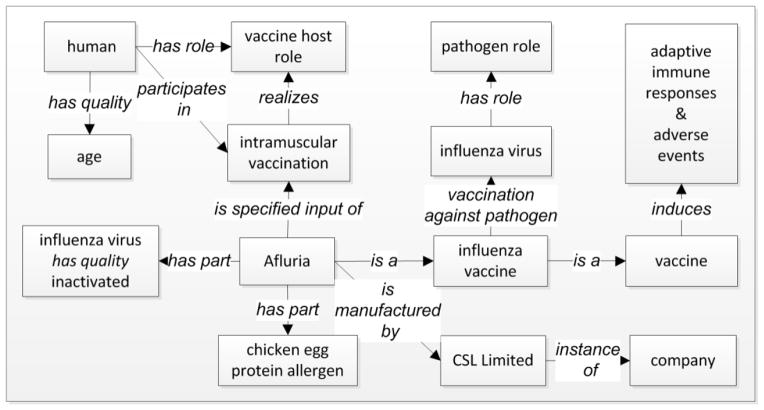
OVAE is an ontology of the AEs known to be associated with the administration of licensed vaccines [57]. OVAE is developed as an extension of OAE and VO. The community-based VO [22–24, 29] is developed to represent and integrate various vaccine and vaccine-related data, particularly those stored in VIOLIN, the largest web-based, comprehensive vaccine database and analysis system [58]. VO includes the information of all licensed human vaccines used in the USA. Figure 3 illustrates how VO logically represents the information of an influenza vaccine (Afluria), including how the vaccine links to the vaccinee, vaccination route, vaccine quality, vaccine allergen, the manufacturer, influenza virus pathogen, and vaccine-induced immune responses. VO has been used in different applications to represent various types of vaccine-related information [22, 25–27, 59]. The VO has also been used for many studies in vaccine literature mining [23, 29, 60–62]. Like OAE, VO is also aligned with the BFO, which facilitates the seamless alignment and extension of the OAE and VO for OVAE generation.
Figure 3. VO representation of vaccine knowledge.
This example ontologically represents the information about Afluria, a killed inactivated influenza vaccine manufactured by the company CSL Limited. In addition to the inactivated influenza virus, the vaccine contains chicken egg protein allergen. The vaccine is administered to humans through intramuscular route. The human’s background such as age can be linked to the vaccination event. The Afluria vaccination has the ability of inducing specific adaptive immunity against virulent influenza infection as well as various adverse events. Note that the AEs that are known to be induced by specific vaccines are primarily represented in OVAE instead of VO.
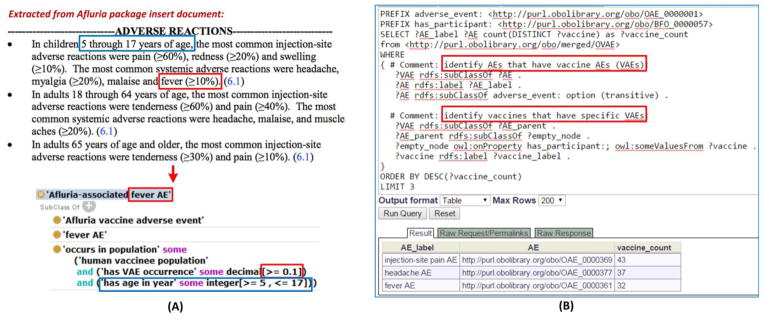
OVAE classifies AEs associated with US-licensed human vaccines [57]. OVAE imports all the licensed vaccines from the VO, related AE terms from OAE, and axioms to link vaccines and AEs with specific details. Figure 4A shows how OVAE is used to represent the fever AE associated with an influenza vaccine Afluria. It is noted that OVAE accurately represents age-specific AE occurrence rates as described in the FDA-approved vaccine package insert documents. Currently, OVAE includes all over 1,300 AEs associated with 63 US-licensed human vaccines [57]. Therefore, OVAE serves as a knowledge base of the AEs known to be associated with the administration of licensed vaccines.
Figure 4. OVAE representation and query of AEs associated with FDA licensed vaccines.
(A) OVAE representing Afluria VAEs reported in FDA vaccine package insert. The top screen shows the adverse reactions of Afluria as recorded in the FDA vaccine package insert document of Afluria. The bottom screen (extracted from a view using the Protégé OWL editor) shows how OVAE represents the >=10% occurrence of ‘Afluria-associated fever AE’ in children 5 through 17 years of age. The OVAE representation matches the information in the FDA vaccine package insert document. (B) SPARQL query of OVAE for the top 3 AEs associated with the highest numbers of vaccines. As shown in (A), ‘Afluria-associated fever AE’ is a cross product of ‘Afluria vaccine adverse event’ and ‘fever AE’, which provides the query strategy for identifying the vaccine (Afluria) and the specific AE (‘fever AE’). The query was performed using the Ontobee SPARQL web program: http://www.ontobee.org/sparql.
The information of the OVAE knowledge base can be queried using SPARQL (a recursive acronym for SPARQL Protocol and RDF Query Language). The Resource Description Framework (RDF) data model makes statements about resources in the form of subject-predicate-object expressions (i.e., triples). The RDF triples can be stored in an RDF triple store and be queried using SPARQL. A previous study has demonstrated that SPARQL is able to retrieve useful information from OVAE such as the top 10 vaccines associated with the highest numbers of VAEs and the top 10 VAEs most frequently observed among all licensed human vaccines [57]. Figure 4B demonstrates how a SPARQL query with a few lines of code can identify useful information from the OVAE stored in a RDF triple store.
The Ontology of Drug Neuropathy Adverse Events (ODNAE)
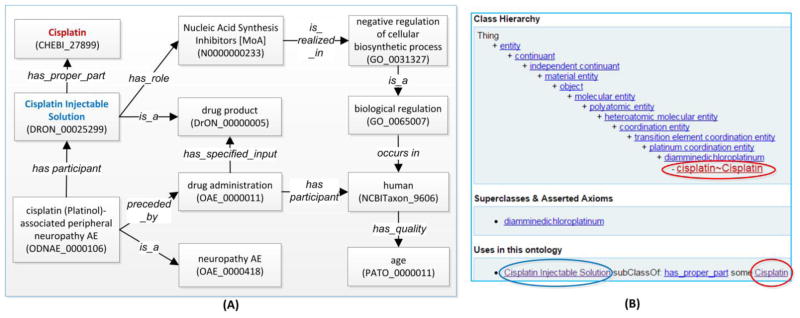
ODNAE is an ontological knowledge base that represents 215 US FDA-licensed drugs able to induce >10 neuropathy AEs and how these drugs are linked to chemicals, human qualities, drug mechanisms of actions, and biological processes [63]. Among several drug ontologies (RxNorm [64], NDF-RT [65], and DrON [30]), we selected DrON as the default ontology for representing drugs, as DrON provides mapping between drugs and ChEBI [66] chemical terms and like ODNAE and OAE, DrON is also aligned with BFO and follows OBO Foundry ontology design principles [53]. In order to enable data integration and data reuse, we added links from the DrON terms to RxNorm and NDF-RT IDs by annotation property rdfs:seeAlso in ODNAE. Figure 5 shows an example of how ODNAE represents drug-associated neuropathy AEs.
Figure 5. Illustration of ODNAE design pattern and representation.
(A) Design pattern for representing cisplatin-associated peripheral neuropathy AE. The drug product Cisplatin Injectable Solution contains the active ingredient of cisplatin. The administration of the drug on a human patient causes a peripheral neuropathy AE. During the bodily process, the drug acts as a nucleic acid synthesis inhibitor, a role that is realized in a ‘negative regulation of cellular biosynthetic process’ that occurs in the patient. Age is the patient’s quality that may affect the AE formation. In addition to ODNAE-specific terms, this model uses terms from other ontologies including ChEBI, DrON, NDF-RT, PATO, GO, and NCBITaxon. (B) Ontological hierarchy of the cisplatin chemical entity. The screenshot was obtained from the Ontobee [56].
As a knowledge base, ODNAE captures verified knowledge obtained from basic biomedical research and clinical practices. Since the ontology is machine readable, ODNAE supports neuropathy AE data representation, exchange, and integration. Furthermore, the integrated ODNAE knowledge base supports computer-assisted advanced analysis. For example, using the SPARQL queried results from the ODNAE, we performed a heatmap analysis to explore the correlation between drug molecular entities and various neuropathy AEs. Our results showed that drug-associated carbon groups, pnictogen, chalcogen and heterocyclic compounds were associated with the highest numbers of AE cases [63]. Among 215 neuropathy-inducing drugs, 127 belong to organic chemical carbon groups, which include 21 organohalogen compounds. We also identified 7 pairs of agonists and antagonists that share targets (e.g., dopamine, serotonin, and sex hormone) [63].
Theories towards understanding adverse event causality mechanisms
A fundamental approach towards understanding AE causality is to propose scientific theories and use these theories to guide our research. Two related theories and their applications in pharmacovigilance studies are introduced below.
The Immune Response Gene Network Theory
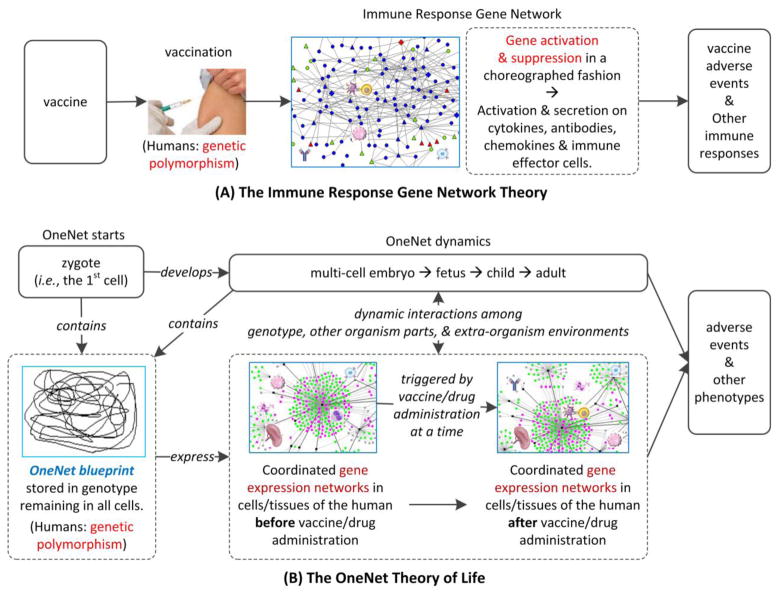
Dr. Gregory A Poland proposed an “Immune Response Gene Network Theory” with an aim to explain the immune response mechanisms induced by vaccinations [32, 33]. This theory states that the responses to a vaccine are the cumulative results of interactions driven by a host of genes and the interactions among these genes [32] (Figure 6A). The interactive and iterative activation and suppression of specific pathway genes occur in a choreographed fashion to generate a coherent immune response to a vaccine. These immune gene activities result in activation and secretion of cytokines, antibodies, chemokines and immune effector cells, leading to innate, humoral, and cell-mediated immune responses, as well as local and systemic adverse events. The basic genetic elements of the theory include key immune response genes, gene polymorphisms and epigenetic modifications, and gene–gene interactions, which may all change the outcomes of host immune responses to a vaccine [32] (Figure 6A).
Figure 6. Graphic illustration of two gene network-based theories for explaining AE mechanisms.
(A) The Immune Response Gene Network Theory. (2) The OneNet Theory of Life. The details about these two theories are described in the text.
The theory has been supported by experimentally identified associations between immune response gene polymorphisms and various antibody and cell-mediated immune responses to many viral vaccines including measles–mumps–rubella vaccine, influenza vaccines, hepatitis B and smallpox vaccines [32]. Many gene polymorphisms have been found to be directly associated with vaccine AEs [67–70]. For example, based on three independent studies, three single nucleotide polymorphisms (SNPs) in two human genes that encode for the methylenetetrahydrofolate reductase (MTHFR) and the interferon regulatory factor-1 (IRF1) were significantly associated with systemic AEs after smallpox vaccination [68].
To understand the immune response gene networks, vaccinomics can be applied that uses immunogenetics and immunogenomics methods to understand the mechanisms of heterogeneity in immune responses to vaccines [71]. Adversomics has also become an emerging field of applying omics and immunogenetics to study AEs including vaccine AEs [72]. The results gained from vaccinomics and adversomics can be used in the rational design and development of new vaccines, leading to the new golden age of “vaccine informatics” [60], “reverse vaccinology” [73], and “predictive vaccinology” [32].
The OneNet Theory of Life and its applications in pharmacovigilance
While the Immune Response Gene Network Theory explains well the mechanisms of vaccine AEs, it does not target the explanation of the AE mechanisms for chemical drug. The Immune Response Gene Network Theory does not focus on the effects of those genes and factors beyond the immune system. Therefore, there is a desire to generate a more systematical theory that traces the root causes of different types of AEs and associated factors. Based on the Immune Response Gene Network Theory and several other theories including the Evolutionary Synthesis Theory [74], the Cell Theory [75], and the “Immune Network Theory” [76], the author has recently proposed a One Network Theory of the Life of an organism (abbreviated as “OneNet theory”) [34]. As described below, the OneNet theory provides an ideal framework to study the molecular mechanisms of causal AEs induced by vaccinations and drug treatments.
The OneNet theory treats the whole process of the life of an individual organism as one single complex and dynamic network (called “OneNet”). The OneNet has four characteristics represented by four tenets:
OneNet Blueprint: The OneNet blueprint is stored in the genotype of the organism;
OneNet Start: The dynamic OneNet process starts to exist at the moment when the first cell of the organism forms;
OneNet Dynamics: The OneNet of temporal interactions between the genetic materials and their environments determines the dynamic phenotype (e.g., morphology, behavior, survival, and reproduction) of the life.
OneNet Effectiveness: An organism with its expressed OneNet profile more adaptive to an environment is advantaged to survive, replicate, and live a better life in the environment.
The first three characteristics were defined and discussed in the original paper [34]. The last OneNet characteristic is newly added to provide a mechanism to measure the effect (or outcome) of the complex OneNet process.
The OneNet theory targets the systematic representation and analysis of the life of one organism (e.g., a human being), with a special focus on the dynamic interactions among genotype, environment, and phenotype along the life process. The theory explores the root cause of an organism’s phenotypes (e.g., AEs after vaccine and drug administrations) (Figure 6B). The genotype of a human is rooted at the genome of the fertilized cell (i.e., zygote) that involves a sperm fusing with an ovum [74]. The zygote is the first cell that contains the intact and complete genotype information of the human. The genotype is the blueprint for the development of an organism [77]. Extending this fact, the genotype would also include the blueprint information of the OneNet mechanism on how the organism interacts with various environments (e.g., drug administration). The life of an organism with different stages is the manifestation of the OneNet blueprint of the organism. Such a OneNet blueprint manifestation is represented by various phenotypes and underlying gene expression profiles out of the dynamic interactions between the genotype and its environments. Different people may have different genotypes (including gene polymorphism) and encountered environments that include intra- and extra-organism environments. The intra-organism environments of the genetic materials include those inside the cells but outside the genetic materials and those outside the cells but inside the human body [34]. All these environmental factors may change the gene expression profiles and phenotypes. Therefore, given various environmental conditions, different people may demonstrate different phenotypes including various AEs following exposure to a vaccine or a drug (Figure 6B).
The OneNet effectiveness tenet is newly added to the OneNet theory and first described in this article. There are different meanings of the terms effectiveness, efficacy, and efficiency in different settings [78–81]. For example, in medicine, effectiveness relates to how well a treatment works in practice, while efficacy measures how well it works in a well-controlled clinical trials or laboratory studies [79, 82, 83]. In physics, an effective theory is a framework intended to explain an observed effect without claiming that the theory correctly models the underlying unobserved processes [80]. In the OneNet theory, the author adopts the definitions of the ontology reference model developed for the uncertainty representation and reasoning evaluation framework (URREF) [81]. In the URREF ontology, the “effectiveness” relates to a system’s capability to produce an effect, and effectiveness includes: (i) “efficiency”: doing a thing in the most economical way, (ii) “efficacy”: getting a thing done (i.e., meeting a target or desire); and (iii) “correctness”: doing a “right” thing, i.e., setting a right target to achieve an overall goal (the effect). Similarly, the OneNet theory uses the term “OneNet effectiveness” to represent the capability of an organism to produce an effect, including the survival, replication, and quality of life in a specific environment. The OneNet effectiveness tenet states that an organism with its own expressed OneNet profile more adaptive to its environment is advantaged (i.e., more effective) to survive, replicate, and live a better life in the environment. Similar to the composition of the effectiveness as defined in the URREF ontology [81], OneNet effectiveness also includes different levels, i.e., OneNet efficiency, efficacy, and correctness.
Both the Immune Response Gene Network Theory and the OneNet theory facilitate the understanding and study of causal pharmacovigilance mechanisms. The Immune Response Gene Network Theory provides a more specific network scenario in the domain of vaccine-induced AEs. This theory informs the importance of studying different immune response gene networks and gene polymorphism to understand fundamental vaccine AE mechanisms. The Immune Response Gene Network Theory fits well with the OneNet theory. The OneNet theory has a broader scope and can be used to study the mechanisms of AEs induced by both vaccines and drugs. In addition, the OneNet theory goes further to lay out the root cause genotype-guided molecular networks and the genotype-environment-phenotype network dynamics where the environment has a broad scope that includes vaccination, drug administration, infection, stress, etc. In a complex dynamic system, prior states will have an influence on present states [84]. Similarly, in the complex OneNet system, the genotype and a patient’s conditions (e.g., age, gender, and previous exposure to the same vaccine or drug) before and during vaccine/drug administration would affect the outcomes of vaccine/drug AE processes.
Ontology-based and theory-guided integrative representation and analysis of AE interaction networks
The standard controlled terminologies, pharmacovigilance-related ontologies, and two network theories are reviewed above. The author has also proposed a new tenet in the OneNet theory to cover the outcome aspect of the OneNet process. Now, two critical questions are how to the ontologies and the theories interact with each other, and how the possible interaction between the theories and ontologies can better support pharmacovigilance research. These two questions are addressed below.
The two network theories and ontologies can closely interact with each other
As described above, the two network theories (the Immune Response Gene Network Theory and the OneNet theory) provide frameworks to study causal AE mechanisms. An ever increasing amount of data has been generated out of intensive research in the pharmacovigilance studies. Although the theories help mechanistic understanding, it has been a huge challenge to consistently and logically represent and analyze the obviously big data associated with the AE complex systems. Ontologies and ontology-based Semantic Web methods naturally provide tools to perform such tasks.
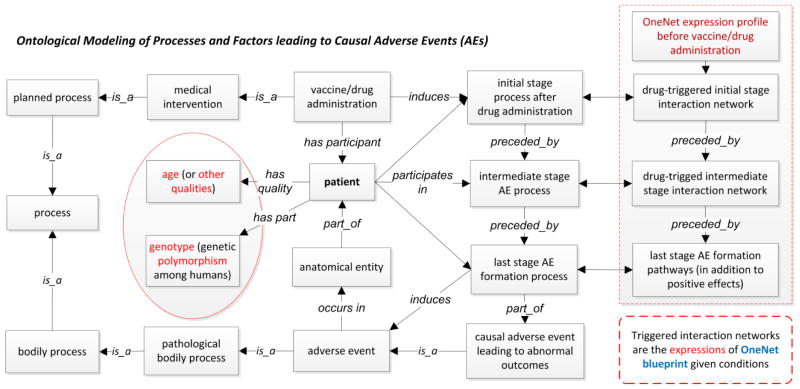
Ontologies support integrative representation of AEs and the processes leading to AEs. Figure 7 outlines an OAE-based ontological model of AE-related processes and related factors. After a vaccine/drug administration, three stages of biological processes occur in the patient (Figure 7) [31]. The initial stage processes involve the vaccine/drug entry and initial host responses. The intermediate stage processes include series of intermediate smaller processes, some eventually leading to positive preventative or therapeutic effect, some leading to noises, and some leading to AEs. The last stage processes are the execution stage processes leading to pathological outcomes [31]. These processes occur in different anatomic locations. Many factors, such as age and gene polymorphism, likely affect the final outcome. In addition to the OAE-based model (Figure 7), the OVAE model (Figure 4) logically represents the knowledge of vaccines and vaccine-induced AEs, and the ODNAE model (Figure 5) ontologically links drugs, drug chemical ingredients, AEs, mechanisms of actions, and biological processes. All the ontologies and models described above cover different granularity of details. A federation of these specific models provides a more comprehensive picture of vaccine- and drug-induced AE mechanisms.
Figure 7. Integrative ontological modeling of the processes and factors leading to adverse events induced by a vaccine or a drug.
The OAE-based model is extended with the knowledge from the OneNet theory. The model logically links the patient’s genotype, qualities (e.g., age), and gene expression profiles before a vaccine/drug administration to the adverse event. See the text for more details.
Ontologies can also support integrative representation of AE-related genetic interaction networks. OVAE and ODNAE can be expanded to represent gene interaction pathways and networks leading to AEs induced by vaccines and drugs. Furthermore, we can use ontologies to represent various human interaction networks and identify those that are relevant to pharmacovigilance. Hundreds of interaction/pathway/network databases exist [85]. For example, Reactome [86], KEGG [87], BioCyc [88], and BioCarta [89] are interaction and pathway databases that cover various biological domains. The Pharmacogenetics Knowledge Base (PharmGKB) contains genomic, phenotype and clinical information collected from various pharmacogenetic studies [90]. Unfortunately, the knowledge in these databases is often overlapped but disintegrated. Extended from a general species-neutral framework of the Interaction Network Ontology (INO) [91], the ongoing Human Interaction Network Ontology (HINO) project is designed to integrate data from different interaction pathway databases [92]. After an expected comprehensive HINO is generated, AE-related genetic interactions and pathways can be extracted from HINO and exported to OVAE and ODNAE to enrich their representation of integrated AE mechanisms.
Since the OneNet theory emphasizes the dynamic interactions among genotype, environment, and phenotype along the life process, it is necessary and critical to ontologically model and represent various environment factors and their interactions with genotype and phenotype. After drug administration, multiple and complex environmental factors may play important roles in the mechanism of AEs through interactions with genotype. There exist several ontologies, including Environment Ontology (EnVO) [93] and the Experimental Factor Ontology (EFO) [94], which represent various environmental and experimental factors. EnVO is a community-led, open ontology for specifying a wide range of environments relevant to life science disciplines [93]. Driven by the critical needs in annotating biological variables in gene expression studies, EFO is developed as an application ontology to represent various experimental factors [94]. In addition, the Gene Ontology (GO) includes a branch of Cellular Components [16], which lays out various levels of cellular components where intracellular molecular interactions may occur. To represent specific interactions between genotypes, environments, and phenotypes, related environmental and experimental factors can first be imported from existing ontologies (e.g., EnVO, EFO, and GO) to a domain ontology (e.g., ODNAE and HINO), and specific interactions can then be represented by building up ontological axioms.
In summary, the network theories and ontologies can closely interact with each other. First, the network theories provide the framework on how to link clinical AE phenotypes and internal causal mechanisms, guide the contents and logics of the entities to be represented in ontologies in pharmacovigilance studies. For example, the integrative ontological modeling of the processes and factors leading to vaccine/drug AEs (Figure 7) is generated based on the guidance of the OneNet theory. Without the theory, it is easy to miss the whole picture of what to include in the ontological model. On the other hand, the ontologies provide standard representations to support the standard description on the two network theories. Without a standard ontological representation, the network theories are hard to describe and be applied in various situations. The integrative relations between the ontologies and theories can be further illustrated when they are combined to analyze fundamental pharmacovigilance questions as exampled below.
Combined theories and ontologies support integrative pharmacovigilance research
Is that possible that the combination of the theories and ontologies can synergistically support integrative pharmacovigilance research? As exemplified below, the author argues that the new theories can be used to generate new and novel hypotheses; and since the large scope and complexity of the hypotheses, the final addressing of the theory-derived hypotheses will require the support of ontologies.
Derived from the OneNet theory, the author hypothesizes that one human host uses one single mechanism to respond to different vaccinations and drug treatments. This hypothesis is based on the fact that one human being has one genotype and a single OneNet blueprint; therefore, the same person would respond to different medical interventions using the same genotype-rooted OneNet mechanism. On the other hand, this hypothesis appears against the obvious experimental observations that human uses different mechanisms against different vaccinations and drug treatments. However, there is no conflict since these are essentially two different types of mechanisms. The OneNet mechanism represents the blueprint design of the sum of interactions, pathways, and networks that occur in the same human given any possible conditions. The experimentally identified mechanisms are those that occur in reality under specific conditions, and they are the manifestations (or expressions) of the OneNet blueprint with specific gene expression and molecular interaction profiles. As a multicellular organism, a human includes trillions of cells organized into tissues, organs, and organ systems. All the human cells contain the same or essentially the same genotype as seen in the zygote. Extending the above hypothesis, the author further hypothesizes that all cells with the same genotype in a human share the same OneNet blueprint mechanism although the blueprint manifestations in specific cells most likely differ, and the OneNet blueprint includes mechanistic design for how different cells interact with each other to form the physical human body and collaboratively respond to various environmental factors. Based on such an OneNet-derived framework, one ultimate goal of our research is to fully identify the comprehensive OneNet blueprint mechanism and use it to predict specific OneNet manifestation profiles given different conditions such as a vaccine or drug administration.
While the above hypothesis targets on each person having a single genotype-rooted OneNet blueprint mechanism, the OneNet mechanisms from different people may share many biological interactions, pathways, and networks. Each person has a single OneNet mechanism due to its own original initiation from one single cell and all cells sharing the same genotype source. Different people originate from different genotypes (except those in monozygotic twins) in different cells, and thus have essentially different OneNet blueprint mechanisms. However, all humans share human-specific genotype contents, suggesting that different people have many shared molecular interactions and pathways. In microbiology, a “pan-genome” represents the union of the gene sets of all the strains of a clade (e.g., species) [95, 96]. Such a pan-genome is composed of a “core genome” containing genes present in all strains, a “dispensable genome” containing genes present in two or more strains, and “unique genes” specific to single strains. Similarly, the human pan-genome is the non-redundant collection of all human DNA sequence present in the entire human population. A recent human pan-genome study has found a large amount of novel sequences that are both population and individual specific [97]. Genetic sequence variations also exist for the same genes and non-coding genomic regions among different human genomes. In molecular biology, an interactome is the whole set of molecular interactions that occur in the cells of an organism (e.g., human) [98–100]. Inspired by the pan-genome and interactome concepts, the author envisions that the OneNet blueprint for a specific organism (e.g., human) population includes a “pan-interactome”, and the human pan-interactome would be composed of a “core interactome” containing interactions present in all humans, a “dispensable interactome” containing interactions present in a population of humans, and “unique interactome” specific to human individuals. The OneNet blueprint mechanism for thewhole human population can be considered as the pan-interactome given all possible environmental conditions, and different interactions then form pathways and networks. The OneNet blueprint mechanism for a person would then include a pan-interactome subset that is specific for this person. This strategy would differentiate and integrate different people’s OneNet mechanisms. One future task is to identify the shared and differential mechanisms among different people, which would facilitate basic mechanism studies and rational therapeutic design in both general and personalized medicines.
Since the above OneNet-derived hypotheses cover a huge and complex scope of the dynamic interactions between genotypes, environments, and phenotypes in one person or different people, it is difficult to study and address these hypotheses for real specific applications without theory-guided ontological representation and knowledge organization. A similar situation occurred when a small number of chemical elements were discovered in early 1800’s. When Dmitri Mendeleev published the first widely recognized Periodic Table of Chemical Elements in 1869, there had been only 63 elements discovered. The generation of such a periodic table led to correct organization of published elements and prediction of new elements and their attributes. Similar to the Periodic Table of Chemical Elements that well-organizes chemical elements, ontologies will allow semantic organization of complex biological entities and interaction networks among these entities. The OneNet theory provides a framework for rational ontology design for modeling and representing the complex biological networks. While the Periodic Table of Chemical Elements is relatively simple, the Table had been used as a very powerful platform for predicting new chemical elements. With much more complex biological ontologies, it is much more difficult to generate new hypotheses. Therefore, new ontology-based algorithms and tools will need to be developed and evaluated for real case studies. Many advanced computational modeling methods and tools for complex systems have been developed [84, 101, 102], which can be combined with existing theories and ontology representations to better understand complex organism systems including pharmacovigilance, leading to precision medicine.
Discussion and Future Perspectives
The contributions of this paper are multiple. First, this paper reviews standard and classical AE terminology systems (including MedDRA, CTCAE, and WHO-ART), and three AE-related ontologies (i.e., OAE, OVAE, and ODNAE). Second, this paper reviews two network theories, (i.e., the Immune Response Gene Network Theory and the OneNet theory) and proposes to use the two theories as the integrative framework to systematically explain and analyze complex network mechanisms in pharmacovigilance. Third, the author proposes and explains a new OneNet effectiveness tenet to cover the outcome of the whole OneNet process of an organism’s life. With this newly added tenet, the OneNet is characterized by four features: blueprint, start, dynamics and effectiveness. Fourth, the author argues that the ontologies and theories can closely interact with each other, and the combination of the theories and ontologies can be used to support integrative pharmacovigilance research. Fifth, to better illustration of such integrative research, the author proposes a novel OneNet-derived hypothesis that differentiates and integrates the single genotype-rooted OneNet blueprint mechanism in a human and various expressions of the blueprint given specific conditions, extends the hypothesis with new hypotheses, and proposes ontology-based systems biology methods to study these hypotheses.
While the standard AE terminologies are widely used to support the evaluation of clinical trials, their shortcomings have promoted the development of new ontologies related to AE classification and analysis. The classical terminologies classify specific AEs and higher level AE types with specified hierarchical structures. However, these terminology systems have their internal drawbacks such as the lack of robust hierarchical structure and logical definitions, and their inability to semantically link to patients’ attributes. As noted by FDA scientists in a prominent Opinion article published in early 2012 [49], although MedDRA has served the public health well, to continue to advance the drug safety, adverse drug reaction (ADR) classification must “evolve beyond relatively simple vocabularies and toward being the knowledge framework for a systematic organization of all ADR-related data and information”, and such evolvement “promises to enable new discoveries, inform researchers and regulators, and create new biosurveillance capabilities”. The OAE and OAE-based OVAE/ODNAE provide such knowledge frameworks for systematic organization, representation, and linkage of AE-related information, including vaccines/drugs, patient, patient quality, anatomic locations, biological mechanisms, biological processes, and gene responses. As demonstrated in the OVAE and ODNAE data analyses [57, 63], such ontology knowledge base representations also support data query and allow the statistical correlations between different factors and meaningful inference from one to the other.
Extending from the ontological knowledge frameworks, the author proposes to use and integrate complex network theories to further study scientific questions in pharmacovigilance. To systematically study molecular mechanisms leading to AEs, two theories are reviewed in this article. The Immune Response Gene Network theory emphasizes the importance of gene-driven activation of immune response pathways, and can be used to explain vaccine AE causality and study gene-AE associations. The OneNet theory focuses more on the root case analysis of the whole life process, and is applicable for studying the mechanisms of vaccine/drug AEs and linking them to different conditions (e.g., genotype, age, and history). The author also proposes to use ontologies to represent and analyze various interaction networks covered by these theories. Furthermore, to demonstrate the usage of the network theories, the author derived a hypothesis on the unified mechanism of each person (and even different people) to respond to different medical treatments. This newly proposed hypothesis differentiates one single OneNet blueprint design in one person and various OneNet manifestation (or expression) profiles given different conditions. Based on the hypothesis, the author proposes an integrative framework of representing the comprehensive OneNet blueprint mechanism using ontology, and predicting OneNet manifestations in specific people given different conditions such as vaccination and drug administration. While such a hypothesis is ambitious to tackle, integrated ontological and computational methodologies provide a feasible strategy to address and study the hypothesis.
Although this article clearly demonstrates the critical role of ontologies in systematic pharmacovigilance study, its efficient usage meets the bottleneck of extensive time-consuming efforts needed for ontology development. To overcome the bottleneck, many software programs have been developed, for example, the Protégé OWL editor [103] and the BioPortal web ontology repository system [104]. In the past years, the author’s laboratory has also developed a collection of web-based “Ontoanimal” tools including OntoFox [105], Ontodog [106], Ontorat [107], Ontobee [56], and Ontobeep [108]. Each tool has specific functions; together, these tools are able to extract ontology subsets, provide ontology community views, generate and edit ontology terms, query and visualize ontology terms, provide statistics of ontologies, and compare ontologies. These “Ontoanimal” tools have been widely used to facilitate the development of various ontologies including the OAE [31], OVAE [57], and ODNAE [63].
Although extensive research has been done, there is still a long way to go to fully understand how a human develops and responds to a vaccine/drug administration. However, the author is optimistic that the development of well-organized, theory-guided ontologies will gradually and firmly increase the semantic representation and organization of complex entities and interaction networks. Furthermore, the theories will be developed and used to generate new and novel hypotheses. Such theoretic and ontological frameworks will lay out a solid foundation for the development of more advanced computational algorithms and the generation of new hypotheses, leading to integrated pharmacovigilance research that better bridges and understands clinical AE phenotypes and fundamental biological mechanisms.
Acknowledgments
The development of OAE, VO, INO, and HINO was supported by NIH-NIAID grant R01AI081062. The author appreciates the editorial review by Ms. Rebecca Racz, Doctor of Pharmacy Candidate (class of 2016) at the University of Michigan, Ann Arbor, Michigan, USA. The author also appreciates three anonymous reviewers and the paper editor for their insightful comments.
Abbreviations
- ADR
Adverse Drug Reaction
- AE
Adverse Event
- BFO
Basic Formal Ontology
- ChEBI
Chemical Entities of Biological Interest
- CTCAE
Common Terminology Criteria for Adverse Events
- DrON
Drug Ontology
- FDA
Food and Drug Administration
- GO
Gene Ontology
- HINO
Human Interaction Network Ontology
- INO
Interaction Network Ontology
- LAIV
Live Attenuated Influenza Vaccine
- MedDRA
Medical Dictionary for Regulatory Activities
- NCBITaxon
National Center for Biotechnology Information Taxonomy Ontology
- NCBO
National Center for Biomedical Ontology
- NDF-RT
National Drug File - Reference Terminology
- OBO
Open Biological and Biomedical Ontologies
- ODNAE
Ontology of Drug Neuropathy Adverse Events
- OGMS
Ontology for General Medical Science
- OWL
Web Ontology Language
- PATO
Phenotypic Quality Ontology
- RDF
Resource Description Framework
- SNP
Single Nucleotide Polymorphism
- SPARQL
SPARQL Protocol and RDF Query Language
- TIV
Trivalent Inactivated Influenza Vaccines
- VAE
Vaccine Adverse Event
- VO
Vaccine Ontology
- WHO
World Health Organization
- WHO-ART
WHO Adverse Reaction Terminology
Footnotes
Competing interests
The author declares no competing interests.
References
- 1.Silvers LE, Ellenberg SS, Wise RP, Varricchio FE, Mootrey GT, Salive ME. The epidemiology of fatalities reported to the vaccine adverse event reporting system 1990–1997. Pharmacoepidemiology and drug safety. 2001;10(4):279–285. doi: 10.1002/pds.619. [DOI] [PubMed] [Google Scholar]
- 2.Zhou W, Pool V, Iskander JK, English-Bullard R, Ball R, Wise RP, Haber P, Pless RP, Mootrey G, Ellenberg SS, et al. Surveillance for safety after immunization: Vaccine Adverse Event Reporting System (VAERS)--United States, 1991–2001. MMWR Surveill Summ. 2003;52(1):1–24. [PubMed] [Google Scholar]
- 3.Sarntivijai S, Xiang Z, Shedden KA, Markel H, Omenn GS, Athey BD, He Y. Ontology-based combinatorial comparative analysis of adverse events associated with killed and live influenza vaccines. PloS one. 2012;7(11):e49941. doi: 10.1371/journal.pone.0049941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salmon DA, Proschan M, Forshee R, Gargiullo P, Bleser W, Burwen DR, Cunningham F, Garman P, Greene SK, Lee GM, et al. Association between Guillain-Barre syndrome and influenza A (H1N1) 2009 monovalent inactivated vaccines in the USA: a meta-analysis. Lancet. 2013;381(9876):1461–1468. doi: 10.1016/S0140-6736(12)62189-8. [DOI] [PubMed] [Google Scholar]
- 5.Why Learn about Adverse Drug Reactions (ADR)? http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/DrugInteractionsLabeling/ucm114848.htm.
- 6.Qing-ping S, Xiao-dong J, Feng D, Yan L, Mei-ling Y, Jin-xiu Z, Shu-qiang Z. Consequences, measurement, and evaluation of the costs associated with adverse drug reactions among hospitalized patients in China. BMC health services research. 2014;14:73. doi: 10.1186/1472-6963-14-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. Jama. 1998;279(15):1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 8. [accessed on May 22 2015];FDA Adverse Event Reporting System (FAERS) [FDA Adverse Event Reporting System (FAERS) http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/default.htm.
- 9.Chen RT, Rastogi SC, Mullen JR, Hayes SW, Cochi SL, Donlon JA, Wassilak SG. The Vaccine Adverse Event Reporting System (VAERS) Vaccine. 1994;12(6):542–550. doi: 10.1016/0264-410x(94)90315-8. [DOI] [PubMed] [Google Scholar]
- 10.Varricchio F, Iskander J, Destefano F, Ball R, Pless R, Braun MM, Chen RT. Understanding vaccine safety information from the Vaccine Adverse Event Reporting System. The Pediatric infectious disease journal. 2004;23(4):287–294. doi: 10.1097/00006454-200404000-00002. [DOI] [PubMed] [Google Scholar]
- 11.He Y, Toldo L, Burns G, Tao C, Abernethy DR. A 2012 Workshop: Vaccine and Drug Ontology in the Study of Mechanism and Effect (VDOSME 2012) Journal of biomedical semantics. 2012;3(1):12. doi: 10.1186/2041-1480-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA) Drug safety. 1999;20(2):109–117. doi: 10.2165/00002018-199920020-00002. [DOI] [PubMed] [Google Scholar]
- 13.Introductory Guide MedDRA Version 18.0. http://www.meddra.org/sites/default/files/guidance/file/intguide_18_0_english.pdf.
- 14. [access on October 7, 2015];The Common Terminology Criteria for Adverse Events (CTCAE) http://evs.nci.nih.gov/ftp1/CTCAE/About.html.
- 15.The WHO Adverse Reaction Terminology – WHO-ART. https://www.umc-products.com/graphics/28010.pdf.
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinkman RR, Courtot M, Derom D, Fostel JM, He Y, Lord P, Malone J, Parkinson H, Peters B, Rocca-Serra P, et al. Modeling biomedical experimental processes with OBI. Journal of biomedical semantics. 2010;1(Suppl 1):S7. doi: 10.1186/2041-1480-1-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng J, Manduchi E, Stoeckert CJ., Jr Development of an Application Ontology for Beta Cell Genomics based On the Ontology for Biomedical Investigations. The 4th International Conference on Biomedical Ontology (ICBO-2013); July 7–12 2013; Montreal, Qc, Canada. 2013. pp. 62–67. CEUR Workshop Proceedings. [Google Scholar]
- 19.Brinkman RR, Courtot M, Derom D, Fostel J, He Y, Lord P, Malone J, Parkinson H, Peters B, Rocca-Serra P, et al. Modeling biomedical experimental processes with OBI. Journal of Biomedical Semantics. 2010 Jun 22;21(Suppl 21):S27. doi: 10.1186/2041-1480-1181-S1181-S1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dugan VG, Emrich SJ, Giraldo-Calderon GI, Harb OS, Newman RM, Pickett BE, Schriml LM, Stockwell TB, Stoeckert CJ, Jr, Sullivan DE, et al. Standardized metadata for human pathogen/vector genomic sequences. PloS one. 2014;9(6):e99979. doi: 10.1371/journal.pone.0099979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarntivijai S, Lin Y, Xiang Z, Meehan TF, Diehl AD, Vempati UD, Schürer TC, Pang C, Malone J, Parkinson H, et al. CLO: The Cell Line Ontology. Journal of biomedical semantics. 2014;5:37. doi: 10.1186/2041-1480-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Y, Cowell L, Diehl AD, Mobley HL, Peters B, Ruttenberg A, Scheuermann RH, Brinkman RR, Courtot M, Mungall C, et al. VO: Vaccine Ontology. Nature Precedings; The 1st International Conference on Biomedical Ontology (ICBO-2009); July 24–26 2009; Buffalo, NY, USA. 2009. [Google Scholar]
- 23.Ozgur A, Xiang Z, Radev DR, He Y. Mining of vaccine-associated IFN-gamma gene interaction networks using the Vaccine Ontology. Journal of biomedical semantics. 2011;2(Suppl 2):S8. doi: 10.1186/2041-1480-2-S2-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y, He Y. Ontology representation and analysis of vaccine formulation and administration and their effects on vaccine immune responses. Journal of biomedical semantics. 2012;3(1):17. doi: 10.1186/2041-1480-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang B, Sayers S, Xiang Z, He Y. Protegen: a web-based protective antigen database and analysis system. Nucleic acids research. 2011;39(Database issue):D1073–1078. doi: 10.1093/nar/gkq944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayers S, Ulysse G, Xiang Z, He Y. Vaxjo: a web-based vaccine adjuvant database and its application for analysis of vaccine adjuvants and their uses in vaccine development. J Biomed Biotechnol. 2012;2012:831486. doi: 10.1155/2012/831486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Racz R, Chung M, Xiang Z, He Y. Systematic annotation and analysis of “virmugens” - virulence factors whose mutants can be used as live attenuated vaccines. Vaccine. 2013;31(5):797–805. doi: 10.1016/j.vaccine.2012.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, Racz R, Sayers S, Lin Y, Todd TE, Hur J, Li X, Patel M, Zhao B, Chung M, et al. Updates on the Web-based VIOLIN Vaccine Database and Analysis System. Nucl Acids Res. 2013:1–9. doi: 10.1093/nar/gkt1133. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hur J, Xiang Z, Feldman EL, He Y. Ontology-based Brucella vaccine literature indexing and systematic analysis of gene-vaccine association network. BMC immunology. 2011;12:49. doi: 10.1186/1471-2172-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanna J, Joseph E, Brochhausen M, Hogan WR. Building a drug ontology based on RxNorm and other sources. Journal of biomedical semantics. 2013;4(1):44. doi: 10.1186/2041-1480-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Y, Sarntivijai S, Lin Y, Xiang Z, Guo A, Zhang S, Jagannathan D, Toldo L, Tao C, Smith B. OAE: The Ontology of Adverse Events. Journal of biomedical semantics. 2014;5:29. doi: 10.1186/2041-1480-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poland GA, Ovsyannikova IG, Jacobson RM, Smith DI. Heterogeneity in vaccine immune response: the role of immunogenetics and the emerging field of vaccinomics. Clin Pharmacol Ther. 2007;82(6):653–664. doi: 10.1038/sj.clpt.6100415. [DOI] [PubMed] [Google Scholar]
- 33.Poland GA, Ovsyannikova IG, Jacobson RM. Application of pharmacogenomics to vaccines. Pharmacogenomics. 2009;10(5):837–852. doi: 10.2217/PGS.09.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y. Ontology-supported research on vaccine efficacy, safety and integrative biological networks. Expert Rev Vaccines. 2014;13(7):825–841. doi: 10.1586/14760584.2014.923762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deans AR, Lewis SE, Huala E, Anzaldo SS, Ashburner M, Balhoff JP, Blackburn DC, Blake JA, Burleigh JG, Chanet B, et al. Finding our way through phenotypes. PLoS biology. 2015;13(1):e1002033. doi: 10.1371/journal.pbio.1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berners-Lee T, Hendler J, Lassila O. The Semantic Web. Scientific American. 2001:29–37. [Google Scholar]
- 37.Schroll JB, Maund E, Gotzsche PC. Challenges in coding adverse events in clinical trials: a systematic review. PloS one. 2012;7(7):e41174. doi: 10.1371/journal.pone.0041174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The Uppsala Monitoring Centre. The WHO Adverse Reaction Terminology – WHO-ART, Terminology for coding clinical information in relation to drug therapy. 2015 http://www.umc-products.com/graphics/28010.pdf.
- 39.Brown SH, Elkin PL, Bauer BA, Wahner-Roedler D, Husser CS, Temesgen Z, Hardenbrook SP, Fielstein EM, Rosenbloom ST. SNOMED CT: utility for a general medical evaluation template. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium; 2006; pp. 101–105. [PMC free article] [PubMed] [Google Scholar]
- 40.Mullooly JP, Donahue JG, DeStefano F, Baggs J, Eriksen E Group VSDDQW. Predictive value of ICD-9-CM codes used in vaccine safety research. Methods of information in medicine. 2008;47(4):328–335. [PubMed] [Google Scholar]
- 41.Iavindrasana J, Bousquet C, Degoulet P, Jaulent MC. Clustering WHO-ART terms using semantic distance and machine learning algorithms. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium; 2006; pp. 369–373. [PMC free article] [PubMed] [Google Scholar]
- 42.Brown EG. Effects of coding dictionary on signal generation: a consideration of use of MedDRA compared with WHO-ART. Drug safety. 2002;25(6):445–452. doi: 10.2165/00002018-200225060-00009. [DOI] [PubMed] [Google Scholar]
- 43.Declerck G, Hussain S, Daniel C, Yuksel M, Laleci GB, Twagirumukiza M, Jaulent MC. Bridging data models and terminologies to support adverse drug event reporting using EHR data. Methods of information in medicine. 2015;54(1):24–31. doi: 10.3414/ME13-02-0025. [DOI] [PubMed] [Google Scholar]
- 44.Reich C, Ryan PB, Stang PE, Rocca M. Evaluation of alternative standardized terminologies for medical conditions within a network of observational healthcare databases. Journal of biomedical informatics. 2012;45(4):689–696. doi: 10.1016/j.jbi.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Nadkarni PM, Darer JD. Determining correspondences between high-frequency MedDRA concepts and SNOMED: a case study. BMC medical informatics and decision making. 2010;10:66. doi: 10.1186/1472-6947-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bodenreider O. Using SNOMED CT in combination with MedDRA for reporting signal detection and adverse drug reactions reporting. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium. 2009;2009:45–49. [PMC free article] [PubMed] [Google Scholar]
- 47.Van Rees R. Clarity in the usage of the terms ontology, taxonomy and classification. CIB REPORT. 2003;284(432):1–8. [Google Scholar]
- 48.Ceusters W, Smith B, Flanagan J. Towards an Electronic Patient Record (TEPR 2003) Boston, MA: 2003. Ontology and medical terminology: Why description logics are not enough. [Google Scholar]
- 49.Zhichkin PE, Athey BD, Avigan MI, Abernethy DR. Needs for an expanded ontology-based classification of adverse drug reactions and related mechanisms. Clin Pharmacol Ther. 2012;91(6):963–965. doi: 10.1038/clpt.2012.41. [DOI] [PubMed] [Google Scholar]
- 50.Schulz S, Balkanyi L, Cornet R, Bodenreider O. From Concept Representations to Ontologies: A Paradigm Shift in Health Informatics? Healthcare informatics research. 2013;19(4):235–242. doi: 10.4258/hir.2013.19.4.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tirmizi SH, Aitken S, Moreira DA, Mungall C, Sequeda J, Shah NH, Miranker DP. Mapping between the OBO and OWL ontology languages. Journal of biomedical semantics. 2011;2(Suppl 1):S3. doi: 10.1186/2041-1480-2-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.W3C. [Accessed on March 20160308];OWL 2 Web Ontology Language document overview (Second Edition) 2012 https://www.w3.org/TR/owl2-overview/
- 53.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature biotechnology. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grenon P, Smith B. SNAP and SPAN. Towards Dynamic Spatial Ontology. Spatial Cognition and Computation. 2004;4(1):69–103. [Google Scholar]
- 55.The Ontology for General Medical Science (OGMS) https://code.google.com/p/ogms/
- 56.Xiang Z, Mungall C, Ruttenberg A, He Y. Ontobee: A linked data server and browser for ontology terms. CEUR Workshop Proceedings; The 2nd International Conference on Biomedical Ontologies (ICBO); 2011; Buffalo, NY, USA. 2013. pp. 279–281. [Google Scholar]
- 57.Marcos E, Zhao B, He Y. The Ontology of Vaccine Adverse Events (OVAE) and its usage in representing and analyzing adverse events associated with US-licensed human vaccines. Journal of biomedical semantics. 2013;4(1):40. doi: 10.1186/2041-1480-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang Z, Todd T, Ku KP, Kovacic BL, Larson CB, Chen F, Hodges AP, Tian Y, Olenzek EA, Zhao B, et al. VIOLIN: vaccine investigation and online information network. Nucleic acids research. 2008;36(Database issue):D923–928. doi: 10.1093/nar/gkm1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Y, Racz R, Sayers S, Lin Y, Todd T, Hur J, Li X, Patel M, Zhao B, Chung M, et al. Updates on the web-based VIOLIN vaccine database and analysis system. Nucleic acids research. 2014;42(1):D1124–1132. doi: 10.1093/nar/gkt1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He Y, Rappuoli R, De Groot AS, Chen RT. Emerging vaccine informatics. J Biomed Biotechnol. 2010;2010:218590. doi: 10.1155/2010/218590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hur J, Ozgur A, Xiang Z, He Y. Identification of fever and vaccine-associated gene interaction networks using ontology-based literature mining. Journal of biomedical semantics. 2012;3(1):18. doi: 10.1186/2041-1480-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Tao C, He Y, Kanjamala P, Liu H. Network-based analysis of vaccine-related associations reveals consistent knowledge with the vaccine ontology. Journal of biomedical semantics. 2013;4(1):33. doi: 10.1186/2041-1480-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo A, Racz R, Hur J, Lin Y, Xiang Z, Zhao L, Jiang G, Zhu Q, Bai J, He Y. Ontology-based collection, representation and analysis of drug-associated neuropathy adverse events. The 4th International Workshop on Vaccine and Drug Ontology Studies (VDOS-2015), in conjunction with the International Conference on Biomedical Ontologies (ICBO-2015); July 27–30 2015; Lisbon, Portugal. 2015. pp. 1–6. [Google Scholar]
- 64.Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. Journal of the American Medical Informatics Association: JAMIA. 2011;18(4):441–448. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carter JS, Brown SH, Bauer BA, Elkin PL, Erlbaum MS, Froehling DA, Lincoln MJ, Rosenbloom ST, Wahner-Roedler DL, Tuttle MS. Categorical information in pharmaceutical terminologies. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium; 2006; pp. 116–120. [PMC free article] [PubMed] [Google Scholar]
- 66.Hastings J, de Matos P, Dekker A, Ennis M, Harsha B, Kale N, Muthukrishnan V, Owen G, Turner S, Williams M, et al. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic acids research. 2013;41(Database issue):D456–463. doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stanley SL, Jr, Frey SE, Taillon-Miller P, Guo J, Miller RD, Koboldt DC, Elashoff M, Christensen R, Saccone NL, Belshe RB. The immunogenetics of smallpox vaccination. The Journal of infectious diseases. 2007;196(2):212–219. doi: 10.1086/518794. [DOI] [PubMed] [Google Scholar]
- 68.Reif DM, McKinney BA, Motsinger AA, Chanock SJ, Edwards KM, Rock MT, Moore JH, Crowe JE. Genetic basis for adverse events after smallpox vaccination. The Journal of infectious diseases. 2008;198(1):16–22. doi: 10.1086/588670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pulendran B, Miller J, Querec TD, Akondy R, Moseley N, Laur O, Glidewell J, Monson N, Zhu T, Zhu H, et al. Case of yellow fever vaccine--associated viscerotropic disease with prolonged viremia, robust adaptive immune responses, and polymorphisms in CCR5 and RANTES genes. The Journal of infectious diseases. 2008;198(4):500–507. doi: 10.1086/590187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin Y, He Y. The ontology of genetic susceptibility factors (OGSF) and its application in modeling genetic susceptibility to vaccine adverse events. Journal of biomedical semantics. 2014;5:19. doi: 10.1186/2041-1480-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poland GA, Ovsyannikova IG, Kennedy RB, Haralambieva IH, Jacobson RM. Vaccinomics and a new paradigm for the development of preventive vaccines against viral infections. OMICS. 2011;15(9):625–636. doi: 10.1089/omi.2011.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poland GA, Kennedy RB, McKinney BA, Ovsyannikova IG, Lambert ND, Jacobson RM, Oberg AL. Vaccinomics, adversomics, and the immune response network theory: individualized vaccinology in the 21st century. Seminars in immunology. 2013;25(2):89–103. doi: 10.1016/j.smim.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rappuoli R. Reverse vaccinology. Curr Opin Microbiol. 2000;3(5):445–450. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 74.Futuyma DJ. Evolutionary Biology. 3. Sinauer Associates; 1997. [Google Scholar]
- 75.Mazzarello P. A unifying concept: the history of cell theory. Nat Cell Biol. 1999;1(1):E13–15. doi: 10.1038/8964. [DOI] [PubMed] [Google Scholar]
- 76.Perelson AS. Immune network theory. Immunol Rev. 1989;110:5–36. doi: 10.1111/j.1600-065x.1989.tb00025.x. [DOI] [PubMed] [Google Scholar]
- 77.Griffiths JF, Miller JH, Suzuki DT, Lewontin RC, Gelbart WM. An Introduction to Genetic Analysis. 7. New York: W. W. Freeman and Company; 2000. [Google Scholar]
- 78.Effectiveness. https://en.wikipedia.org/wiki/Effectiveness.
- 79.Flay BR. Efficacy and effectiveness trials (and other phases of research) in the development of health promotion programs. Preventive medicine. 1986;15(5):451–474. doi: 10.1016/0091-7435(86)90024-1. [DOI] [PubMed] [Google Scholar]
- 80.Wells JD. Effective Theories in Physics: From Planetary Orbits to Elementary Particle Masses: SpringerBriefs in Physics. 2012 [Google Scholar]
- 81.Costa PCG, Laskey KB, Blasch E, Jousselme A-L. Towards unbiased evaluation of uncertainty reasoning: The URREF ontology. The 15th International Conference on Information Fusion; 9–12 July 2012; Singapore. pp. 2301–2308. [Google Scholar]
- 82.Heather N. The efficacy-effectiveness distinction in trials of alcohol brief intervention. Addiction science & clinical practice. 2014;9:13. doi: 10.1186/1940-0640-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weinberg GA, Szilagyi PG. Vaccine epidemiology: efficacy, effectiveness, and the translational research roadmap. The Journal of infectious diseases. 2010;201(11):1607–1610. doi: 10.1086/652404. [DOI] [PubMed] [Google Scholar]
- 84.Boccaletti S, Latora V, Moreno Y, Chavez M, Hwang DU. Complex networks: Structure and dynamics. Physics Reports. 2006;424:175–308. [Google Scholar]
- 85.HarshaRani GV, Vayttaden SJ, Bhalla US. Electronic data sources for kinetic models of cell signaling. J Biochem. 2005;137(6):653–657. doi: 10.1093/jb/mvi083. [DOI] [PubMed] [Google Scholar]
- 86.Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic acids research. 2011;39(Database issue):D691–697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic acids research. 2004;32(Database issue):D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Latendresse M, Paley S, Karp PD. Browsing metabolic and regulatory networks with BioCyc. Methods in molecular biology. 2012;804:197–216. doi: 10.1007/978-1-61779-361-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.BioCarta Apoptosis pathway. http://www.biocarta.com/genes/Apoptosis.asp.
- 90.Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics. 2010;11(4):501–505. doi: 10.2217/pgs.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hur J, Ozgur A, Xiang Z, He Y. Development and application of an interaction network ontology for literature mining of vaccine-associated gene-gene interactions. Journal of biomedical semantics. 2015;6:2. doi: 10.1186/2041-1480-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He Y, Xiang Z. HINO: a BFO-aligned ontology representing human molecular interactions and pathways. 2013 arXiv. arXiv:1311.3355. [Google Scholar]
- 93.Buttigieg PL, Morrison N, Smith B, Mungall CJ, Lewis SE Consortium E. The environment ontology: contextualising biological and biomedical entities. Journal of biomedical semantics. 2013;4(1):43. doi: 10.1186/2041-1480-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Malone J, Holloway E, Adamusiak T, Kapushesky M, Zheng J, Kolesnikov N, Zhukova A, Brazma A, Parkinson H. Modeling sample variables with an Experimental Factor Ontology. Bioinformatics. 2010;26(8):1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15(6):589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 97.Li R, Li Y, Zheng H, Luo R, Zhu H, Li Q, Qian W, Ren Y, Tian G, Li J, et al. Building the sequence map of the human pan-genome. Nature biotechnology. 2010;28(1):57–63. doi: 10.1038/nbt.1596. [DOI] [PubMed] [Google Scholar]
- 98.Sanchez C, Lachaize C, Janody F, Bellon B, Roder L, Euzenat J, Rechenmann F, Jacq B. Grasping at molecular interactions and genetic networks in Drosophila melanogaster using FlyNets, an Internet database. Nucleic acids research. 1999;27(1):89–94. doi: 10.1093/nar/27.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bonetta L. Protein-protein interactions: Interactome under construction. Nature. 2010;468(7325):851–854. doi: 10.1038/468851a. [DOI] [PubMed] [Google Scholar]
- 100.Bouveret E, Brun C. Bacterial interactomes: from interactions to networks. Methods in molecular biology. 2012;804:15–33. doi: 10.1007/978-1-61779-361-5_2. [DOI] [PubMed] [Google Scholar]
- 101.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393(6684):440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 102.Arenas A, Diaz-Guilera A, Kurths J, Moreno Y, Zhou C. Synchronization in complex networks. Physics Reports. 2008;469:93–153. [Google Scholar]
- 103.The protege ontology editor. http://protege.stanford.edu/
- 104.Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, Musen MA. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic acids research. 2011;39(Web Server issue):W541–545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiang Z, Courtot M, Brinkman RR, Ruttenberg A, He Y. OntoFox: web-based support for ontology reuse. BMC Res Notes. 2010;3:175, 1–12. doi: 10.1186/1756-0500-3-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zheng J, Xiang Z, Stoeckert CJ, Jr, He Y. Ontodog: a web-based ontology community view generation tool. Bioinformatics. 2014;30(9):1340–1342. doi: 10.1093/bioinformatics/btu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xiang Z, Zheng J, Lin Y, He Y. Ontorat: Automatic generation of new ontology terms, annotations, and axioms based on ontology design patterns. Journal of biomedical semantics. 2015;6(1):4, 10. doi: 10.1186/2041-1480-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiang Z, He Y. IDO extensions alignment using Ontobeep. Infectious Disease Ontology (IDO) Workshop; 2010: December 8–9; Baltimore, MD, USA. 2010. [Google Scholar]