Abstract
Forskolin (FSK) induces activation of protein kinase A (PKA). This activation protects specifically some cancer cells from death induced by glucose starvation. Cell effects upon FSK treatment prompted us to investigate in detail the physiological role of PKA in the activation of pro-survival mechanisms in glucose starvation. In this regard we performed a microarray analysis of normal NIH3T3 and transformed NIH3T3-K-ras mouse fibroblasts cultured at 1 mM glucose and daily treated or not with 10 μM FSK until 72 h of growth, when the samples were collected. The microarray is deposited into Gene Expression Omnibus under Series GSE68266. The microarray data revealed that the activation of PKA regulates the expression of genes involved in metabolic, stress-response and pro-survival processes, like glutamine metabolism, autophagy and unfolded protein response, preventing cancer cell death in glucose starvation. Altogether these findings suggest that PKA activation, by inducing a complex transcriptional program, leads to cancer survival in nutrient stress, a typical feature of developing tumor. These transcriptional data, identifying this important role of PKA, will be useful to identify novel target in cancer therapy.
Keywords: Cancer metabolism, Protein kinase A (PKA), Glucose starvation, Cancer cell resistance
Specifications
Where applicable, please follow the Ontology for Biomedical Investigations: http://obi-ontology.org/page/Main_Page
| Organism/cell line/tissue | NIH3T3 mouse fibroblasts and transformed NIH3T3-K-ras |
| Sex | NA |
| Sequencer or array type | Mouse Genechip MoGene-1_0 -st-v1 Arrays (Affymetrix) |
| Data format | Raw data: CEL files, normalized data: SOFT, MINIML, TXT |
| Experimental factors | Immortalized NIH3T3 mouse fibroblasts vs. transformed NIH3T3-K-ras mouse fibroblasts cultured at 1 mM glucose and daily treated with 10 μM Forskolin |
| Experimental features | Transcriptome profiling of genes modulated by PKA in immortalized and transformed mouse fibroblasts. Cells were cultured in medium with 1 mM glucose and then treated with FSK. The samples for the microarray analysis were collected at 72 h of culture in low glucose after daily treatments with FSK. |
| Consent | Data are publicly available |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Cell culture and reagents
The immortalized NIH3T3 and the transformed NIH3T3-K-ras cell lines were routinely maintained in DMEM medium (Life Technologies) with 10% new born calf serum (Life Technologies), 25 mM glucose and other supplements as described previously [1], [2]. All cells were cultured at 37 °C in 5% CO2. To perform the array experiments the cells were seeded at 3000 cells/cm2 in complete growth medium. After 16 h, the cells were washed twice with phosphate-buffered saline and incubated in medium (time 0) supplemented with 1 mM glucose instead of 25 mM (Fig. 1). To induce the increase of cAMP levels and hence PKA activation, the cells were daily treated with 10 μM forskolin (FSK) (Sigma-Aldrich) starting from 24 h after medium replacement. FSK-treated and untreated (DMSO) cells were then collected for array analyses at 72 h of culture in low glucose medium, which corresponds to 48 h of FSK-treatment (Fig. 1). The basal and the FSK-induced cAMP levels as well as the PKA activation in both cell lines are described in [1], [2].
Fig. 1.
Experimental scheme.
2.2. Microarray and data analysis
Cells were lysed in TRIZOL (Invitrogen, Carlsbad, CA) and total RNA was further purified with the Rneasy kit (QIAGEN, Hilden, Germany) following manufacturer instructions. RNA was quantified on the NanoDrop ND-1000. RNA quality and integrity was determined by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) on RNA Nano Chips. RNA integrity numbers (RIN) were > 8.5 indicating high RNA quality. Single strand biotinylated cDNA was generated as follows: 100 ng of total RNA were subjected to 2 cycles of cDNA synthesis with the Ambion WT expression Kit (Life Technologies). The first cycle - first strand synthesis is performed using an engineered set of random primers that exclude rRNA-matching sequences and include the T7 promoter sequences. After second-strand synthesis, the resulting cDNA is in vitro transcribed with the T7 RNA polymerase to generate a cRNA. This cRNA is subjected to a second cycle - first strand synthesis in the presence of dUTP in a fixed ratio relative to dTTP. Single strand cDNA is then purified and fragmented with a mixture of uracil DNA glycosylase and apurinic/apirimidinic endonuclease 1 (Affymetrix) in correspondence of incorporated dUTPs. DNA fragments are then terminally labeled by terminal deoxynucleotidyl transferase (Affymetrix) with biotin. The biotinylated DNA was hybridized to the Mouse Genechip MoGene-1_0 -st-v1 Arrays (Affymetrix), containing almost 23000 genes selected from Mus musculus genome databases RefSeq, ENSEMBL and GenBank. Chips were washed and scanned on the Affymetrix Complete GeneChip Instrument System, generating digitized image data (DAT) files. Data where then normalized using a Global Normalization approach with the Affymetrix Gene Expression Console software, generating the .CEL files
2.3. Microarray and gene expression analysis
Microarray analysis was performed on biological triplicate samples. Labeling of cDNA and hybridization to Mouse MoGene-1_0 -st-v1 Arrays were performed at the IRGS, Biogem, Ariano Irpino (AV), Italy. The samples included (see Table 1): 1) GSM1666885, immortalized NIH3T3 mouse fibroblasts replicate 1, 2) GSM1666886, immortalized NIH3T3 mouse fibroblasts replicate 2, 3) GSM1666887, immortalized NIH3T3 mouse fibroblasts replicate 3, 4) GSM1666888, NIH3T3-K-ras transformed mouse fibroblasts replicate 1, 5) GSM1666889, NIH3T3-K-ras transformed mouse fibroblasts replicate 2, 6) GSM1666890, NIH3T3-K-ras transformed mouse fibroblasts replicate 3, 7) GSM1666891, immortalized NIH3T3 mouse fibroblasts treated with FSK replicate 1, and 8) GSM1666892, immortalized NIH3T3 mouse fibroblasts treated with FSK replicate 2, 9) GSM1666893, immortalized NIH3T3 mouse fibroblasts treated with FSK replicate 3, 10) GSM1666894, NIH3T3-K-ras transformed mouse fibroblasts treated with FSK replicate 1, 11) GSM1666895, NIH3T3-K-ras transformed mouse fibroblasts treated with FSK replicate 2, 12) GSM1666896, NIH3T3-K-ras transformed mouse fibroblasts treated with FSK replicate 3. All samples were normalized through the Robust Multi-array Average (RMA) method [3] with background correction and quantile normalization at core genes level as implemented by the R/Bioconductor package “oligo”. Gene name annotations have been downloaded from ENSEMBL using Biomart on Jan. 22, 2015. Probes not mapping to an Entrez gene have been discarded. The resulting dataset was composed of 22,035 probes mapping to 17,665 unique genes.
Table 1.
Sample identity.
| Samples | Parameter |
GEO identification number | |
|---|---|---|---|
| Phenotype | Treatment (FSK) | ||
| NIH3T3 | Normal | − | GSM1666885 |
| NIH3T3 | Normal | − | GSM1666886 |
| NIH3T3 | Normal | − | GSM1666887 |
| NIH3T3-K-ras | Transformed | − | GSM1666888 |
| NIH3T3-K-ras | Transformed | − | GSM1666889 |
| NIH3T3-K-ras | Transformed | − | GSM1666890 |
| NIH3T3 | Normal | + | GSM1666891 |
| NIH3T3 | Normal | + | GSM1666892 |
| NIH3T3 | Normal | + | GSM1666893 |
| NIH3T3-K-ras | Transformed | + | GSM1666894 |
| NIH3T3-K-ras | Transformed | + | GSM1666895 |
| NIH3T3-K-ras | Transformed | + | GSM1666896 |
Differential expression analysis was performed using a linear model approach with least-square fitting and False Discovery Rate correction as provided by the R/Bioconductor package “limma” [4]. Two differential expression tests (contrasts) were performed: untreated normal cells vs. FSK-treated normal cells - denoted as NF/N - and untreated transformed cells vs. FSK-treated transformed cells - denoted as TF/T. Only the probe of each gene having the minimum average p-value in the two contrasts has been kept. There were 496 genes (probes) with absolute log fold-change at least 1 for contrast NF/N and 208 genes for contrast TF/T.
To gain a better understanding of the biological implications of these expression profiles, we performed a comprehensive gene set analysis of the differential expression results using the implementation provided by the R/Bioconductor package “piano” [5]. The analysis used as gene sets the 202 KEGG pathways classified as metabolic or signaling as downloaded on Jan. 22, 2015. The pathways contain 6885 unique genes of which 6010 are included in the analysis dataset (approx. 34% of the genes).
Package “piano” provides the implementation of several gene set analysis tests and of a consensus methodology to summarize the results obtained with the different tests. This approach should be less sensitive to possible hidden biases of the single tests and allows, for example, the integration of sources of gene expression data with other types of high throughput data like proteomic data obtained by 2-DIGE [2]. The following tests have been performed:
-
a)
Mean
-
b)
Median
-
c)
Sum
-
d)
Maxmean
-
e)
Gene set enrichment analysis (GSEA)
-
f)
Parametric analysis of gene set enrichment (PAGE)
-
g)
Reporter features
-
h)
Wilcoxon rank-sum test
-
i)
Tail strength.
Tests (a)–(f) used t-values computed in the differential expression analysis while tests (g)–(i) used the corresponding p-values (and log fold changes, for devising the directionality). Tests (f), (g), (h) computed gene set statistics significance via the theoretical null distribution, while the remaining ones via gene (re)sampling.
Each test may return distinct p-values for each gene set (i.e., pathway) in the following directionality classes:
-
–
Distinct-directional (up or down), indicated as Dist(up) and Dist(down), which tests if the expression of the gene set as a whole is regulated in a distinct direction (either up or down). By definition, a gene set cannot be significant in both classes as regulations in opposite directions will cancel out the effect.
-
–
Mixed-directional (up or down), indicated as Mix(up) and Mix(down), which tests if a significant subset of its genes are coordinately up- or down-regulated. A gene set can be significant in both classes.
-
–
Non-directional, indicated as NonDir, which tests if the expression of the gene set as a whole is significantly perturbed, discarding any information about directionality.
A total of 35 p-value vectors over the 45 possible combinations for each contrast have been obtained since not all tests returned results for all the directionality classes. The combined analysis of the p-values obtained by each gene set in the various directionality classes provides a clear picture of the (possible) regulation of that gene set.
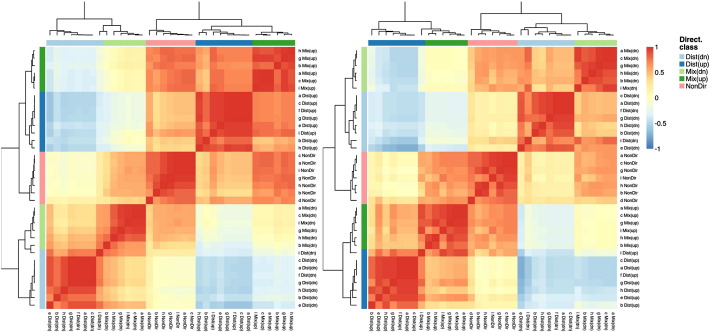
Consistency of the results has been empirically verified by computing the pairwise correlation between the p-value vectors. Fig. 2 depicts the correlation matrices of the p-value vectors of contrast NF/N on the left and of contrast TF/T on the right. Directionality classes of the p-value vectors are indicated by the annotation bars on the top and on the left of the two plots, while letters refer to the different gene set analysis tests as per the previous list. According to the plots, p-value vectors of the same directionality class (obtained with different tests) have strong correlation (see, for example, the class Dist(dn)), p-value vectors of similar classes have a non-negligible correlation (see, for example, Mix(dn) and Dist(dn)) and p-value vectors of opposite classes are inversely correlated (see, for example, Dist(dn) and Dist(up)). Further confirmation is provided by the dendograms depicted on the sides, showing that tests of the same directionality class cluster together. The only exception is test (i) “Tail strength” for the distinct down-directional class on both contrasts. Despite that, the p-values of the gene sets on these classes still appear to be in accordance (albeit less correlated) with the rest of the results and represent only a small perturbation that the final consensus analysis should easily accommodate.
Fig. 2.
Correlation among p-value vectors obtained for each gene set analysis test (indicated by letters from a to i) and each directionality class (annotated by colors on top and left of each plot) for contrast NF/N (left) and contrast TF/T (right).
Principal Component Analysis (PCA) of the same set of vectors provides a second empirical support to the consistency of the results computed by the different tests. Fig. 3 depicts each p-value vector (represented as the corresponding letter, see the list above) for contrast NF/N (left) and contrast TF/T (right) in the space of its first three principal components (x- and y-axes and size). Ideally, each directionality class (represented by colors) should cluster together in the resulting space. Almost all the vectors clustered as expected.
Fig. 3.
Principal Component Analysis (PCA) of the p-value vectors obtained for each gene set analysis test (indicated by letters from a to i) and each directionality class (colors) for contrast NF/N (left) and contrast TF/T (right). Top: the space resulting after PCA in its first three dimensions (x- and y-axes and letter size). Bottom: the proportion of the variance explained by each PCA dimension (only those > 1% have been shown).
The most evident outliers are those computed by test (i) for the distinct down-directional class, that are placed near the vectors of the mixed down-directional class. This confirms the observation made before for Fig. 2 and, again, this should not represent an issue for the consensus analysis.
Consensus aggregation as implemented in the R/Bioconductor package “piano” computes the overall consensus rank by first ranking each gene set in each directionality class for each method according to the predicted p-value. These ranks are then aggregated according to the Borda rule (a single-winner election method). Fig. 4 depicts the aggregated ranks obtained by the gene sets that ranked in the first 5 positions in at least one directionality class. Directionality classes are represented as columns while numbers and colors represent the position of each gene set (row) in the aggregated rank of that column for both contrast NF/N (left block) and contrast TF/T (right block). In general, a pathway can be considered significantly regulated in a particular directionality class if it ranks in the first positions (albeit a general cut-off does not exist). As expected, pathways ranking high (i.e., small numbers) in a directionality class (see, for example, “Ribosome biogenesis in Eukaryotes”) are ranked low (large numbers) in the opposite directionality class. Such a representation provides a clear and concise comparison of the regulation of pathways in the two contrasts, highlighting common regulation of pathways in normal and transformed cells, like “Ribosome biogenesis in Eukaryotes”, down-regulated in both NF/N and TF/T, and specific pathway regulations, different for the two cell lines, like the up-regulation of pathway “Steroid biosynthesis” detected in TF/T but not in NF/N.
Fig. 4.
Aggregated ranks computed by the consensus methodology implemented by “piano” for contrast NF/N (left) and TF/T (right). Each column represents the aggregated ranks of the gene sets (listed on the left) in a particular directionality class (listed at the bottom).
3. Conclusion
The microarray data and the “piano” computational approach here presented, have allowed to the identification of significant pro-survival processes, otherwise hidden, regulated by PKA in glucose deprivation [2]. A part from our approach, these data could be further used to uncover other mechanisms of cancer resistance mediated by PKA, whose role in cancer progression has assumed great relevance in recent years [6], [7], [8].
Conflict of interest
The authors declare no conflict of interests.
Acknowledgments
This work was supported by a grant to F.C. from Italian Government # 2014-ATE-0179 (FAR). F.C. is supported by a grant from Associazione Italiana per la Ricerca sul Cancro (AIRC # 15364). R.P. is supported by a fellowship from MIUR. F.R. is supported by a doctoral fellowship from MIUR. Y.P. was partially supported by Cariplo Foundation grant 2013-0955.
Contributor Information
Ferdinando Chiaradonna, Email: ferdinando.chiaradonna@unimib.it.
Yuri Pirola, Email: yuri.pirola@disco.unimib.it.
Francesca Ricciardiello, Email: f.ricciardiello@campus.unimib.it.
Roberta Palorini, Email: roberta.palorini@unimib.it.
References
- 1.Palorini R., De Rasmo D., Gaviraghi M., Sala Danna L., Signorile A., Cirulli C. Oncogenic K-ras expression is associated with derangement of the cAMP/PKA pathway and forskolin-reversible alterations of mitochondrial dynamics and respiration. Oncogene. 2013 Jan 17;32(3):352–362. doi: 10.1038/onc.2012.50. [DOI] [PubMed] [Google Scholar]
- 2.Palorini R., Votta G., Pirola Y., De Vitto H., De Palma S., Airoldi C. Protein kinase A activation promotes cancer cell resistance to glucose starvation and anoikis. PLoS Genet. 2016 Mar 15;12(3) doi: 10.1371/journal.pgen.1005931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003 Feb 15;31(4) doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015 Apr 20;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varemo L., Nielsen J., Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013 Apr;41(8):4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armaiz-Pena G.N., Allen J.K., Cruz A., Stone R.L., Nick A.M., Lin Y.G. Src activation by beta-adrenoreceptors is a key switch for tumour metastasis. Nat. Commun. 2013;4:1403. doi: 10.1038/ncomms2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beristain A.G., Molyneux S.D., Joshi P.A., Pomroy N.C., Di Grappa M.A., Chang M.C. PKA signaling drives mammary tumorigenesis through Src. Oncogene. 2015 Feb 26;34(9):1160–1173. doi: 10.1038/onc.2014.41. [DOI] [PubMed] [Google Scholar]
- 8.McKenzie A.J., Campbell S.L., Howe A.K. Protein kinase A activity and anchoring are required for ovarian cancer cell migration and invasion. PLoS One. 2011;6(10):e26552. doi: 10.1371/journal.pone.0026552. [DOI] [PMC free article] [PubMed] [Google Scholar]