Abstract
The 2008 World Health Organization (WHO) classification of tumors of hematopoietic and lymphoid tissues introduced a category for myeloid and lymphoid neoplasms with eosinophilia and abnormalities of PDGFRA, PDGFRB or FGFR1. Many of these patients are responsive to tyrosine kinase inhibitor (TKI) therapy. In this case report, we report a unique case of chronic eosinophlic leukemia with novel t(5;12) (q23–31;p13)/ETV6-ACSL6 gene fusion, in which patient was resistant to TKI therapy. This important finding is a novel addition to the above entity in WHO 2008 classification. The ACSL6 gene encodes a long-chain acyl-CoA synthetase, an enzyme that plays an essential role in lipid metabolism and ATP generation pathways in cells. The EBV6-ACSL6 rearrangement is present in diverse types of hematopoietic malignancies. As yet, it is not clear how ACSL6, a gene involved in fatty acid synthesis, contributes to clonal expansion of myeloid progenitor cells. Therefore, elucidating the contribution of ACSL6 to leukemogenesis may allow the development of novel treatment for those resistant to TKI therapy.
Keywords: Chronic eosinophilic leukemia, NOS, Novel ETV6-ACSL6 gene fusion, Myeloid proliferative neoplasm with eosinophilia
Introduction
Clonal eosinophilia is frequently associated with myeloid neoplasms. Cytogenetic analysis has identified four distinct recurrent breakpoint clusters that target PDGFRA at 4q12, PDGFRB at 5q31–33, FGFR1 at 8p11–12 and JAK2 at 9p24. As a consequence of balanced reciprocal translocations or rare insertions or complex translocations, fusion genes similar to FIP1L1-PDGFRA are created, including ETV6-PDGFRB in t(5;12)(q31–33;p13), ZNF198-FGFR1 in t(8;13)(p11;q12) or PCM1-JAK2 in t(8;9)(p11;p24). The 2008 World Health Organization (WHO) classification of tumors of hematopoietic and lymphoid tissues introduced a new category for myeloid and lymphoid neoplasms with eosinophilia and abnormalities of PDGFRA, PDGFRB or FGFR1. Many of these cases present as a myeloproliferative neoplasm, usually with eosinophilia [1].
Translocations involving band 12p13 are one of the most common chromosomal abnormalities in leukemia and myeloproliferative/myelodysplastic disorders. These translocations frequently result in rearrangements of the ETV6 gene. ETV6 fuses to diverse (at least 30 partner) genes in myeloid neoplasms [2]. One of the partner genes is ACSL6 (ACS2 - acyl-CoA synthetase long-chain family member 6), with which ETV6 can rearrange with and lead to cell transformation and leukemogenesis. The ACSL6 gene encodes a long-chain acyl-CoA synthetase that catalyzes the formation of acyl-CoA from fatty acids, ATP, and CoA [3]. ACSL6 is predominantly expressed in the bone marrow (BM), fetal liver and brain [4], and it plays an essential role in lipid metabolism [3, 5]. Several transcript variants encoding different isoforms have been identified for this gene [5].
The t(5;12)(q31;p13) has been described as a recurrent translocation inducing an ETV6-ACSL6 fusion gene and occurring in variety of myeloid malignancies, often associated with eosinophilia [4, 6, 7]. Almost 10 years ago, six cases of t(5;12)(q23–31;p13) with ETV6-ACSL6 gene fusion were summarized in Leukemia, including acute myeloid leukemia (AML), chronic myeloid leukemia (CML), acute eosinophilic leukemia (AEL) and polycythemia vera (PV) [7]. Here, we report a case of t(5;12) (q31;13) translocation with ETV6-ACSL6 rearrangement in a patient with chronic eosinophilic leukemia (CEL).
Case report
A 52-year-old man presented with progressive eosinophilia for two years. His initial eosinophilia was mild, with absolute eosinophils of 2,400/mm3, but the absolute eosinophil count increased to 13,300/mm3 two years later. This was associated with mild normocytic anemia, normal platelet count and no neutropenia or monocytosis. The patient complained of fatigue and muscle pain. Physical examination and imaging studies showed no organomegaly or lymphadenopathy. An extensive workup for infectious diseases was negative for trichinosis, strongyloides, toxoplasma, HIV, schistosomiasis and parasitic infection. The peripheral blood smear showed leukocytosis with marked eosinophilia, mild basophilia and mild normocytic, normochromic anemia (Figure 1a). Bone marrow biopsy revealed a hypercellular (~90% cellularity) marrow with marked eosinophilia. No increase in immature myeloid precursors, blasts or basophils was detected. There was no overt dysplasia in the remaining trilineage hematopoiesis (Figure 1b). No abnormal mast cells were detected by staining of CD117, CD2, CD25 and tryptase immunostains. Florescence in situ hybridization (FISH) for MDS markers [−5/del(5q), −7/del(7q), +8 and del(20q)], t(9;22)(q34;q11)/BCR/ABL1, t(15;17)(q24;q21)/PML-RARA, t(8;21)(q22;q22)/RUNX1T1-RUNX1, and inv(16)(p13.3q22)/CBFB were negative. FISH was also negative for rearrangements of FIP1L1/CHIC2/PDGFRA, 5p33.1/PDGFRB, 8p11/FGFR1 and 16q22/CBFB (Figure 1c). No c-KIT (D816V) mutation or T cell receptor gamma gene rearrangement was detected. Metaphase cytogenetic analysis on the bone marrow aspirate detected a t(5;12)(q31;p13) translocation in 9/20 (45%) metaphases (Figure 1d). ETV6-ACSL6 gene fusion was further confirmed by FoundationOne™ Heme assay (Foundation Medicine, Cambridge MA), a next-generation sequencing (NGS) based assay. The current assay utilizes DNA sequencing to interrogate 405 genes as well as selected introns of 31 genes involved in rearrangements, in addition to RNA sequencing of 265 genes. Targeted RNA-seq found 246 reads supporting the fusion event: fusion:5′-ETV6(ex1-1 NM_001987)-ACSL6(ex2-21 NM_015256). No other fusion transcripts were identified. Therefore, a diagnosis of CEL, NOS with t(5;12)(q31;p13) resulting in a ETV6-ACSL6 gene fusion was rendered. Pending the NGS data, the patient was treated for 3 months with imatinib, but developed progressive anemia without improvement in the leukocytosis and eosinophilia. After confirming the ETV6-ACSL6 fusion, therapy was changed to hydroxyurea, which led to an improvement in eosinophils, anemia and clinical symptoms. Allogeneic stem cell transplant is being considered.
Figure 1.
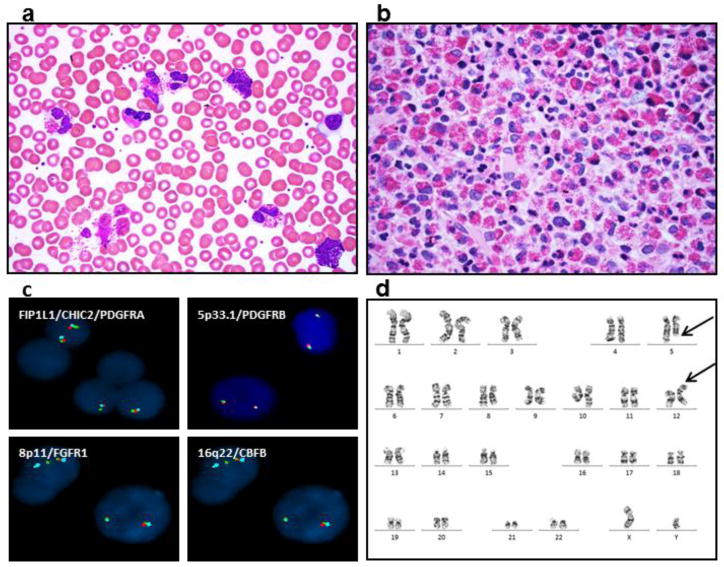
Marked eosinophilia is detected in patient’s peripheral blood smear (a) and bone marrow core biopsy (b). Fluorescence in situ hybridization analysis (FISH) of rearrangements of FIP1L1/CHIC2/PDGFRA, 5p33.1/PDGFRB, 8p11/FGFR1 and 16q22/CBFB are negative (c). Representative G-banded karyotyping reveals translocation of chromosomes 5 and 12 from peripheral blood cells at metaphases. The breakpoints are identified as 5q31 and 12p13 (arrows) (d).
Discussion
Yagasaki et al. identified an ETV6-ACSL6 chimeric gene in three patients with t(5;12)(q31;p13), including refractory anemia with excess blasts (RAEB) with basophilia, AML and AEL [4]. Different fusion genes were identified by cytogenetic and FISH analysis in these patients: a short in-frame fusion of exon 1 of ETV6 to the 3′UTR of ACSL6, an out-of-frame fusion of exon 2 of ETV6 to exon 11 of ACSL6, and an out-of-frame fusion of exon 1 of ETV6 to exon 1 of ACSL6 [4]. FISH with bacterial artificial chromosomes (BACs) specific probes for the ETV6 and ACSL6 genes were performed on two patients with t(5;12)(q31;p13)-associated PV and demonstrated the involvement of ETV6 and the 5′ region of the ACSL6 in the translocation [7]. In the current case, FISH for t(5;12)(q31;p13) was negative, most likely due to PDGFRB (5p33.1) being many kb distal (toward the telomere end) to ACSL6 (5q31). Therefore, detection of the rearrangement was missed by this probe. RNA-seq showed the current rearrangement resulted in an out-of-frame fusion of ETV6 exon 1 and ACSL6 exons 2–21. This particular rearrangement to our knowledge is novel and has not been previously reported. This out-of-frame fusion would most likely lead to a frameshift with truncated fusion product. Similar rearrangements involving out-of-frame fusions of ETV6 exon 1 to ACSL6 have been reported in the above five cases [4, 7]. These observations suggest that the disruption of ETV6 and ACSL6 may lead to the pathogenesis of hematological malignancies with t(5;12)(q31;p13).
Generation of out-of-frame fusions, as in ETV6-ACSL6, can lead to functional consequences. Molecular analysis of these fusion genes leads to multiple proposed mechanisms of leukemogenesis including loss of function of the fusion gene, affecting ETV6 and/or ACSL6 gene [4], and, loss of the untranslocated ETV6 and activation of a proto-oncogene in the vicinity of a chromosomal translocation [7]. An example is interleukin 3 (IL-3, which is located near the breakpoint at 5q31). When activated, IL3 promotes proliferation and differentiation of various hematopoietic cell lineages. Constitutive activation and dysregulated expression of IL3 lead to a marked cell proliferative state, which can explain the eosinophilia observed in the majority of cases with t(5;12)(q31;p13) [8, 9, 10, 16], including the current case. The ETV6-ACSL6 rearrangement can be the driving mutation of leukemogenesis of CEL although detailed molecular mechanisms have not been determined. Besides the ETV6-ACSL6 fusion, other genomic alterations were identified including BRIP1 (DNA repair/chromatin stability), NF1 (tumor suppressor gene), U2AF1 (RNA splicing) (Table 1). The findings further confirm that leukemia may develop as the consequence of an array of molecular alterations that disrupts many facets of hematopoietic precursor cell development to induce leukemia transformation. These processes may include the regulation of cell proliferation, differentiation, self-renewal, survival, cell cycle checkpoint control, DNA repair and chromatin stability, and cell dissemination.
Table 1.
Genomic alteration detected in this patient using clinical next-generation sequencing (NGS) by FoundationOne™ Heme.
| Genomic Alteration detected | Normal Gene Function | Hematological disorders caused | Other tumors caused |
|---|---|---|---|
|
BRIP1 W1217* |
DNA repair Chromosome stability maintenance |
Fanconi Anemia [11] | Breast, ovarian and cervical cancer |
|
NF1 I526S |
Key regulator of the RAS signaling pathway | Juvenile myelomonocytic leukemia MDS |
Sarcoma, glioma, breast tumor and NET |
|
U2AF1 S34F |
Encodes a pre-mRNA splicing factor required for accurate 3′ splice site selection | MDS, AML, CMML and leukemic transformation of MPN [12] | Lung adenocarcinomas |
|
ETV6 ETV6-ACSL6 fusion |
Encodes an ETS family transcription factor required for hematopoiesis | Various hematopoietic malignancies including leukemia and MDS [13] | Secretory breast cancer, papillary thyroid carcinomas associated with radiation [14, 15] |
MDS: myelodysplastic syndrome
MPN: myeloproliferative neoplasms
AML: acute myeloid leukemia
CMML: chronic myelomonocytic leukemia
NET: Neuroendocrine tumor
In conclusion, we have reported a case of chronic eosinophilic leukemia, NOS, with a t(5;12)(q31;p13) resulting in a novel ETV6-ACSL6 gene fusion. Myeloid neoplasms with PDGFRA, PDGFRB or FGFR1 abnormalities are the most important groups of diseases in the differential diagnosis of eosinophilia-associated myeloproliferative neoplasms, as some of these patients may be responsive to tyrosine kinase inhibitor (TKI) therapy. Our current report and the five cases published previously suggest that ETV6 plays a pivotal role in the regulation of myeloid hematopoiesis, especially at the eosinophilic progenitor level and therefore, these patients will not be responsive to TKI. This may represents a distinctive entity to the previous 2008 WHO classification. Further molecular studies are needed to clarify the underlying mechanism of ACSL6 in leukemogenesis and how aberrant expression of a gene involved in fatty acid synthesis can result in clonal expansion.
Acknowledgments
The study was partially support by Collaborative Institutional Research Pilot Grant for Investigators in Oncology, the Cancer Research Coordinating Committee (CRCC) and academic senate grants from University of California (MC) and NIH K12 CA138464 (BAJ). We thank Dr. Ralph Green for critical reading this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST
The authors have no relevant conflict of interest to disclose.
AUTHOR CONTRIBUTIONS
RJS and MC designed the study, analyzed the data and wrote the first draft of the manuscript. BAJ, JW and JPG contributed to the interpretation of clinical data and editing the manuscript. All authors reviewed, provided input and approved the final version of the manuscript.
References
- 1.Bain BJ, Gilliland DG, Horny H-P, Vardiman JW. Myeloid and lymphoid neoplasms with eosinophilia and abnormalities of PDGFRA, PDGFRB and FGFR1. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissue. 4th. International Agency for Research and Cancer (IARC); Lyon: 2008. pp. 68–73. [Google Scholar]
- 2.De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk Res. 2012;36:945–61. doi: 10.1016/j.leukres.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Malhotra KT, Malhotra K, Lubin BH, Kuypers FA. Identification and molecular characterization of acyl-CoA synthetase in human erythrocytes and erythroid precursors. Biochem J. 1999;344:135–143. [PMC free article] [PubMed] [Google Scholar]
- 4.Yagasaki F, Jinnai I, Yoshida S, Yokoyama Y, Matsuda A, Kusumoto S, et al. Fusion of TEL/ETV6 to a novel ACS2 in myelodysplastic syndrome and acute myelogenous leukemia with t(5;12)(q31;p13) Genes Chromosomes Cancer. 1999;26:192–202. doi: 10.1002/(sici)1098-2264(199911)26:3<192::aid-gcc2>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 5.Soupene E, Kuypers FA. Multiple erythroid isoforms of human long-chain acyl-CoA synthetases are produced by switch of the fatty acid gate domains. BMC Mol Biol. 2006;7:21–32. doi: 10.1186/1471-2199-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katsura Y, Suzukawa K, Nanmoku T, Nemoto N, Machino T, Obara N, et al. Myelodysplastic syndrome accompanied by basophilia and eosinophilia with t(5;12)(q31;p13) Cancer Genet Cytogenet. 2007;178:85–88. doi: 10.1016/j.cancergencyto.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 7.Murati A, Adélaïde J, Gelsi-Boyer V, Etienne A, Rémy V, Fezoui H, et al. t(5;12)(q23–31;p13) with ETV6-ACSL6 gene fusion in polycythemia vera. Leukemia. 2006;20:1175–1178. doi: 10.1038/sj.leu.2404194. [DOI] [PubMed] [Google Scholar]
- 8.Chang JM, Metcalf M, Lang RA, Gonda TJ, Johnson GR. Nonneoplastic hematopoietic myeloproliferative syndrome induced by dysregulated multi-CSF (IL-3) expression. Blood. 1989;73:1487–1497. [PubMed] [Google Scholar]
- 9.Perkins A, Kongsuwan K, Visvader J, Adams JM, Cory S. Homeobox gene expression plus autocrine growth factor production elicits myeloid leukemia. Proc Natl Acad Sci USA. 1990;87:8398–8402. doi: 10.1073/pnas.87.21.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weller PF. The immunobiology of eosinophils. N Engl J Med. 1991;324:1110–1118. doi: 10.1056/NEJM199104183241607. [DOI] [PubMed] [Google Scholar]
- 11.Khincha PP, Savage SA. Genomic characterization of the inherited bone marrow failure syndromes. Semin Hematol. 2013;50(4):333–347. doi: 10.1053/j.seminhematol.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Przychodzen B, Jerez A, Guinta K, Sekeres MA, Padgett R, Maciejewski JP, et al. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood. 2013;122(6):999–1006. doi: 10.1182/blood-2013-01-480970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wlodarska I, Mecucci C, Baens M, Marynen P, van den Berghe H. ETV6 gene rearrangements in hematopoietic malignant disorders. Leuk Lymphoma. 1996;23(3–4):287–295. doi: 10.3109/10428199609054831. [DOI] [PubMed] [Google Scholar]
- 14.Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2(5):367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 15.Leeman-Neill RJ, Kelly LM, Liu P, Brenner AV, Little MP, Bogdanova TI, et al. ETV6-NTRK3 is a common chromosomal rearrangement in radiation-associated thyroid cancer. Cancer. 2014;120(6):799–807. doi: 10.1002/cncr.28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cools J, Mentens N, Odero MD, Peeters P, Wlodarska I, Delforge M, et al. Evidence for position effects as a variant ETV6-mediated leukemogenic mechanism in myeloid leukemias with a t(4;12)(q11–q12;p13) or t(5;12)(q31;p13) Blood. 2002;99:1776–1784. doi: 10.1182/blood.v99.5.1776. [DOI] [PubMed] [Google Scholar]


