Main text
The unfolded protein response (UPR) is a 3-pronged signaling axis charged with correcting toxic protein misfolding stresses within the endoplasmic reticulum. The ER-localized bifunctional enzyme IRE1α, which possesses both kinase and endoribonuclease activity, is the most evolutionarily conserved and best understood UPR branch. IRE1α primarily functions by inducing an mRNA reading frame shift in the protein XBP1 via a unique cytoplasmic RNA splicing event, thereby generating a potent transcription factor that upregulates a wide variety of chaperones, glycosylases, and oxidative protein folding enzymes.
Numerous studies with have delineated critical roles for XBP1 and IRE1α in the maturation and survival of professional secretory cells such as salivary acinar cells, which must repeatedly synthesize large quantities of protein. However, the role of XBP1 in cells exposed to temporally limited bursts of protein production, as often occur during major state transitions like differentiation, was unknown. To address this question, we chose granulocyte differentiation as our model system since granulocytes dramatically reduce protein synthesis and overall synthetic capacity after completing their developmental secretory granule formation.1
By studying mice with hematopoietic-restricted deletion of Xbp1 (Vav1-Cre), we showed that the vast majority of hematopoietic cell types readily compensated for loss of Xbp1. Unexpectedly, we identified a remarkably specific and complete requirement for XBP1 and its upstream activator IRE1α in terminal eosinophil differentiation and eosinophil progenitor survival to the exclusion of highly developmentally related granulocytes such as neutrophils, mast cells and basophils2 (and unpublished observations).
To better understand this phenotypic selectivity, we then evaluated Xbp1 splicing in all relevant hematopoietic progenitors. Murine eosinophil development progresses linearly through the hematopoietic stem cell (LSK), common myeloid progenitor (CMP), granulocyte macrophage progenitor (GMP), eosinophil progenitor (EoP), immature eosinophil and terminally mature eosinophil. Unexpectedly, although only eosinophils were selectively eliminated from Xbp1vav1 mice, Xbp1 splicing was broadly and robustly induced during physiological myelopoiesis, including CMPs, GMPs, EoPs, and immature eosinophils. This lengthy timeframe for IRE1α activation was additionally surprising given that previous reports had demonstrated that robust IRE1α activation during chemically induced ER stress was only maintained for 8-12 hours. Furthermore, we failed to observe concomitant induction of IRE1α-independent target genes during myelopoiesis, suggesting that, like TLR-signaling in macrophages,3 myelopoiesis involves branch-specific UPR activation.
We next explored whether XBP1 was a cell-intrinsic or cell-extrinsic requirement for eosinophil differentiation. Crossing the Xbp1f/f line onto an eosinophil-specific Cre4 (Xbp1EoCre) significantly reduced peripheral eosinophil numbers, and surviving eosinophils had escaped Xbp1 deletion. This suggested that XBP1 was likely required after commitment to the eosinophil lineage rather than for the GMP to EoP transition. We additionally fate mapped eosinophils in Xbp1EoCre mice by crossing them to the mT/mG reporter strain, in which Cre-expressing cells also express GFP. All GFP+ cells in the Xbp1EoCre mice were Siglec-F+ non-deleting eosinophils, indicating that knocking out Xbp1 after commitment to the eosinophil lineage did not divert these cells to alternative hematopoietic lineages (unpublished observations). Mixed bone marrow chimera experiments and in vitro bone marrow-derived eosinophil cultures corroborated this cell-intrinsic function, and development could be rescued by retrovirally overexpressing the spliced Xbp1 isoform, but not an unspliceable form of Xbp1. Collectively these experiments mapped the function of XBP1 to the committed EoP.
Unbiased transcriptomic and bioinformatic analyses on GMPs revealed that Xbp1 deficiency comprehensively reduced the adaptive protein folding capacity of the ER without inducing overt ER stress or affecting fundamental eosinophil developmental signaling networks (Fig. 1). To overcome the intractable technical hurdles involved in sorting sufficient endogenous EoPs from Xbp1vav1 mice, we modeled very early eosinophil differentiation by overexpressing GATA2 in GMPs to enforce eosinophil lineage commitment.5 Through a combination of transcriptomic, ultrastructural, and biochemical techniques we found that the initial ER vulnerabilities present in the GMPs led to massive defects in post-translational maturation of key granule proteins required for survival, and these unresolvable structural defects fed back to indirectly suppress transcription of the critical lineage determining factor Gata1. Suppression of Gata1 was likely responsible for the observed concomitant downregulation of many indispensable granule protein genes such as Prg2, Epx, and various eosinophil-associated ribonucleases. We further observed that bone marrow-derived Xbp1vav1 eosinophils failed to stain with the amyloid-binding dye Thioflavin T (unpublished observations), demonstrating a profound loss of characteristic eosinophil intra-granule amyloid structures6 and corroborating the fundamental defects in granule protein packaging.
Figure 1.
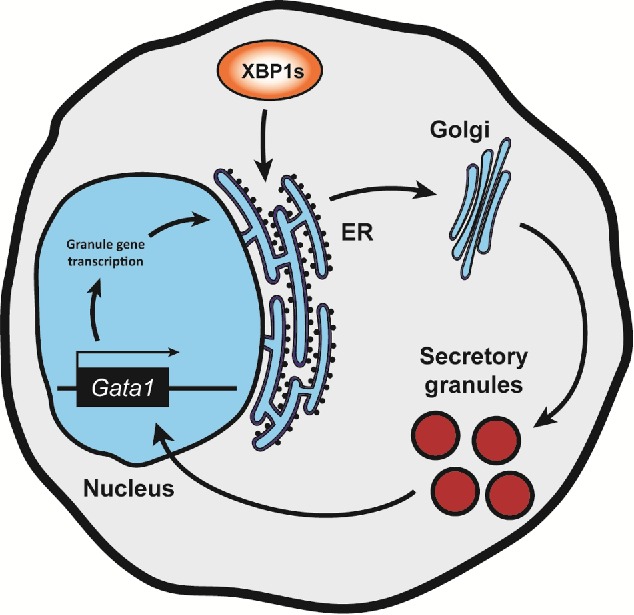
A hypothetical model of the secretory pathway-Gata1 circuit. Gata1 expression is modulated under conditions of suboptimal secretory pathway function and/or inefficient granule formation, as happens in the absence of Xbp1 during eosinophil differentiation. This feedback loop may operate to balance the rate of differentiation with the need to protect the cell from secretory pathway dysfunction.
Based on these observations we postulate that the status of granule formation itself likely act as a rheostat for Gata1 expression in eosinophils, and that this loop is independent from previously reported HSP70-mediated post-transcriptional regulatory mechanisms.7 Future studies are needed to mechanistically dissect how this feedback loop operates, and whether the observed links between granule formation and transcriptional identity are generalizable to varying degrees in other cell types. Taken together, our study presents the first evidence that granulocyte subsets can be distinguished by their differential sensitivities to perturbations in XBP1-mediated secretory pathway function and granule formation. Disentangling this selectivity represents a key challenge for future work, and may yield novel cell biological vulnerabilities that could be exploited therapeutically. Furthermore, this work implicates the IRE1α/XBP1 signaling axis as a potential therapeutic target for eosinophil-mediated diseases such as eosinophilic leukemias and certain subtypes of asthma.
Disclosure of potential conflicts of interest
L.H.G. is on the board of directors of and holds equity in Bristol Myers Squibb Pharmaceutical Company. No other potential conflicts of interest were disclosed.
References
- [1].Bainton DF, et al.. J Cell Biol 1970; 45:54-73. PMID:2108001; http://dx.doi.org/ 10.1083/jcb.45.1.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bettigole SE, et al.. Nat Immunol 2015; 16:829-837. PMID:4577297; http://dx.doi.org/ 10.1038/ni.3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Martinon F, et al.. Nat Immunol 2010; 11:411-418. PMID:3113706; http://dx.doi.org/ 10.1038/ni.1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Doyle AD, et al.. J Leukocyte Biol 2013; 94:17-24. PMID:3685019; http://dx.doi.org/ 10.1189/jlb.0213089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Iwasaki H, et al.. Gen Dev 2006; 20:3010-3021. PMID:1620021; http://dx.doi.org/ 10.1101/gad.1493506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Soragni A, et al.. Mol Cell 2015; 57:1011-1021; http://dx.doi.org/ 10.1016/j.molcel.2015.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Arlet JB, et al.. Nature 2014; 514:242-246; PMID: 25156257; http://dx.doi.org/10.1038/nature13614 [DOI] [PubMed] [Google Scholar]


