Abstract
An emerging strategy for the treatment of monogenic diseases uses genetic engineering to precisely correct the mutation(s) at the genome level. Recent advancements in this technology have demonstrated therapeutic levels of gene correction using a zinc-finger nuclease (ZFN)-induced DNA double-strand break in conjunction with an exogenous DNA donor substrate. This strategy requires efficient nucleic acid delivery and among viral vectors, recombinant adeno-associated virus (rAAV) has demonstrated clinical success without pathology. However, a major limitation of rAAV is the small DNA packaging capacity and to date, the use of rAAV for ZFN gene delivery has yet to be reported. Theoretically, an ideal situation is to deliver both ZFNs and the repair substrate in a single vector to avoid inefficient gene targeting and unwanted mutagenesis, both complications of a rAAV co-transduction strategy. Therefore, a rAAV format was generated in which a single polypeptide encodes the ZFN monomers connected by a ribosome skipping 2A peptide and furin cleavage sequence. On the basis of this arrangement, a DNA repair substrate of 750 nucleotides was also included in this vector. Efficient polypeptide processing to discrete ZFNs is demonstrated, as well as the ability of this single vector format to stimulate efficient gene targeting in a human cell line and mouse model derived fibroblasts. Additionally, we increased rAAV-mediated gene correction up to sixfold using a combination of Food and Drug Administration-approved drugs, which act at the level of AAV vector transduction. Collectively, these experiments demonstrate the ability to deliver ZFNs and a repair substrate by a single AAV vector and offer insights for the optimization of rAAV-mediated gene correction using drug therapy.
Keywords: Zinc-finger nuclease, adeno-associated virus, double-strand break repair, gene targeting, proteasome inhibitor, homologous recombination
INTRODUCTION
Gene therapy is fundamentally aimed at using nucleic acids as reagents to alter cellular behavior and thereby cure disease. The recognition that the phenotype of cells could be readily altered by the introduction of new DNA fragments, made the prospect of gene therapy seem feasible. Over the last several decades, significant work has been invested in translating this research tool into clinical benefit for patients with monogenic diseases as disparate as severe combined immunodeficiency,1 adenosine deaminase deficiency,2 Wiskot--Aldrich syndrome3 and Leber’s congenital amaurosis.4 Such experiments rely on gene addition approaches in which the therapeutic cDNA is expressed from a non-native promoter and is maintained extrachromosomal or integrated into a random chromosomal site. An emerging strategy is to correct the mutant gene at the endogenous locus via homologous recombination, which preserves the integrity/regulation imparted by the native locus (herein referred to as gene targeting).
The spontaneous frequency of gene targeting using plasmid vectors is on the order of 10−6, a frequency too low to translate to clinical use.5,6 However, reports demonstrate that sequence-specific endonucleases can increase the efficiency of gene targeting by 100–50 000-fold,7–13 a frequency that has clinical relevance to a variety of diseases. Currently, there are three major classes of nucleases that can be used to create sequence-specific DNA double-strand breaks (DSBs): homing endonucleases, TALE nucleases and zinc-finger nucleases (ZFNs).14–17 In particular, ZFNs are engineered proteins that fuse zinc finger DNA-binding domains to a C-terminal nuclease domain derived from the type IIS FokI restriction endonuclease.18 A pair of ZFNs bind their cognate recognition sites in a specific orientation thereby allowing the nuclease domain to dimerize and enzymatically create the DNA DSB. Theoretically, a pair of ZFNs can be designed to bind and cleave nearly any site in the human genome including sites of mutated genes that cause disease. Practically, ZFNs have been engineered to a wide range of gene targets and have been shown to stimulate specific gene modification in a wide range of cell types.19 –24
Despite the ‘hit and run’ nature of nuclease-mediated genome modification, the issue of how to introduce the components necessary for gene targeting (the nuclease and donor fragment) remains a challenge, particularly in cell types that are transfected at low efficiency. Moreover, if one were to consider using gene targeting for in vivo therapy, physical, chemical and electrical means of transfection are currently not practically viable options. The use of non-integrating viral vectors for delivery of the gene targeting components is one strategy to overcome the barrier of inefficient transfection. Lombardo et al.22 have demonstrated that integration-defective lentivirus can be an effective method to deliver the gene targeting components as they demonstrated high frequencies of gene targeting in a range of human cell types. A potential alternative to integration-defective lentivirus, is the use of recombinant adeno-associated virus (rAAV) for delivery, which demonstrates enhanced transduction in vitro and in vivo and is currently used in human trials.
rAAV capsids are packaged with a single-strand DNA molecule <5 kb containing transgenic DNA flanked by the viral inverted terminal repeats. The most widely studied serotype is AAV2, but currently there are well over 10 natural described serotypes, with hundreds of variants, which have a broad tropism to transduce dividing or non-dividing cell types both ex vivo and in vivo.25,26 One of the fascinating, and still not fully explained, properties of rAAV is that high efficiencies of gene targeting (up to 1%) have been reported without the induction of a gene-specific DSB using rAAV DNA as the repair substrate.27 In addition, rAAV has been shown to be an effective method of delivery to stimulate gene targeting using homing endonucleases. Both Miller et al.28 and Porteus et al.29 demonstrated that rAAV2 could be used to deliver the homing endonuclease I-SceI and the donor fragment via co-transduction in human cell lines to stimulate high frequencies of gene targeting (>1%). Recently, it was shown that using I-SceI in a different gene targeting system, in different cells (U2OS), could produce targeting frequencies up to 65% using rAAV2 for delivery.30 It was also recently reported that gene targeting was stimulated in cells provided with ZFNs linked by a 2A peptide by plasmid transfections.31,32
In this work, we show that high frequencies of gene targeting (~1%) can be achieved in a human cell line and in mouse model derived fibroblasts using a single AAV6 vector for delivery of both ZFNs and a repair substrate. To do this we demonstrated efficient polypeptide cleavage to discrete ZFNs using a previously described 2A peptide and furin cleavage sequence in a rAAV context.33 This genetic arrangement accommodated a 750 nucleotide DNA repair substrate for a total vector size at the AAV packaging capacity of 4.7 kb. Experiments using a supplementary donor vector suggest that the repair substrate is limiting to a point, at which the efficiency decreases, presumably due to vector transduction competition. Finally, we show that the frequency of gene targeting mediated by rAAV6 transduction, but not plasmid transfection, can be increased by proteasome or histone deacetylase (HDAC) inhibitors. Additionally, an additive effect on gene targeting is observed when cells are treated with both drug regimens.
RESULTS
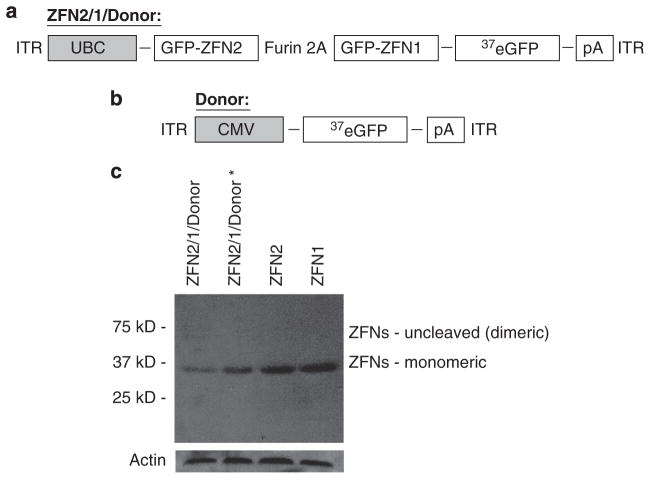
Initially, we created two AAV plasmids for these experiments investigating ZFN-induced gene targeting by rAAV vectors (Figures 1a and b). The first, ‘ZFN2/1/donor’, contains an expression cassette for green fluorescent protein (GFP)-ZFN1 and GFP-ZFN2(refs 20,34) driven by the human ubiquitin C promoter (UBC) in which the two ZFNs are linked by a 2A peptide sequence and a furin cleavage site.33 In addition, this vector contains an ~750 base pair eGFP DNA fragment (truncated to eGFP nucleotide 37; Figure 1 ‘donor’), 5′ of the BgH poly-adenylation sequence, that serves as the donor template for the homologous recombination machinery to correct our previously described defective GFP reporter system.5 The final vector size is ~4.7 kb, which is about the size of the wild-type AAV genome. In addition to this ZFN2/1donor vector, we also created an AAV construct containing only the repair substrate (donor; Figure 1b).
Figure 1.
Schematic representations of the genomic contents of AAV6 ZFN2/1/D and AAV6 donor. (a) The genome of the AAV6 viral vector ZFN2/1/D, which contains DNA encoding two GFP ZFNs separated by a 2A peptide driven by the ubiquitin C promoter, and includes the eGFP gene truncated at nucleotide 37 as a donor substrate in gene targeting by homologous recombination. (b) The genome of the AAV6 viral vector donor, which contains the same truncated, non-functional form of eGFP as in (a), and is driven by the cytomegalovirus promoter. (c) Western blot from calcium phosphate transfection of ZFN2/1/D construct in the AAV backbone (lane 1), the ZFN2/1/donor* construct in a lentiviral backbone (lane 2), the ZFN2 construct in a lentiviral backbone (lane 3) and the ZFN1 construct in a lentiviral backbone (lane 4). Note that most of the protein from lane 1 is present at the monomeric weight (37 kD marker).
The ZFN co-synthesis peptide cleavage system should allow for production of two ZFNs from a single polypeptide, thus poly-peptide cleavage into discrete ZFNs was investigated by western blotting (Figure 1c). Transfection of two different viral vector plasmids containing the ZFN expression cassette demonstrated that the ZFNs were efficiently processed into monomeric units, while no unprocessed molecules were observed (Figure 1b). Next, we produced rAAV particles harboring the described ZFN +donor genome. AAV serotype 6 was used in these experiments as we found in general, this capsid is most efficient for primary cell transduction (Ellis et al., submitted). We then investigated the ability of the produced rAAV6 particles to stimulate gene targeting in two different cell types. The first is a human HEK 293 cell line ‘293/GFP*’ that contains a single chromosomal integrated copy of a mutated GFP gene.5 The other cell type investigated herein is a primary mouse fibroblast, immortalized by using the standard NIH 3T3 protocol, which contains the same defective GFP gene correction reporter integrated into the ROSA26 locus.20 We chose to use mouse fibroblasts that are heterozygous for the mutated GFP transgene cassette (GFP*) to allow a better comparison of targeting with our single copy 293 integrant line (theoretically the same copy number of the GFP* reporter). In these systems, gene targeting results in the correction of a small insertion in the integrated GFP* gene and converts the cell from being phenotypically GFP negative to GFP positive. The frequency of correction can then be quantitated by flow cytometry.
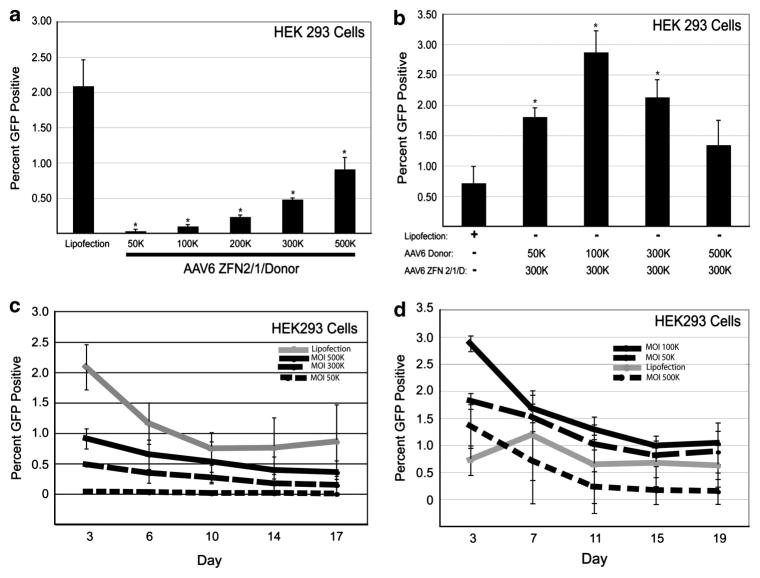
Initially, we infected the 293/GFP* cells with increasing amounts of rAAV6-ZFN2/1/donor and found a dose-dependent increase in gene targeting up to 0.91% (Figure 2a) at a multiplicity of infection (MOI) of 500 000 vector genomes per cell ‘500K’. Although, this MOI seems extremely high, it is important to note these particles use the AAV6 capsid, which is 5–10-fold deficient for transduction on this cell type compared with AAV2. Control experiments demonstrated that ZFN expression in the absence of the repair substrate, or the repair substrate alone, does not produce GFP-positive cells, which we consistently see using these ZFNs and the GFP* reporter model5,29 (Supplementary Figure S1). Previously, we have found by transfection that targeting frequencies can diminish with excessive ZFN expression and with limiting DNA donor fragment amounts.35 Here, we found that if we fix the MOI of rAAV6 ZFN2/1/donor at 300K and supplement the infection with increasing amounts of the rAAV6 donor virus (an MOI from 50 to 500K), we increased the frequency of gene targeting several fold to ~3% of treated cells (Figure 2b). Importantly, we performed this experiment in the presence of donor virus alone at increasing concentrations and did not observe a single gene targeting event in over 100 000 analyzed cells, again providing evidence of the necessity of a DNA DSB to stimulate gene targeting (Figure 1b, Supplementary Figure S1). In this particular experiment the targeting frequency is higher than that achieved by transfection, however, it is comparable to what we routinely achieve by transfection in other experiments in this cell line (Figures 2a and b, data not shown). Thus, supplementing rAAV6 ZFN2/1/donor transduction with rAAV6 donor vectors resulted in increased gene targeting to a point, then the efficiency decreased thereafter, possibility as a result of transduction competition of the ZFN2/1/donor and donor only vectors.
Figure 2.
ZFN-mediated gene targeting in HEK 293 cells delivered by AAV6. (a) Gene targeting in HEK 293 cells with increasing MOIs of AAV6 ZFN 2/1/D virus, analyzed on day 3 for GFP by flow cytometry. (b) Gene targeting in HEK 293 cells with increasing MOIs of AAV6 donor virus and a constant MOI of 300K for the AAV6 ZFN 2/1/D virus, analyzed on day 3 for GFP by flow cytometry. (c) Kinetic analysis of gene targeting in HEK 293 cells with increasing MOIs of AAV6 ZFN 2/1/D virus analyzed by flow cytometry at the indicated time points post infection. (d) Gene targeting in HEK 293 cells with increasing MOIs of AAV6 donor virus and a constant MOI of 300K for the AAV6 ZFN 2/1/D virus, analyzed at the indicated time points for GFP by flow cytometry. *Significantly different compared with the lipofected sample. n = 3, P < 0.05 (for c and d: only evaluated at the last time point between lipofection and the infected population closest to the lipofected value).
To determine the toxicity of rAAV6-mediated ZFN-stimulated gene targeting, we performed a time course in which the percentage of GFP-positive cells was determined over time (Figures 2c and d). When high MOIs of the rAAV6 ZFN2/1/donor virus were used, there was a greater fall in the number of GFP-positive cells over time, suggesting greater toxicity (Figure 2c). We also examined the persistence of GFP-positive cells over time with an MOI of 100 and 200K and found the same trend (data not shown). A similar drop in the percentage of GFP-positive cells was observed after transfection, suggesting that the toxicity was not entirely the result of rAAV6 infection. We examined the persistence of GFP-positive cells when we supplemented rAAV6 ZFN2/1/donor with rAAV6 donor and found a similar decrease in GFP-positive cells over time (Figure 2d). The maximal targeting efficiency was achieved, however, at both short and long time points using rAAV6 ZFN2/1/donor at an MOI of 300K supplemented with rAAV6 donor at 100K (Figure 2b). For purposes of clarity the time course of rAAV6 donor at an MOI of 300K was left out, but the trend was similar (data not shown). When we supplemented with a high MOI of rAAV6 donor, which does not express a nuclease, we observed increased toxicity, suggesting that high a MOI of rAAV6 itself has some degree of toxicity in HEK 293 cells. In summary, by using two AAV6 vectors simultaneously, gene targeting efficiencies at both early and later time points can be achieved that are similar to the values obtained with standard transfection techniques.
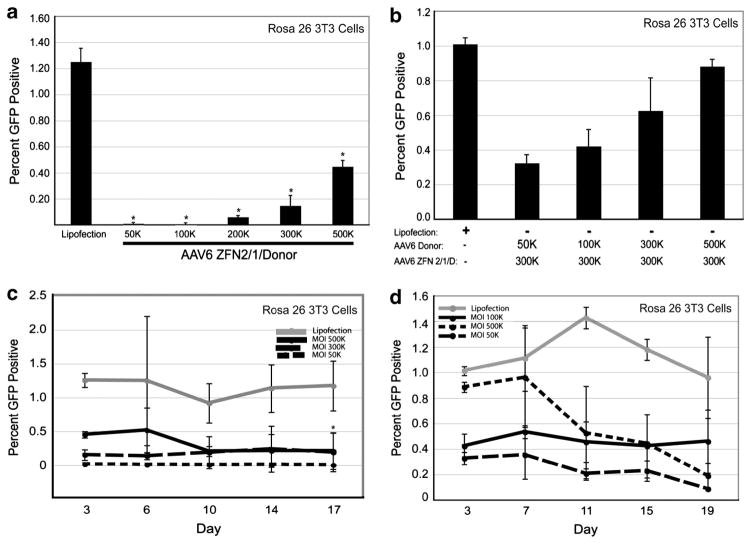
To determine whether rAAV6 delivery of the gene targeting components also stimulated DSB repair in a primary cell type, we used the same rAAV6 viral vectors to transduce immortalized fibroblasts isolated from the mouse model.20 These cells were infected with increasing amounts of rAAV6 ZFN2/1/donor and a dose-dependent increase in gene targeting was observed with a maximal frequency of 0.45% at an MOI of 500K (Figure 3a). Again the repair substrate was deemed limiting as a 10-fold supplementation with the rAAV6 donor vector increased gene targeting threefold for a targeting frequency of ~0.9% (Figure 3b). This value was nearly equivalent to the efficiency of gene targeting achieved by plasmid transfection (Figure 3b). In the time course experiments in the immortalized mouse fibroblast GFP* cells, we found that the percentage of GFP-positive cells did not change significantly over time after transfection by lipofectamine (Figures 3c and d). In contrast, we found that the percentage of GFP-positive cells decreased over time after infection with rAAV6 vectors (Figures 3c and d). The decrease in GFP-positive cells directly correlated to the vector MOI. In particular, the with the rAAV donor virus, suggesting that something about rAAV6 transduction of this cell type, rather than the in the viral vector, was causing some toxicity.
Figure 3.
ZFN-mediated gene targeting in Rosa 26 3T3 cells delivered by rAAV6. (a) Gene targeting in Rosa 26 3T3 cells with increasing MOIs of rAAV6 ZFN 2/1/D virus, analyzed on day 3 for GFP by flow cytometry. (b) Gene targeting in Rosa 26 3T3 cells with increasing MOIs of rAAV6 donor virus and a constant MOI of 300K for the rAAV6 ZFN 2/1/D virus, analyzed on day 3 for GFP by flow cytometry. (c) Kinetic analysis of gene targeting in Rosa 26 3T3 cells with increasing MOIs of rAAV6 ZFN 2/1/D virus, analyzed for GFP by flow cytometry. (d) Kinetic analysis of gene targeting in Rosa 26 3T3 cells with increasing MOIs of rAAV6 donor virus and a constant MOI of 300K for the rAAV6 ZFN 2/1/D virus. GFP-positive cells were then analyzed at the indicated time points by flow cytometry.*Significantly different compared with the lipofected sample. n = 3, P < 0.05 (for c and d: only evaluated at the last time point between lipofection and the infected population closest to the lipofected value).
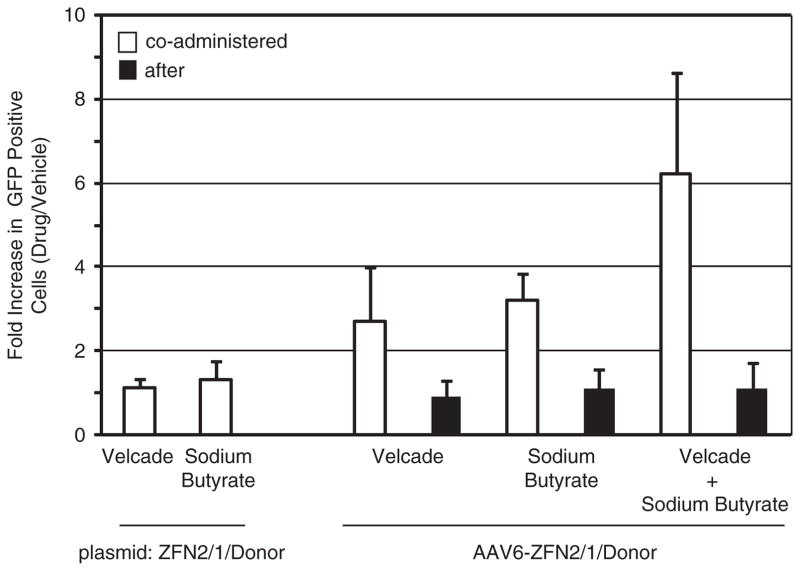
It is well established that proteasome inhibition increases transduction in different cell types in vitro and in mouse dog models in vivo.36 – 43 The mechanism by which inhibition increases rAAV transduction remains unknown, ever, evidence suggests that AAV particle trafficking to nucleus is more efficient. In addition, HDAC inhibitors also been reported to increase rAAV transduction by an mechanism.35 To determine whether proteasome or inhibitors also increase rAAV-mediated gene targeting, performed transduction experiments using ZFN2/1/donor in HEK 293/GFP* cell line in the presence of the bortezomib sodium butyrate. The results demonstrate a two- to increase in GFP +cells in the presence of either drug (Figure Addition of bortezomib or sodium butyrate 18 h following administration had no significant effect on the percentage GFP +cells in accordance with the notion that the drugs particle trafficking (Figure 4).36 Likewise, these drugs had no on plasmid-mediated gene targeting, which is consistent with earlier report testing the effect of proteasome inhibitors on targeting (Figure 4).35 Next, the proteasome and HDAC were added in combination to the cells at the time of administration. An additive sixfold increase in percentage GFP +cells was noted 48 h post treatment (Figure 4). collective results suggest that methods to enhance rAAV transduction also result in increased rAAV-mediated gene ing. In addition, the additive effect noted with the drug indicates that these drugs likely target different steps or mechanisms to enhance rAAV transduction.
Figure 4.
Enhancement of gene targeting using Food and Drug Administration-approved drugs. HEK 293 cells were transduced with rAAV6 ZFN 2/1/donor at 100 000 viral genomes per cell. At the time of vector addition (co-administered) or 18 h post vector addition, cells were treated with the indicated drug (bortezomib 2 μM; sodium butyrate 1.5 μM). Two days following vector addition, cells were harvested and GFP +cells were quantitated by flow cytometry. The value determined in the presence of the drug was divided by the value determined in the absence of the drug and the data are presented as a fold change. Plasmid experiments relied on the same general strategy, however, in these cases the drug was given 4 h pos-transfection.
DISCUSSION
Using homologous recombination to correct disease-causing mutations or to precisely target transgene insertion, theoretically, is an ideal gene therapy approach. The beneficial aspects of this strategy, to correct deleterious mutations thus making the cell genetically wild type, include the following: (1) the proper copy number of the gene is maintained, (2) endogenous gene regulation is preserved and (3) elimination of the concerns of the insertional activation of an oncogene or the insertional inactivation of a tumor suppressor gene by a viral integration. This strategy falls under the general category of ‘non-viral’ gene therapy because no viral remnants are left in the cell when the process is completed. However, low levels of rAAV integration can occur,44–46 however, thus far, rAAV has shown safety in clinical trials.47 –52 To achieve therapeutically relevant frequencies of gene targeting by homologous recombination in the human genome, at least two different components need to be introduced into cells: (1) a donor DNA molecule that serves as the repair template for the homologous recombination machinery and (2) a nuclease to create a site-specific genomic DNA DSB to stimulate homologous recombination at (or near) that site. In this work, we present a generally applicable context in which all of these genetic elements, as well a promoter and a poly-adenylation signal, can be packaged in a single AAV vector. Consistently, we have demonstrated that the ZFN2/Furin/T2A/ZFN1 expression cassette is efficiently processed to monomeric ZFNs, which are functional for gene targeting using the provided repair substrate. This methodology guarantees that every cell receiving ZFNs also receives the desired repair molecule and avoids concerns associated with multiple particle transduction, especially when relatively high particle numbers are necessary for efficient gene targeting.
In regard to nucleic acid delivery, there are several methods that can be used to deliver the gene targeting components. Chemical transfection and electroporation have been used in a variety of different cell types by multiple groups to deliver genes encoding nucleases and to stimulate homologous recombination.21,53,54 Although chemical- and electroporation-based techniques have been widely used and can accommodate large genetic payloads, they can be ineffective or highly toxic in primary cell types (data not shown). An alternative strategy is to use viral vectors to deliver the components necessary for gene targeting by homologous recombination. Viruses have evolved to efficiently transduce a wide variety of different primary cell types with minimal cytotoxic effect and have demonstrated success for gene delivery in vitro, ex vivo and in vivo. Deleterious clinical consequences using viral vectors that rely on host chromosome integration have emphasized the importance of using non-integrating vectors for delivery. Integration-defective lentivirus vectors have been successfully used to deliver the gene targeting components and have demonstrated stimulation of gene targeting in a variety of different cell types.22,55,56 An alternative to using integration-defective lentivirus is to use rAAV as a viral vector for delivery, which comparatively, is more efficient for transduction in vivo. Previously, high frequencies of gene targeting were achieved using the rAAV2 to deliver I-SceI and the donor template to cells either as distinct single-strand or as self-complementary genomes.5,29,57 An inherent problem with these previous approaches is the possibility of single vector transduction, for instance some cells are transduced by the donor only vector, which is inefficient for gene targeting (Supplementary Figure S1). In addition, transduction only by the endonuclease vector is a more serious problem as induced DSBs are most often repaired by non-homologous end joining, which is associated with unwanted deletions and mutations. In this work, we eliminated these concerns by engineering a single AAV6 vector that effectively delivers both the endonuclease and repair substrate into mouse and human cells to stimulate ZFN-mediated gene targeting.
Regarding the efficiency of gene targeting, we found that titrating the amount of AAV6 used to transduce cells was important for maximizing the frequency of gene correction. For instance, in both cell types the number of GFP +cells directly correlated to the amount of AAV-ZFN2/1/donor used for infection. As seen with most ZFN-mediated targeting experiments, the efficiency of the gene correction decreased to a stable level over time.35 This result herein, which was more obvious in the human cell line (Figure 2), could be explained in several ways, as well as a by a combination of effects including: (i) ZFN toxicity and (ii) rAAV-induced toxicity at high particle titers. This trend, a loss of GFP +cells over time, was also noted for the transfection experiments in the human cells (Figure 2). In addition to titration of the AAV-ZFN2/1/donor cassette, we also noted that the use of additional AAV-donor particles increased gene correction in both cell types about threefold. This result suggests that the amount of repair DNA is limiting in these gene-targeting experiments. However, too much AAV-donor actually decreased the gene targeting efficiency, an effect we speculate is due to the inherent problem with co-transduction strategies: AAV vector competition (Figure 2b). The data herein demonstrate that, in general, increased amounts of the AAV-ZFN 2/1/donor vector resulted in increased targeting and that supplemental AAV-donor transduction further increases rAAV transduction until, perhaps, particle competition becomes inhibitory. This interpretation is further supported by no inhibitory effect on gene targeting using AAV-donor vector supplementation in the mouse model derived cells, which are not efficiently transduced by AAV6.
Our finding also highlights the importance of the intracellular metabolism of the AAV6 vector and noted that the frequency of targeting was increased 2- to threefold in the presence of either proteasome or HDAC inhibitors. In previous work, we have demonstrated that proteasome inhibition increases rAAV trans-duction in cell culture and in small and large animal models.36,37 Although the mechanism by which proteasome inhibition increases rAAV transduction remains unknown, it appears to elicit its effect on the nuclear trafficking of rAAV particles.36 Consistently, we noticed no effect of the proteasome inhibitor when the drug was administered 18 h post transduction or on transfected plasmid DNA. Only a few reports demonstrate increased rAAV vector transduction in the presence of sodium butyrate and again, the mechanism by which this occurs remains elusive.58 Similar to the results with the proteasome inhibitor, sodium butyrate did not alter gene targeting levels when added 18 h post transduction or when using transfected plasmid components. Thus, the enhancement of gene targeting by either of the drug inhibitors is specific to the AAV vector and is likely elicited at the level of rAAV particle trafficking to, and perhaps, within the nucleus. Given this, the demonstration of an additive effect on gene targeting in the presence of both drugs is interesting, and suggests that at least partially different AAV transduction pathways exist and are influenced separately by each drug. This observation is currently under further investigation.
The objective of this work was to devise and validate a safer and more efficient rAAV-mediated gene targeting strategy at 2 levels: (i) a universal single rAAV vector capable of inducing a specific DNA DSB and repairing it with a provided DNA substrate via HR and (ii) optimization of rAAV transduction conditions using clinical drugs that also, secondarily, result in increased targeted gene repair. Both of these levels of optimization are applicable to dividing primary cell cultures as well as particular tissues in vivo. These enhancements become particularly important for transduction of less permissive cells or a great number of cells (that is, CD34 +cells or the liver, respectively), in which instances of multiple vector transduction of a single cell are unlikely. The transition of these vector enhancements to ex vivo gene targeting is underway and was the rationale for using the AAV6 capsid herein, which is generally efficient for stem cell transduction (Ellis et al., submitted). However, several complications exist preventing simple data interpretation in these experiments. For example, our unpublished data of rAAV-mediated gene targeting in mouse mesenchymal stem cells reveal that gene correction does occur, however, it is not clear if naïve mesenchymal stem cells, or partially differentiated mesenchymal stem cell derivatives, undergo the event. To further complicate result interpretation, a significant amount of toxicity was observed posing the question of whether naïve mesenchymal stem cells would rather tolerate and repair a DSB or induce apoptosis, a decision that likely reflects the recognition specificity of the endonuclease among other factors. Currently, we are in the process of elucidating the DSB repair capacity of multiple primary cell types at varying levels of differentiation.
In summary, we demonstrate for the first time ZFN-mediated gene targeting using rAAV transduction in both human and mouse cells. To do this, we engineered a universal vector format to include all of the necessary components in a single vector by using a 2A peptide sequence, which is significant because the use of an internal ribosome entry site sequence or multiple promoters would have put the genome above the packaging restriction size for AAV capsids. AAV vectors based on this format eliminate the associated safety concerns of a multiple transduction strategy and are applicable to both in vitro and in vivo applications.59 In addition, this is the first report to demonstrate that Food and Drug Administration-approved drugs can be used solely, or in combination, to increase AAV-mediated gene targeting up to sixfold. These collective enhancements on rAAV vector design and application decrease the relatively high required vector dose for safe and efficient gene targeting. However, several concerns remain, such as the efficiency of stem/dividing cells to undergo homologous recombination (instead of apoptosis), that should be addressed before the transition of technology for human therapy.
MATERIALS AND METHODS
DNA manipulations and cloning
AAV constructs
The ZFN1/2/donor construct was made by PCR of the UbC promoter from pUB6/V5-His A (Invitrogen, Carlsbad, CA, USA) in which the 5′-primer hybridized with the 5′-end of the UbC and contained a NotI site (5′-AGGACCGCGGCCGCGGCCTCCGCGCCGGGTTTTGG-3′) and the 3′-primer hybridized with he 3′ end of the UbC promoter and contained a BamHI site (5′-AGGACCGGATCCGAGCTCGGTACCAAGCTTCGT-3′). This PCR fragment was then digested with NotI and BamHI and ligated into the pAAVMCS (Stratagene, La Jolla, CA, USA) construct digested with BamHI and partially digested with NotI. This construct already contained the GFP-ZFN2 2A GFPZFN1(ref. 37)-eGFP fragment, which was generated using by PCR/cloning of a previously described furin (RNRR) T2A linker sequence using the following primer sequences: M480-A 5′-AGGCCCTGCAGGGGGG CCGGGGTTCTCCTCCACGTCGCCGCAGGTCAGCAGGCTGCCCCTG-3′, M480-B 5′-GTCAGCAGGCTGCCCCTGCCCTCGCCGCTGCCGCTGCGGCGCTTGCGAAAG TTTATCTCGCCGTTATT-3′, M480-C 5′-AGGCCTCGAGCTAAAAGTTTATCTCGC CGTTATT-3′, SNP21-F 5′-TCGATCGGATCCTAGGGATAACAGGGTAATATGGA CTACAAAGACGATGAC-3′, SNP38-F 5′-TCGATCCCTGCAGGCGACTACAAAGA CGATGAC-3′ (details available upon request.33 Briefly, the GFP-ZFN2 2A GFP-ZFN1(ref. 37)-eGFP was cloned in using BamHI and XhoI sites of the pAAVMCS vector. The donor construct was made by digestion of a plasmid containing a truncated form of eGFP (37-eGFP) discussed previously, and ligated into the multiple cloning site of the pAAVMCS vector with BamHI and XhoI.
Cell culture
Cell culture experiments were performed in either stably transfected HEK 293 cells containing the GFP gene targeting construct described previously5 or in 3T3 cells derived from a transgenic mouse containing the GFP gene targeting construct also described previously20. As previously reported, the GFP gene targeting construct contains a 35 nucleotide insertion within the egfp coding sequence: 5′-CGACGGCAACTACAAG ACCTAAGCTCTCGAGATTACCCTGTTATCCCTAAGCTTCGCGCCGAGGTGAAGT-3′ (italics are egfp coding sequence flanking the insertion sequence, the I-SceI site is underlined). The cells were cultured in Dulbecco’s modified Eagle’s medium (Media Tech, Manassas, VA, USA) supplemented with 10% bovine growth serum (Hyclone, Logan, UT, USA), 2 mM L-glutamine, 100 IU ml−1 penicillin and 100 mg ml−1 streptomycin. The cultures were grown in a humidified incubator at 37 °C with 5% CO2.
Western blot
About 100 000 HEK 293T cells were calcium phosphate transfected with 100 ng of the ITR-UBC-ZFN2/1/donor-ITR, LTR-UBC-ZFN2/1/donor-LTR, LTR-UBC-ZFN2-LTR or LTR-UBC-ZFN1-LTR construct in triplicate. Transfection efficiency was about 40% determined by separate transfection with eGFP and analyzed by flow cytometry on a fluorescence-activated cell sorting Calibur (Becton-Dickerson, San Jose, CA, USA). Cells were harvested at 48 h post transfection and triplicates were combined. Equal amounts of total lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, wet transferred to polyvinylidene difluoride membranes and incubated with specific antibodies. ZFNs were detected with an anti-Flag M2 monoclonal antibody (1:10 000, Sigma-Aldrich, St Louis, MO, USA), and β-actin was detected with a rabbit anti-actin antibody (1:5 000, Sigma-Aldrich). The blots were then incubated with horseradish peroxidase-conjugated secondary antibodies and visualized using western blotting luminal reagent (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Virus production
rAAV production
Single-strand rAAV was generated using the triple transfection method60 in plated HEK 293T cells. All transfections for rAAV6 production contained an adenovirus helper plasmid (pXX680) and the pXR6 plasmid containing the AAV Rep2 and capsid six genes.60 In addition, a third plasmid was included that contains the desired AAV genome flanked by the inverted terminal repeats of AAV2. For the experiments herein, two such plasmids were used, pZFN2/1/donor and pdonor, which are described in the DNA manipulations and cloning section above. Three days post transfection, nuclei were purified from harvested cells, lysed by sonication and applied to a cesium chloride gradient as described.59,60 Following an overnight spin at 65 000 r.p.m, gradient fractions were investigated for AAV particles containing the desired genome. rAAV peak fractions were then pooled, dialyzed in phosphate-buffered saline and stored at −80 °C. The number of genome containing particles was determined by quantitative PCR using the following GFP primer set: forward primer, 5′-AGCAGCACGACTTCTTCAAGTCC-3′ and the reverse primer 5′-TGTAGTTGTACTCCAGCTTGTGCC-3′.
Gene targeting by infection or lipofection
For all gene targeting experiments, at hour 0, 10 000 cells per well were split into a 24-well plate in 500 μl of media and cells were infected at the indicated MOI. For lipofection experiments, Lipofectamine 2000 (Invitrogen) was added to the cells with either 800 ng eGFP (transfection control) or 100 ng of ZFN2/1/D and 700 ng of donor (a typical amount and ratio for optimum targeting frequency). The cells were then maintained in the 37 °C incubator until time of analysis. If a time course was done, of the cells were analyzed for GFP expression and of the cells were replated for further expansion. For gene targeting experiments, media were changed at hour 6. For the gene targeting experiments with the proteasome inhibitor MG-132 (Sigma-Aldrich), drug was resuspended in dimethyl sulfoxide for a stock concentration of 1 mM and was then added to each well to the indicated final concentration at the time of infection. In the case of the MG-132 lipofection experiments, drug was not added until hour 2 of the experiment. In all MG-132 experiments the media were changed after 4 h of exposure and drug was not added back.
Measurement of gene targeting using the GFP system
For all gene-targeting experiments, both with HEK 293 cells and 3T3 cells, the cells were harvested and then analyzed on a fluorescence-activated cell sorting Calibur (Becton-Dickerson) for GFP expression. Each condition was analyzed in triplicate. Typical transfection efficiency by Lipofectamine 2000 was 50–70%.
Supplementary Material
Acknowledgments
We thank Kelley Ellis for her helpful review and comments. This work was funded by the Northwest Genome Engineering Consortium (NGEC) to support MH. Work in the Porteus lab is supported by funds from the state of Texas, a career development award from the Burroughs-Wellcome fund, the Amon Carter Foundation and grant PN2EY018244 supporting a Nanomedicine Development Center through the Director’s Common Fund.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Fischer A, Hacein-Bey-Abina S, Cavazzana-Calvo M. 20 years of gene therapy for SCID. Nat Immunol. 2010;11:457– 460. doi: 10.1038/ni0610-457. [DOI] [PubMed] [Google Scholar]
- 2.Cappelli B, Aiuti A. Gene therapy for adenosine deaminase deficiency. Immunol Allergy Clin North Am. 2010;30:249– 260. doi: 10.1016/j.iac.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Qasim W, Gaspar HB, Thrasher AJ. Progress and prospects: gene therapy for inherited immunodeficiencies. Gene Therapy. 2009;16:1285– 1291. doi: 10.1038/gt.2009.127. [DOI] [PubMed] [Google Scholar]
- 4.Liu MM, Tuo J, Chan CC. Gene therapy for ocular diseases. Br J Ophthalmol. 2011;95:604– 612. doi: 10.1136/bjo.2009.174912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 6.Sedivy JM, Dutriaux A. Gene targeting and somatic cell genetics--a rebirth or a coming of age? Trends Genet. 1999;15:88– 90. doi: 10.1016/s0168-9525(98)01689-8. [DOI] [PubMed] [Google Scholar]
- 7.Brenneman M, Gimble FS, Wilson JH. Stimulation of intrachromosomal homologous recombination in human cells by electroporation with site-specific endonucleases. Proc Natl Acad Sci USA. 1996;93:3608– 3612. doi: 10.1073/pnas.93.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968– 1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070– 4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096– 8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sargent RG, Brenneman MA, Wilson JH. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267– 277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smih F, Rouet P, Romanienko PJ, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012– 5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taghian DG, Nickoloff JA. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol Cell Biol. 1997;17:6386– 6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757– 761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143– 148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 16.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther. 2007;7:49– 66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 17.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967– 973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 18.Smith J, Berg JM, Chandrasegaran S. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 1999;27:641– 681. doi: 10.1093/nar/27.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll D, Beumer KJ, Trautman JK. High-efficiency gene targeting in Drosophila with zinc finger nucleases. Methods Mol Biol. 2010;649:271– 280. doi: 10.1007/978-1-60761-753-2_17. [DOI] [PubMed] [Google Scholar]
- 20.Connelly JP, Barker JC, Pruett-Miller S, Porteus MH. Gene correction by homologous recombination with zinc finger nucleases in primary cells from a mouse model of a generic recessive genetic disease. Mol Ther. 2010;18:1103– 1110. doi: 10.1038/mt.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839– 847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298– 1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 23.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808– 816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646– 651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 25.Michelfelder S, Trepel M. Adeno-associated viral vectors and their redirection to cell-type specific receptors. Adv Genet. 2009;67:29– 60. doi: 10.1016/S0065-2660(09)67002-4. [DOI] [PubMed] [Google Scholar]
- 26.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073– 1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 27.Hendrie PC, Russell DW. Gene targeting with viral vectors. Mol Ther. 2005;12:9– 17. doi: 10.1016/j.ymthe.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Miller DG, Petek LM, Russell DW. Human gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol Cell Biol. 2003;23:3550– 3557. doi: 10.1128/MCB.23.10.3550-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porteus MH, Cathomen T, Weitzman MD, Baltimore D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol Cell Biol. 2003;23:3558– 3565. doi: 10.1128/MCB.23.10.3558-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gellhaus K, Cornu TI, Heilbronn R, Cathomen T. Fate of recombinant adeno-associated viral vector genomes during DNA double-strand break-induced gene targeting in human cells. Hum Gene Ther. 2010;21:543– 553. doi: 10.1089/hum.2009.167. [DOI] [PubMed] [Google Scholar]
- 31.Dekelver RC, Choi VM, Moehle EA, Paschon DE, Hockemeyer D, Meijsing SH, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133– 1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orlando SJ, Santiago Y, Dekelver RC, Freyvert Y, Boydston EA, Moehle EA, et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S, Cohen CJ, Peng PD, Zhao Y, Cassard L, Yu Z, et al. Development of optimal bicistronic lentiviral vectors facilitates high-level TCR gene expression and robust tumor cell recognition. Gene Therapy. 2008;15:1411– 1423. doi: 10.1038/gt.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16:707– 717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- 35.Pruett-Miller SM, Reading DW, Porter SN, Porteus MH. Attenuation of zinc finger nuclease toxicity by small-molecule regulation of protein levels. PLoS Genet. 2009;5:e1000376. doi: 10.1371/journal.pgen.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson JS, Samulski RJ. Enhancement of adeno-associated virus infection by mobilizing capsids into and out of the nucleolus. J Virol. 2009;83:2632– 2644. doi: 10.1128/JVI.02309-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monahan PE, Lothrop CD, Sun J, Hirsch ML, Kafri T, Kantor B, et al. Proteasome inhibitors enhance gene delivery by AAV virus vectors expressing large genomes in hemophilia mouse and dog models: a strategy for broad clinical application. Mol Ther. 2010;18:1907– 1916. doi: 10.1038/mt.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denby L, Nicklin SA, Baker AH. Adeno-associated virus (AAV)-7 and -8 poorly transduce vascular endothelial cells and are sensitive to proteasomal degradation. Gene Therapy. 2005;12:1534– 1538. doi: 10.1038/sj.gt.3302564. [DOI] [PubMed] [Google Scholar]
- 39.Douar AM, Poulard K, Stockholm D, Danos O. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J Virol. 2001;75:1824– 1833. doi: 10.1128/JVI.75.4.1824-1833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan D, Yue Y, Yan Z, Yang J, Engelhardt JF. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J Clin Invest. 2000;105:1573– 1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn JD, Hui D, Downey HD, Dunn D, Pien GC, Mingozzi F, et al. Proteasome inhibitors decrease AAV2 capsid derived peptide epitope presentation on MHC class I following transduction. Mol Ther. 2010;18:135– 142. doi: 10.1038/mt.2009.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jennings K, Miyamae T, Traister R, Marinov A, Katakura S, Sowders D, et al. Proteasome inhibition enhances AAV-mediated transgene expression in human synoviocytes in vitro and in vivo. Mol Ther. 2005;11:600– 607. doi: 10.1016/j.ymthe.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 43.Yan Z, Zak R, Luxton GW, Ritchie TC, Bantel-Schaal U, Engelhardt JF. Ubiquitina-tion of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J Virol. 2002;76:2043– 2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 45.Kay MA, Nakai H. Looking into the safety of AAV vectors. Nature. 2003;424:251. doi: 10.1038/424251b. [DOI] [PubMed] [Google Scholar]
- 46.Nakai H, Montini E, Fuess S, Storm TA, Grompe M, Kay MA. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat Genet. 2003;34:297– 302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
- 47.Kaplitt MG, Feigin A, Tang C, Fitzsimons HL, Mattis P, Lawlor PA, et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson’s disease: an open label, phase I trial. Lancet. 2007;369:2097– 2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- 48.Maguire AM, High KA, Auricchio A, Wright JF, Pierce EA, Testa F, et al. Age-dependent effects of RPE65 gene therapy for Leber’s congenital amaurosis: a phase 1 dose-escalation trial. Lancet. 2009;374:1597– 1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr, Mingozzi F, Bennicelli J, et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N Engl J Med. 2008;358:2240– 2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muramatsu S, Fujimoto K, Kato S, Mizukami H, Asari S, Ikeguchi K, et al. A phase I study of aromatic L-amino acid decarboxylase gene therapy for Parkinson’s disease. Mol Ther. 2010;18:1731– 1735. doi: 10.1038/mt.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simonelli F, Maguire AM, Testa F, Pierce EA, Mingozzi F, Bennicelli JL, et al. Gene therapy for Leber’s congenital amaurosis is safe and effective through 1. 5 years after vector administration. Mol Ther. 2010;18:643– 650. doi: 10.1038/mt.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun X, Pawlyk B, Xu X, Liu X, Bulgakov OV, Adamian M, et al. Gene therapy with a promoter targeting both rods and cones rescues retinal degeneration caused by AIPL1 mutations. Gene Therapy. 2010;17:117– 131. doi: 10.1038/gt.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Kingston RE, Chen CA, Okayama H. Calcium phosphate transfection. Curr Protoc Immunol. 2001;Chapter 10(Unit 10 13):10.13.1–10.13.9. doi: 10.1002/0471142735.im1013s31. [DOI] [PubMed] [Google Scholar]
- 54.Rao NM. Cationic lipid-mediated nucleic acid delivery: beyond being cationic. Chem Phys Lipids. 2010;163:245– 252. doi: 10.1016/j.chemphyslip.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 55.Cornu TI, Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol Ther. 2007;15:2107– 2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- 56.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200– 1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 57.Hirsch ML, Green L, Porteus MH, Samulski RJ. Self-complementary AAV mediated gene targeting and enhances endonuclease delivery for double-strand break repair. Gene Therapy. 2010;19:1175– 1180. doi: 10.1038/gt.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong X, Tian W, Wang G, Dong Z, Sen W, Zheng G, et al. Establishment of AAV reverse infection-based array. PloS One. 2010;5:e13479. doi: 10.1371/journal.pone.0013479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217– 221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grieger JC, Choi VW, samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc. 2006;1:1412– 1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.