Abstract
Lipid droplets (LDs) are ubiquitous intracellular structures whose formation, growth, and maintenance are highly regulated (Wang et al., 2013; Ranall et al., 2011; Goodman, 2009). Lipid metabolism and droplet dynamics are of considerable interest to agriculture, biofuel production, viral pathology, nutrition, and cancer biology (Walther and Farese, 2009; Liu et al., 2010). Accumulation of fatty acids and neutral lipids in nonadipose tissues is cytotoxic (Kourtidis et al., 2009). BODIPY 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene) is the standard dye to study LDs within adipocytes. BODIPY 493/503 contains a nonpolar structure that, upon binding to neutral lipid, emits a green fluorescence signal with a narrow wavelength range, making it an ideal fluorophore for multi-labeling experiments. The hydrophobic nature of the dye molecules promotes rapid entry into the nonpolar environment of LDs (Listenberge and Brown, 2007). Gocze and Freeman showed that the lipid fluorescent variability is significantly lower when using BODIPY493/503 compared to Nile Red, suggesting that it may be more specific for the LD (Gocze and Freeman, 1994). Here, we describe a BODIPY 493/503 assay for the detection of neural fat stores in cultured cells (Figure 1) (Wang et al., 2013).
Materials and Reagents
Cell line suitable for testing, MCF7 (ATCC, catalog number: HTB-22™) cells perform well as a staining control
Dulbecco’s Modified Eagle’s Medium (DMEM) (high glucose with L-glutamine) (Thermo Fisher Scientific, catalog number: SH3024301) or other media appropriate for MCF7 cell culture
Fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: F4135)
Phosphate buffered saline (PBS) (e.g. HyClone, catalog number: SH30258-02) or Dulbecco’s phosphate buffered saline (with Ca2+ and Mg2+) (DPBS) (e.g. Sigma-Aldrich, catalog number: D1283)
Sodium palmitate, (used as positive control) (Sigma-Aldrich, catalog number: P9767)
Dimethyl sulfoxide (Sigma-Aldrich, catalog number: D8418)
BODIPY 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene) (Life Technologies, catalog number: D-3922)
37% formaldehyde (Thermo Fisher Scientific, catalog number: F79)
Hoechst 33342 (Life Technologies, catalog number: H21492)
500× BODIPY 493/503 stock solution (see Recipes)
10,000× Hoechst 33342 stock solution (see Recipes)
Equipment
96-well tissue culture plate suitable for imaging, (e.g. Corning, Costar®, catalog number: 3603)
Cell culture incubator at 37 °C with 5% CO2
Fluorescence microscope or automated imaging system (e.g. IN Cell Analyzer, GE Healthcare)
Procedure
Plate 10,000-20,000 cells per well in 100 μl DMEM high glucose with L-glutamine, 10% FBS, in the 96-well plate. Doing a serial dilution can be helpful to determine the effect of cell number on BODIPY intensity.
Incubate the cells at 37 °C overnight.
Dilute the sodium palmitate stock solution in pre warmed DMEM high glucose with L-glutamine, 10% FBS. A range of 50-500 μM is useful to demonstrate BODIPY-based lipid content relative quantification. Prepare appropriate vehicle controls.
Carefully aspirate medium from the 96-well plate and replace with treatment medium (use 100 μl per well).
Incubate the cells at 37 °C overnight.
Prepare a 5% formaldehyde solution in PBS. Add 100 μl per well directly into the tissue culture medium to achieve a final formaldehyde concentration of 2.5%.
Incubate for 15 min at room temperature.
Prepare the staining solution; dilute the BODIPY and Hoechst stock solutions in PBS to a working concentration of 10 μg/ml BODIPY and 1 μg/ml Hoechst. Calculate 100 μl per well. Both dyes can be added in parallel, there is no need for sequential staining.
Carefully remove the formaldehyde from the cells and wash twice with 100 μl of PBS per well.
Add 100 μl staining solution per well.
Incubate for 30 min at room temperature, in the dark.
Wash twice with 100 μl of PBS per well.
Remove PBS, add 130 μl of PBS per well (only required for IN Cell Analyzer).
BODIPY and nuclei Hoechst fluorescence are imaged and quantified using fluorescence microscopy with appropriate filter sets. Our laboratory makes extensive use of the INCell Analyzer 2200 and INCell Investigator software for these measurements. Fluorescence intensity per cell is proportional to the neutral lipid content in the cell.
Notes
Solubilize sodium palmitate in DMSO at 75 mM immediately before usage, do not store.
If many cells are lost from the plate during washing with regular PBS, use DPBS for washing and preparing working solutions of BODIPY and Hoechst.
Care should be taken to stain at approximately equal cell densities when comparing different cells or culture conditions.
Recipes
-
500× BODIPY 493/503 stock solution
Solubilize BODIPY 493/503 in DMSO at 5 mg/ml
Aliquot and stored at −20 °C in the dark
-
10,000× Hoechst 33342 stock solution
Solubilize Hoechst 33342 in DMSO at 10 mg/ml
Aliquot and stored at −20 °C in the dark
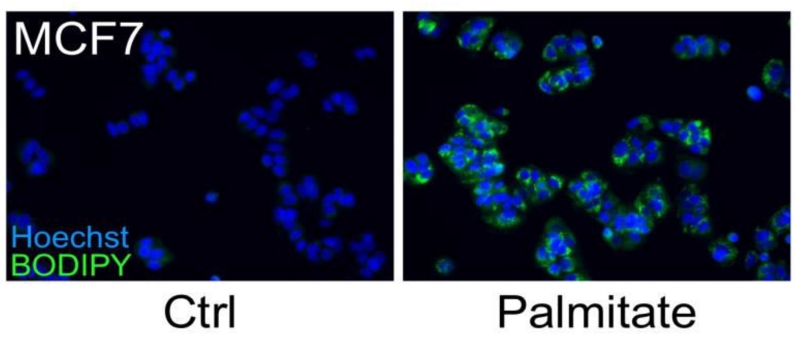
Figure 1. MCF7 cells were treated with 250 μM palmitate or vehicle control for 24 h.
A number of breast cancer cells possess a lipogenic metabolic phenotype that makes them especially sensitive to the addition of physiological concentrations of exogenous saturated fatty acids, such as palmitate. Although palmitate supplementation induces cell death in HER2/neu-positive cells, other breast cancer sub-types, including MCF-7 cells, accumulate the fatty acid which leads to significant increases in intracellular triglyceride fat stores. Cells were fixed and stained for fat stores with BODIPY 493/503 (green). Hoechst 33342 (blue) was used for nuclei staining.
Acknowledgments
Supported by U.S. Army Medical Research Acquisition Activity W8IWXH-04-1-0474 and NCI 1R01CA136658 to DSC.
References
- 1.Gocze PM, Freeman DA. Factors underlying the variability of lipid droplet fluorescence in MA-10 Leydig tumor cells. Cytometry. 1994;17(2):151–158. doi: 10.1002/cyto.990170207. [DOI] [PubMed] [Google Scholar]
- 2.Goodman JM. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J Lipid Res. 2009;50(11):2148–2156. doi: 10.1194/jlr.R001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kourtidis A, Srinivasaiah R, Carkner RD, Brosnan MJ, Conklin DS. Peroxisome proliferator-activated receptor-gamma protects ERBB2-positive breast cancer cells from palmitate toxicity. Breast Cancer Res. 2009;11(2):R16. doi: 10.1186/bcr2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Listenberger LL, Brown DA. Fluorescent detection of lipid droplets and associated proteins. Curr Protoc Cell Biol. 2007 doi: 10.1002/0471143030.cb2402s35. Chapter 24: Unit 24 22. [DOI] [PubMed] [Google Scholar]
- 5.Liu H, Liu JY, Wu X, Zhang JT. Biochemistry, molecular biology, and pharmacology of fatty acid synthase, an emerging therapeutic target and diagnosis/prognosis marker. Int J Biochem Mol Biol. 2010;1(1):69–89. [PMC free article] [PubMed] [Google Scholar]
- 6.Ranall MV, Gabrielli BG, Gonda TJ. High-content imaging of neutral lipid droplets with 1,6-diphenylhexatriene. Biotechniques. 2011;51(1):35–36. 38–42. doi: 10.2144/000113702. [DOI] [PubMed] [Google Scholar]
- 7.Walther TC, Farese RV., Jr. The life of lipid droplets. Biochim Biophys Acta. 2009;1791(6):459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Sun Y, Wong J, Conklin DS. PPARgamma maintains ERBB2-positive breast cancer stem cells. Oncogene. 2013;32(49):5512–552. doi: 10.1038/onc.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]