Abstract
Heat-labile enterotoxins (LT) of enterotoxigenic Escherichia coli (ETEC) are structurally and functionally related to cholera toxin (CT). LT-I toxins are plasmid-encoded and flanked by IS elements, while LT-II toxins of type II ETEC are chromosomally encoded with flanking genes that appear phage related. Here, I determined the complete genomic sequence of the locus for the LT-IIa type strain SA53, and show that the LT-IIa genes are encoded by a 51 239 bp lambdoid prophage integrated at the rac locus, the site of a defective prophage in E. coli K12 strains. Of 50 LT-IIa and LT-IIc, 46 prophages also encode one member of two novel two-gene ADP-ribosyltransferase toxin families that are both related to pertussis toxin, which I named eplBA or ealAB, respectively. The eplBA and ealAB genes are syntenic with the Shiga toxin loci in their lambdoid prophages of the enteric pathogen enterohemorrhagic E. coli. These novel AB5 toxins show pertussis-toxin-like activity on tissue culture cells, and like pertussis toxin bind to sialic acid containing glycoprotein ligands. Type II ETEC are the first mucosal pathogens known to simultaneously produce two ADP-ribosylating toxins predicted to act on and modulate activity of both stimulatory and inhibitory alpha subunits of host cell heterotrimeric G-proteins.
Keywords: heat-labile enterotoxin, prophage, pertussis-toxin-like toxin, ETEC, ADP-ribosyltransferase
Two novel pertussis-toxin-like toxins are also present in the genome of the prophage that also encodes the LT-II enterotoxin genes in type II enterotoxigenic Escherichi coli.
Graphical Abstract Figure.

Two novel pertussis-toxin-like toxins are also present in the genome of the prophage that also encodes the LT-II enterotoxin genes in type II enterotoxigenic Escherichi coli.
INTRODUCTION
Enterotoxigenic Escherichia coli (ETEC) are the main cause of morbidity and mortality due to diarrhea in travelers to and children under five in the developing world (World Health Organization 2006), producing a profuse watery diarrhea due mainly to the effect of secreted enterotoxins, either or both of a large multimeric heat-labile toxin or the smaller heat-stable peptide toxin. The plasmid-encoded heat-labile enterotoxin (LT-I) of classical ETEC is structurally, functionally and immunologically related to cholera toxin (CT) of Vibrio cholerae, the causative agent of cholera (Sack 2011). A second group of ETEC (type II ETEC) possesses chromosomally-encoded heat-labile enterotoxins (LT-II) that are structurally and functionally related, but immunologically non-cross-reactive to CT and LT-I (Guth et al. 1986). Together CT, LT-I and LT-II form the heat-labile enterotoxin family. The chromosomal locus encoding LT-II toxins is currently undefined. Diagnosis of ETEC requires specialized tests not routinely performed in clinical microbiology laboratories (Sjöling et al. 2007), and as LT-II toxins bind different ganglioside receptors and are genetically divergent from LT-I, even these techniques (e.g. GM1-ELISA or PCR) will not detect LT-II strains, and their contribution to diarrheal disease in both humans and animals is currently unknown. Type II ETEC were originally identified by biological assays for enterotoxin activity that was not neutralizable by CT or LT-I antisera (Green et al. 1983), and have been found in human clinical isolates (Seriwatana et al. 1988), diseased bovines (Pohl et al. 1989) and from foodstuffs such as hamburger and milk (Franco et al. 1991). Initial studies identified two strains that produced LT-IIa (Pickett et al. 1986) or LT-IIb (Pickett et al. 1989) variants. Recently, a third variant (LT-IIc) was described (Nawar et al. 2010) in a strain isolated from sick ostriches (Nardi et al. 2005), and the LT-II family now consists of three related subgroups – LT-IIa, LT-IIb and LT-IIc enterotoxins. The contributions of LT-II toxins to disease has received little study, while initial studies showed that purified LT-IIa did not cause fluid accumulation in rabbit intestinal loops (Holmes, Jobling and Connell 1995), a recent study showed that purified LT-IIa and to a lesser extent LT-IIb did so in intestinal loops of calves, and that cloned copies of LT-IIa and LT-IIb alone, present in an otherwise non-toxigenic porcine E. coli strain, were capable of causing severe diarrhea and weight loss in a neonatal pig model (Casey et al. 2012). The first genomic analysis of a large collection of type II enterotoxin strains (Jobling and Holmes 2012) showed that (i) the prototype LT-IIa and LT-IIb toxin types are rare (found in only 6% and 2% of isolates, respectively), that (ii) six subgroups of LT-IIc toxin genes are found the majority of the type II isolates, and that (iii) all LT-II toxin genes are flanked 5′ and 3′ by phage-related genes, suggesting that they may be prophage-encoded. All genes immediately 5′ of the LT-II genes encode very similar phage lysozyme-like proteins (Pei and Grishin 2005) of the glycosyl hydrolase 108 family (Stojković and Rothman-Denes 2007), while the immediate 3′ phage-related genes are conserved within but differ between the LT-IIa, LT-IIb and LT-IIc families (Jobling and Holmes 2012).
All members of the heat-labile enterotoxin family are encoded by two genes encoding precursor polypeptides of the enzymatic A and receptor-binding B subunits. These are secreted to the periplasm with concomitant removal of their signal sequences, where they assemble into the latent holotoxin consisting of a single A subunit and a homo-pentameric B subunit. The B pentamers confer specificity on the toxin by binding to specific host cell gangliosides that differ between family members. The first step in activation of the latent holotoxin is proteolytic nicking (by either bacterial or host proteases) of the A subunit within a disulfide-distended loop that generates the A1 and A2 subunits. Reduction of the disulfide bond within the host cell is required for the ultimate release of the catalytically active A1 subunit into the host cell cytoplasm, where it binds and hydrolyzes NAD, transferring the ADP-ribose moiety to an arginine in the stimulatory alpha subunit (Gsα) of heterotrimeric G-protein complex. This constitutively activated ADP-ribosylated Gsα permanently activates host adenylate cyclase, leading to increased cAMP levels that in intestinal cells ultimately lead to increased chloride secretion and production of profuse watery diarrhea characteristic of cholera and ETEC infections.
Gα proteins are also targets of another ADP-ribosylating toxin (pertussis toxin, PT) produced by the airway mucosal pathogen, Bordetella pertussis, the causative agent of whooping cough (Carbonetti 2015). PT is also an A-B toxin with a single enzymatic subunit (S1) and a heteropentameric-binding subunit consisting of S2-S3, S2-S4 and S5 monomers, all encoded by a five gene operon. The B-pentamer preferentially binds to sialic-acid-containing host cell glycoproteins and delivers the S1 subunit to the target cell. The crystal structure of PT shows that the S1 polypeptide shares a common fold with the A1 subunits of the LT family (Stein et al. 1994b), and it similarly uses NAD to ADP-ribosylate a cysteine in the carboxyl terminus of the inhibitory alpha subunit (Giα) of the heterotrimeric G-protein complex. This prevents its interaction with G-protein coupled receptors and its direct inhibitory interaction with host adenylate cyclase. These effects lead to inhibition of all signaling via Giα-coupled receptors (Katada 2012) and increased levels of cAMP in target cells in response to other stimuli (Hsia et al. 1984; Melsom et al. 2014).
The LT family of toxins and PT each have major effects on the hosts immune response to infection; CT and LT are potent immunomodulators that both suppress an inflammatory response (Fullner Satchell 2003; Glenn, Francis and Danielsen 2009), enhance colonization (Pierce et al. 1985; Allen, Randolph and Fleckenstein 2006) and modulate cytokine production by epithelial cells (Soriani, Bailey and Hirst 2002). PT similarly promotes airway colonization (Carbonetti et al. 2007) and contributes to significant immunomodulatory effects observed during infection (Connelly, Sun and Carbonetti 2012). All these toxins are potent mucosal adjuvants for coadministered antigens (Wilson et al. 1995; Liang and Hajishengallis 2010), effects that are attributable to both their enzymatic activities and receptor-binding properties (Schneider, Weiss and Miller 2007; Liang and Hajishengallis 2010). Thus, these toxins have multiple effects on the host beyond the major disease symptom that also contribute to the pathogenesis of the disease.
The objectives of this study were to characterize in detail the chromosomal locus for the prototype LT-IIa and LT-IIb toxins, and compare them with the recently discovered LT-IIc loci. I also describe the cloning, expression, purification and functional characterization of two novel PT-like toxins that I found closely linked to the LT-IIa and LT-IIc loci.
METHODS
Bacterial strains and specific type II ETEC isolates are described in Jobling and Holmes (2012), except that NADC2044 (LT-IIc3) was erroneously listed as LT-IIc2, and strains 35548-3 (adult cow, diarrhea, Belgium, 1984; Pohl et al. 1989) and NADC557 (calf, diarrhea, USA, 1964; Casey et al. 2012), both LT-IIc4, were unintentionally omitted from the list of 50 LT-II positive isolates. LT-IIa isolate KH655 was obtained from Dr Firdausi Qadri (ICDDRB, Bangladesh). Escherichia coli TE1 (Jobling and Holmes 2000) was used for routine cloning and expression of toxin genes. Routine molecular biology methods are described in (Sambrook, Fritsch and Maniatis 1989). PCR was done with 0.2 mM dNTPs and 0.2 μM primers using DreamTaqGreen (Thermo Fisher) or with recombinant H6-tagged Pfu (Lu and Erickson 1997) in (NH4)2SO4-based PCR buffer (Thermo Fisher) if the product was to be cloned. Primers for PCR and DNA sequencing, synthesized by Integrated DNA Technologies, Inc (Iowa), were used without further purification and are listed in Table S1 (Supporting Information). Mutations were introduced either directly by subcloning of SOE-PCR fragments (Horton et al. 1990) or by QuickChange mutagenesis (Agilent Technologies) using a mutagenic primer and its reverse complement.
Clone construction, expression and Talon chromatography
EplBIIah6 was made by first adding an ApaI restriction site 5′ of the his6 tag by PCR, using ApasmaH6F and a reverse vector primer on a cloned his6 tag in pT7his6 (Tripet et al. 2004), followed by cloning an ApaI-SacI-digested PCR fragment into pTSKII- (Ichihara and Kurosawa 1993) to make pApaH6. EplBIIa was amplified with EplBIIaKpnF and EplBIIaApaR and cloned as a KpnI-ApaI fragment into pApaH6 to make pAEplBgpgph6, where the coding sequence for EplB is joined directly to a gly-pro-gly-pro-his6 linker. The EplAIIa gene was then amplified with EplAIIaAatF and EplAIIaSacR to add AatII and SacI flanking sites and cloned 3′ of EplBgpgph6 in pEplBh6, to make the IPTG-inducible expression clone pAEplBgpgpH6EplA. The EalAB operon from LT-IIa isolate P393-F10 was cloned by adding an SphI site at the start of the coding sequence for the mature EalA subunit and an ApaI site at the end of the EalB gene by PCR with EalASphF and EalBApaR primers, followed by cloning into a derivative of the LT-IIb signal sequence vector pLDR5 (Jobling et al. 1997) that also has the ApaH6 tag. EplABgpgpH6 and EalABgpgpH6 holotoxins were purified by Talon affinity chromatography (as otherwise described by the manufacturer) from 1% Elugent (Calbiochem) extracts in Talon extraction buffer (0.3M NaHxP04, pH 8.0, 0.5 M NaCl) of cells obtained from log-phase cultures induced with 0.4 mM IPTG and grown overnight at 30°C. Extracts were passed over a Talon resin column and washed with extraction buffer with 5 mM imidazole until the absorbance at A280 nm returned to baseline; bound proteins were eluted with Talon buffer with 150 mM imidazole and fractions analyzed by SDS-PAGE. Toxin-containing fractions were pooled and dialyzed into 50 mM Tris-HCl, pH 8.0 (buffer A) and further purified by HQ10 anion exchange chromatography on an AKTA system (GE Health Sciences) and eluted with a 0–60% gradient of buffer B (buffer A + 1M NaCl). Holotoxin (A + B subunits) eluted first, followed by a second peak that contained only tagged EplB subunits.
Protein cross-linking
Purified EplB-gpgpH6 B subunits (0.9 mg mL−1, buffer exchanged into 0.2 M triethanolamine, pH 8.0) were cross-linked by treatment with 20 mM imidoester cross-linkers targeting primary amines (Thermo Scientific), with variable spacer length (DMA, DMP or DMS – dimethyl adipimidate, pimelimidate or suberimidate, respectively) for 1 hour at room temperature prior to analysis by SDS-PAGE.
Tissue culture and toxin assays
CHO-K1 cells (ATCC CCL-61) and CHO-15B (obtained from J. Jay, U. Montana; a ricin-resistant derivative of CHO-K1 cells (Gottlieb, Skinner and Kornfeld 1974) that specifically lacks Neu5Ac-Galβ1-4GlcNAc modifications of glycoproteins (Gottlieb, Baenziger and Kornfeld 1975) and is also resistant to PT (Brennan et al. 1988)) were cultured in Hams F12 medium with 10% fetal calf serum (ATCC). Toxin containing extracts were serially diluted in 50 μl of medium in 96-well plates, and seeded with 150 μl of freshly harvested cells in serum-containing medium at 104 cells per well, followed by 2 days growth, after which they were observed microscopically (200x magnification) for clustering and/or elongation relative to control (untreated) wells. Toxin neutralization was done by incubating cell extracts for 20 minutes prior to tissue culture assay with 1:20 dilutions of rabbit antisera, raised against purified LT-IIa (Pickett et al. 1986)], or against his-tagged EplBA or EalAB purified in this study, antisera produced by Pocono Rabbit Farm & Laboratory (PRF&L, Canadensis, PA).
DNA sequencing and data analysis
Clone Manager Professional v 9.3 (Science and Educational software) was used for assembly of DNA sequences obtained from the UCD sequencing core facility. The assembled LT-IIa prophage sequence was first annotated by the RAST server (http://rast.nmpdr.org/) (Aziz et al. 2008) then manually edited to remove conflicts and improve accuracy, and submitted with accession number KU052037; the LT-IIb locus sequence was assigned as KU052038. Representative toxin-encoding loci were manually annotated by homology matches and submitted to NCBI with accession numbers KU052039-KU052043.
RESULTS AND DISCUSSION
Type II ETEC isolates producing LT-IIa and LT-IIc also encode a novel PT-like toxin closely linked to the LT-II toxin locus
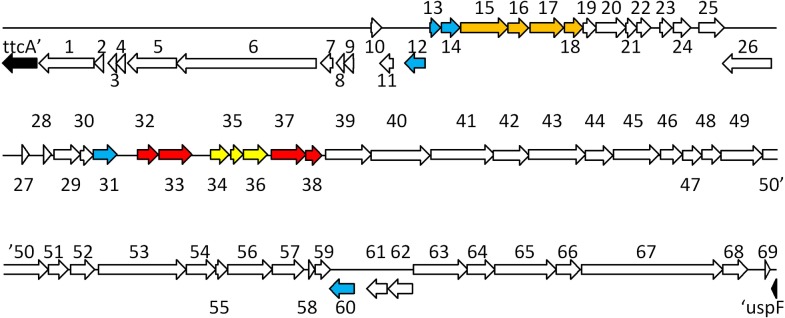
I previously identified the genomic locus for the first type LT-IIc2 toxin in strain 442/2 by inverse PCR on a self-ligated PstI digest of chromosomal DNA. This 4 kb amplicon was used to determine the LT-IIc2 and immediate flanking gene sequences (Jobling and Holmes 2012). Here I determined the complete sequence of the inverse PCR product. This produced a further 2.3 kb of sequence that, in the genome, is 5′ of the LT-IIc A gene (Fig. 1). Intriguingly, this sequence encoded the 3′ end of another novel ADP-ribosylating toxin gene pair – the PstI site was internal to a partial open reading frame followed by a gene that encodes a homolog of the S1 catalytic subunit of PT (Fig. 1). The closest BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi ) match was to the PltBA (Sty1891-1890) PT-like toxin locus of Salmonella enterica serovar Typhi (Spano, Ugalde and Galan 2008). PltBA was discovered as a pair of linked genes required for delivery of the cytotoxin CdtB to target cells, and with CdtB form a tripartite toxin (Song, Gao and Galan 2013). They are part of a small pathogenicity islet that appears to be phage-derived (Hodak and Galan 2013). I have designated this novel type II ETEC toxin locus eplBA, for E. coli pltBA-like toxin. This novel LT-IIc-associated toxin locus was followed by a three gene phage lysis cassette (holin-holin-lysin) the last of which encoded the gene previously identified as a lysin immediately 5′ of the LT-IIc operon (Fig. 1) (Jobling and Holmes 2012). Since all LT-II loci also have homologous lysin genes upstream of their LT-II operons, it was possible that other LT-II isolates also encoded similar novel toxin loci. I have determined the complete sequence of the largest available plasmid subclone of the LT-IIa locus (pCP3727 containing a 3 kb EcoRI fragment; Fig. 1) (Pickett et al. 1986; Pickett, Weinstein and Holmes 1987). The cloned sequence begins with a similar three-gene lysis cassette that shows 90% identity to the LT-IIc sequence, but does not encompass the region that in LT-IIc encodes the novel toxin operon. I therefore used three sequential primer walks upstream of the LT-IIa genes in the original LT-IIa cosmid clone, A38 (Pickett et al. 1986), to determine 3 kb of sequence 5′ of the holin genes, which showed that the LT-IIa locus also encodes a complete syntenic eplBA gene pair that are located 3′ of a predicted phage antiterminator Q-gene (Fig. 1).
Figure 1.
Organization of novel pertussis toxin like toxin locus linked to the LT-IIa enterotoxin genes and comparison of A and B subunit polypeptides. A, Graphical map of the dual toxin loci from SA53 (LT-IIa) cosmid A38, subclone pCP3727 and inverse-PCR fragment from 442/2 (LT-IIc2). Upper line, the 4 kb inverse PCR of PstI-self-ligated chromosomal DNA from 442/2. Blue arrows show primer sites and point 5′ to 3′. Primers 1 and 2, inverse PCR primers; primer 3, RacatermF; and primer 4, RacholinR. Lower solid line shows insert sequenced from pCP3727, and dotted line shows region sequenced by primer walking into cosmid A38. Novel genes identified in this study are show as solid red arrows; open arrows show genes identified in Jobling and Holmes (2012). B and C, Comparison of the amino acid sequences of EplB with PltB from S. enterica serovar Typhi and SubB from EHEC (B) and EplA with PltA or PT-S1 (C). Residues identical to the upper sequence are shown as dots, with differences shaded in light blue. Dashes show gaps introduced to maximized alignments. Red boxes denote the predicted signal sequences. Red circles identify key conserved catalytic resides in PltA that differ from EplA and PT-S1. Blue box identifies the catalytic glutamate fully conserved in all toxins.
Homologs of these Q-like and lysis genes are found in several lambdoid prophages, the closest match being with a putative prophage inserted into the potABCD locus in the prototype ETEC strain H10407 (FN649414). The LT-IIa and H10407 Q and holin gene sequences are 70% and 79% identical to their homologs, respectively, and are separated by similar distance (2.2 and 2.4 kb), although the intervening DNA sequence and encoded genes are completely different. A primer pair designed to highly conserved regions of the Q and holin genes (RacatermF and RacholinR, respectively) amplified the corresponding region from the LT-IIc2 locus, whose sequence showed that the LT-IIc locus also encodes a complete eplBA operon similarly located 3′ of a Q-like gene. The organization of these novel eplBA loci in the LT-IIa and LT-IIc genomes are shown graphically in Fig. 1, The complete nucleotide sequence including translations of open reading frames for the LT-IIa RacatermF-RacholinR PCR product is shown in Fig. S1 (Supporting Information).
The LT-IIa and LT-IIc eplBA loci encode highly related polypeptides. The predicted LT-IIa and LT-IIc EplB proteins differ at only seven of 156 residues, and intriguingly also differ in the number of repeats of the four residue sequence DVSS immediately following the predicted signal sequence (EplB from the LT-IIa locus, with two repeats, is shown in Fig. 1). Comparing the mature polypeptides (excluding the predicted signal sequences and the DVSS repeats) EplB shows 38% identity to PltB (44/121 residues, Fig. 1), but is slightly more related (43% identity over 120 residues) to the B subunit of subtilase toxin, which is encoded by a plasmid in Shigatoxigenic E. coli (Paton et al. 2004). The predicted mature EplA polypeptides are 97% identical, differing at only five of 229 residues, and are 53% and 27% identical to the mature PltA and PT-S1 polypeptides, respectively (Fig. 1).
Both EplBA and LT-IIa toxins are biologically functional and simultaneously expressed from native promoters
The location of the eplBA toxin loci following Q-like genes suggests that their expression may be induced during lytic replication of the encoding-phage. Indeed, the LT-IIc Q-gene is followed by a dyad sequence predicted to be a strong transcriptional terminator (FindTerm server, www.softberry.com), ∼80 bp distal to the Q-gene stop codon and ∼300 bp upstream of the eplBA loci. However both eplBA loci also have predicted promoters (see Fig. S1, Supporting Information; Bprom server at www.softberry.com) within the 300 bp 5′ proximal region that could drive transcription of the toxin loci independent of lytic phage transcription.
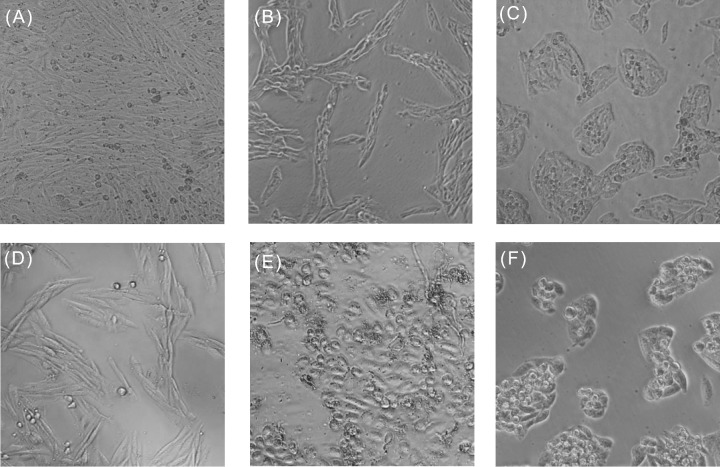
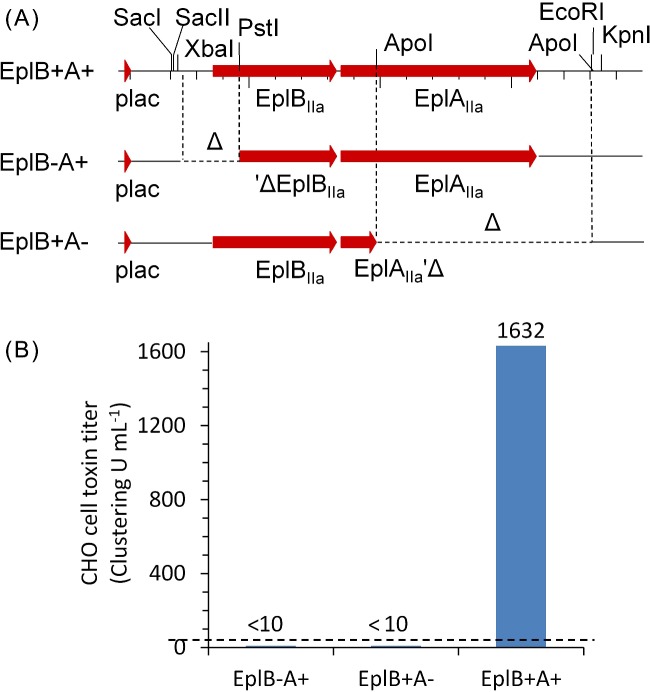
In the A38 cosmid clone that encodes EplBA and LT-IIa, expression of the cosmid-encoded genes is predicted to occur in their native context (see later) albeit at slightly increased copy number compared to the chromosomal locus, as the copy number of cosmid clones in this vector is estimated to be between 5 and 10 (Hohn and Collins 1980). Toxin gene expression independent of phage replication is supported by detection of EplBA and LT-II activity in extracts of overnight cultures of TE1 [A38] when these are added to CHO cells in tissue culture (Fig. 2). CHO cells normally grow as uniform monolayers (Fig. 2A), but respond to heat-labile enterotoxins by elongating (Guerrant et al. 1974) and to PT by forming tight clusters (Hewlett et al. 1983). Cosmid cell extract treated CHO cells grew in tight clusters with elongated morphology, showing both PT-like and LT-II activity (Fig. 2B). Pre-incubation of extract with toxin-specific rabbit-anti-serum neutralized the toxins and resulted in monolayers that formed clusters only (Fig. 2C, when pre-incubated with anti-LT-IIa), elongated cells only (Fig. 2D, when pre-incubated with anti-EplBA) or normal morphology when pre-incubated with both antisera (Fig. 2E). This demonstrates that the morphological changes are specific to each toxin; their effects are independent of each other, and that both toxins are naturally produced by cells carrying the cosmid clone. The CHO clustering phenotype is indistinguishable from that seen for CHO cells treated with purified PT (Fig. 2F; see also Hewlett et al. 1983). Hewlett et al. 1983 also showed that simultaneous addition of PT and CT to CHO cells causes them to both cluster and elongate, indicating that the effect of the two toxins was additive. Additionally, the Hewlett group found that treatment with both PT and CT had a synergistic effect on the amount of cAMP produced by intoxicated cells (Hewlett et al. 1982), and further that treatment with both toxins was required to observe a maximal cAMP response (Hsia et al. 1984). However, as these two toxins (PT and CT) are produced by different pathogens that colonize distinct mucosal environments (B. pertussis in the lung and Vibrio cholerae in the gut, respectively), this effect was not at the time biologically relevant. Here, I have shown for the first time that an enteric pathogen encodes and can simultaneously produce both types of ADP-ribosylating toxins that, together, are predicted to completely inactivate G-protein dependent control over cAMP production in target cells, by ADP-ribosylating and constitutively activating the stimulatory Gsα subunit (by LT-II toxin) and ADP-ribosylating and inactivating the inhibitory Giα subunit (by PT-like toxins). Concurrent production of both toxins thus has the potential to increase disease severity by maximizing cAMP production leading to greater net fluid secretion (Sears and Kaper 1996).
Figure 2.
Periplasmic extracts of E. coli K12 transformed with EplBA and LT-IIa-encoding cosmid A38 produce distinct morphological changes to Chinese hamster ovary (CHO) cell monolayers. Freshly seeded CHO cells were incubated for 2 days with periplasmic extracts of E. coli cells carrying cosmid A38, and photographed at 200 × magnification. A, Untreated cells (normal confluent monolayer); B–F, toxin-treated cells. B, treatment with cosmid extract causes cells to cluster (Cl) and elongate (El). C–F, morphological effects are specific for each toxin, shown by preincubating cosmid extracts with specific rabbit antisera (α-) before adding to CHO cells; observed phenotype is noted in parentheses. C, extract + α-LT-IIa (cells Cl only, El activity neutralized); D, extract + α-EplBA (cell El only, Cl activity neutralized); F, extract + α-LT-IIa and α-EplBA (normal morphology, both Cl and El activities neutralized); F, positive control, purified pertussis toxin, 10 ng (show Cl only). A–C and D–F are from independent experiments.
The EplBA and LT-II toxin loci are encoded by a full-sized lambdoid prophage
As our original characterization of the LT-II loci family indicated that the flanking genes were phage-related (Jobling and Holmes 2012), and the above data extend this determination to include the Q-like and holin genes flanking the eplBA loci, I undertook to determine the full genomic context of the EplBA/LT-II encoding genes. By analyzing the sequences at the cosmid vector-insert borders of original LT-IIa cosmid clone (A38), using vector-specific primers that flank the BamHI insertion site in the tet gene (Hohn and Collins 1980) I found that one end showed homology to predicted phage structural gene in a prophage-containing contig from an Arsenophonus genome (FN545185), and the other end of the A38 cosmid insert was homologous to the zntB gene located at 1.4 Mb in the MG1655 genome. ZntB is ∼5 kb from the integration site for the defective Rac prophage of K12 (Kaiser and Murray 1979). The rac (recombination activation) locus was originally defined by mutations that suppressed the recombination deficiency phenotypes of recBC mutations (Low 1973), by activation of the recE gene encoded within the Rac prophage (Kaiser and Murray 1980). This data indicated that the LT-II locus is contained within a lambdoid-type prophage integrated at the same site as the Rac prophage in E. coli K12. Sequencing the complete insert in cosmid A38, showed that it begins with 5 kb of chromosomal DNA 99% identical to the K12 chromosomal genes 5′ of the rac prophage locus—zntB, ydaN, dpbA and ttcA. Following the integration site in ttcA is 30 kb of a lambdoid-type prophage. This appears to be a portion of a complete lambdoid prophage inserted at the K12 rac locus, but lacking ∼15 kb of the right end that should encode many of the phage structural genes and the right chromosomal junction. As this project progressed, several complete E. coli genomes became available that encoded full-sized, but not toxin-encoding, prophages at the rac locus, and some of these (e.g. E. coli W, NC_0117664) had high homology to the right end of the cosmid insert. By a combination of long-range PCR and primer walking, I completed the sequence for the 16 kb missing region and confirmed that in the prototype LT-IIa strain SA53, the LT-IIa locus is chromosomally encoded within a 51 kb prophage inserted in the 5′ end of the ttcA gene, in an identical location to the defective Rac prophage of K12 strains (Fig. 3).
Figure 3.
Graphical map of the genes identified in the DNA sequence of the LT-IIa prophage locus. Filled black arrows denote chromosomal genes flanking the prophage (ttcA and uspF); prophage genes (numbered from left to right) are shown as open white arrows; blue arrows are predicted regulatory genes, orange arrows show replication-related genes; red arrows show toxin genes and yellow arrows show genes in the lysis cassette; arrrows point in the direction of transcription. Table 1 gives a detailed description of each gene.
The LT-IIa Rac prophage appears largely intact and is organized in typical lambdoid manner (Oppenheim et al. 2005; Casjens and Hendrix 2015). It has homologs of many of the genes still present in the defective Rac prophage in K12 strains (Casjens 2003). Predicted genes are annotated in Table 1 and shown graphically in the context of the prophage in Fig. 3. The majority of the genes in the first 9.5 kb of the prophage are encoded on the reverse strand and encode genes predicted to be involved in the lysis/lysogeny decision—these loci begins with the predicted phage repressor (gene 12, most closely related to the dicA repressor gene of the K12 defective Qin prophage), followed by a kil homolog (Conter, Bouche and Dassain 1996) and predicted genes 6 and 5 involved in recombination (recE/recT). Predicted xis and integrase genes are the last before the chromosomal insertion site att, located within ttcA. The majority of the rest of the prophage genome is encoded on the sense strand and begins the early right operon with genes 13 and 14 encoding dicA-divergent transcriptional regulators (dicC and ydaT), followed in order by a group of phage replication genes (15–18) and a collection of likely non-essential genes (like the lambda Nin gene loci). This region ends with a functional though genetically unrelated homolog (pfam 06576, DUF1133, gene 31) of the lambda Q antiterminator gene. Following the Q-gene, beginning the phage late operon, are the toxin morons [beneficial regions, often encoding toxins, present in certain phages but absent in other related phages, (Hendrix et al. 2000; Juhala et al. 2000; Cumby, Davidson and Maxwell 2012a)] encoding EplBA and LT-IIa enterotoxins, separated by a three-gene lysis cassette 5′ of the LT-IIa genes. The last 28 kb of the prophage genome is predicted to encode mostly structural proteins for packaging DNA and forming the phage particles. Finally, the second att site creates a pseudo gene ttcC 5′ of the chromosomal gene uspF both on the reverse strand.
Table 1.
Predicted genes in LT-IIa Rac-like prophage of type II ETEC isolate SA53.
| Gene | Name | No. of codons | Coding region (complementary strand, C) | E. coli K12 product, functional match or BLAST match |
|---|---|---|---|---|
| ttcA′ | ttcA | - | C 771-1 | chromosomal gene, tRNA 2-thiocytidine biosynthesis protein |
| 1 | intR | 823 | C 2058-823 | Rac integrase |
| 2 | ydaO | 72 | C 2275-2060 | Rac Xis putative excisionase |
| 3 | ydaC | 63 | C 2563-2375 | Rac putative double-strand break reduction protein |
| 4 | ralR | 70 | C 2765-2556 | Rac restriction alleviation |
| 5 | recT | 368 | C 3917-2814 | Rac recombination |
| 6 | recE | 1049 | C 7063-3917 | Rac exoVIII recombination |
| 7 | racC | 92 | C 7440-7165 | Rac unknown function |
| 8 | ydaE | 59 | C 7691-7515 | Rac prophage conserved |
| 9 | kilR | 74 | C 7906-7685 | Rac inhibitor of FtsZ, killing protein |
| 10 | HPa | 78 | 8304-8537 | No CDDb match |
| 11 | HP | 102 | 8804-8499 | No CDD match |
| 12 | dicA | 154 | C 9517-9066 | Transcriptional regulator; RacR repressor in Rac prophage |
| 13 | dicC | 83 | 9625-9873 | DicC transcriptional regulator; YdaS in Rac prophage |
| 14 | ydaT | 141 | 9884-10306 | Rac uncharacterized protein—possible cII functional homolog |
| 15 | ydaU | 357 | 10319-11389 | Rac primosomal replication protein |
| 16 | ydaV | 154 | 11382-11843 | Rac replication |
| 17 | ydaW | 254 | 11878-12639 | Rac replication |
| 18 | lygF | 141 | 12656-13078 | Salmonella defective prophage gene homolog (linked to ydaO gene) |
| 19 | HP | 102 | 13075-13380 | No CDD match |
| 20 | ead | 226 | 13367-14044 | Lambda Ead/P22 Ea22 protein |
| 21 | HP | 85 | 14041-14295 | No CDD match |
| 22 | HP | 104 | 14288-14599 | No CDD match |
| 23 | IS1Δ | 91 | 14806-15078 | 5′ end of IS1 transposase frameshifted; -1 translational slippage into gene 24 frame |
| 24 | ΔIS1 | 132 | 15105-15500 | 3′ end of IS1 transposase; with gene 23 creates functional InsAB transposase protein |
| 25 | cII | 193 | 15680-16258 | P4 protein |
| 26 | beta | 366 | C 17315-16128 | P4 beta protein |
| 27 | mok/hok | 71 | 17816-18028 | Host addiction module toxin/antitoxin (hok starts 17873, 52 codons); Pfam 08848 |
| 28 | rem | 59 | 18353-18529 | Qin K12 defective prophage no known function |
| 29 | irsA | 200 | 18589-19188 | Increased resistance to stress Salmonella phage; DUF1367 |
| 30 | ybcO | 97 | 19188-19748 | DLP12 K12 defective prophage; DUFc1364; Pfam07102 |
| 31 | Q | 179 | 19475-20011 | Phage antiterminator DUF1133; Pfam06576 |
| 32 | EplB | 157 | 20466-20936 | PT-like B-binding subunit; subtilase toxin B subunit |
| 33 | EplA | 248 | 20954-21697 | PT-like S1 subunit; ADP-ribosyltransferase |
| 34 | holin | 131 | 22191-22583 | Phage lysis gene |
| 35 | holin | 93 | 22573-22851 | Phage lysis gene |
| 36 | lysin | 182 | 22853-23398 | Phage endolysin |
| 37 | LT-IIa A | 260 | 23488-24267 | Heat-labile enterotoxin type IIa A subunit; ADP-ribosyltransferase |
| 38 | LT-IIa B | 124 | 24257-24628 | Heat-labile enterotoxin type IIa B binding subunit |
| 39 | endo-terS | 343 | 24713-25741 | endolysin-terminase small subunit fusion; pfam03592 |
| 40 | terL | 443 | 25731-27059 | Terminase large subunit; pfam03237; DUF1073 |
| 41 | HP | 429 | 27078-28514 | No CDD match |
| 42 | HP | 278 | 28459-29292 | No CDD match |
| 43 | tolA | 431 | 29273-30565 | NUDIX domain; DUF2213; COGd3566 |
| 44 | HP | 206 | 30588-31175 | No CDD match |
| 45 | HP | 343 | 31190-32218 | No CDD match |
| 46 | HP | 166 | 32275-32748 | No CDD match |
| 47 | HP | 147 | 32745-33185 | No CDD match |
| 48 | HP | 147 | 33182-33622 | No CDD match |
| 49 | HP | 315 | 33609-34553 | No CDD match |
| 50 | HP | 446 | 34553-35890 | Duf3383; Pfam11863 |
| 51 | HP | 148 | 35902-36345 | No CDD match |
| 52 | HP | 206 | 36342-36959 | No CDD match |
| 53 | Lytic | 658 | 37023-38996 | lytic transglycosylase, catalytic |
| 54 | HP | 223 | 39000-39668 | No CDD match |
| 55 | HP | 89 | 39665-39931 | No CDD match |
| 56 | HP | 336 | 39931-40938 | No CDD match |
| 57 | bppV | 238 | 40938-41551 | Putative phage baseplate protein V |
| 58 | HP | 46 | 41757-41894 | No CDD match |
| 59 | HP | 116 | 41902-42249 | No CDD match |
| 60 | ant | 186 | C 42782-42225 | Putative phage antirepressor AntA; COG3561; Pfam08346 |
| 61 | HP | 155 | C 43526-43062 | No CDD match |
| 62 | HP | 183 | C 44093-43545 | No CDD match |
| 63 | HP | 409 | 44115-45341 | No CDD match |
| 64 | HP | 209 | 45325-45951 | No CDD match |
| 65 | tail | 469 | 45948-47330 | Phage tail fibre; COG5301 |
| 66 | tail | 181 | 47333-47875 | Caudovirales phage tail fiber assembly protein; Pfam02413 |
| 67 | tail | 1062 | 47899-51084 | Phage tail fiber repeat protein; Pfam 08400; Pfam03406; COG5301 |
| 68 | tail | 188 | 51084-51647 | DUF4376; Pfam14301 |
| ttcC | ttcC | 17 | C 52010-51957 | Pseudogene created by prophage insertion into ttcA |
| 69 | HP | 39 | 52034-52150 | No CDD match |
| ′uspF | uspF | – | C 52290-52186 | Chromosomal gene, stress-induced protein, ATP-binding protein |
HP, hypothetical protein.
CDD, conserved domain database.
DUF, domain of unknown function.
COG, clustered orthologous group.
The location of the toxin morons suggests they are the first of the phage late genes to be expressed following antitermination of the early right transcript, once the Q-gene is induced—when the prophage is committed to lytic development (Yang, Goliger and Roberts 1989). Here, eplBA are syntenic with the Shiga toxin (stx) genes in their enterohemorrhagic (EHEC) prophages, and in this context stx gene expression is upregulated during lytic induction of lysogens (Neely and Friedman 1998a,b). In EHEC infections this can occur during treatment with certain antibiotics in patients (Wong et al. 2000) and in animal models (Matsushiro et al. 1999; Zhang et al. 2000), paradoxically triggering increased toxin expression by SOS-induction of prophages. This can lead to more severe disease or development of hemolytic uremic syndrome. I predict that production of both EplBA and LT-II toxins is similarly increased during lytic replication, and I am currently characterizing potential mechanisms of Rac-like prophage and toxin induction in type II ETEC. Expression data from the cosmid above and preliminary data with toxin 5′ sequences cloned into promoter probe vectors indicates that even without prophage induction, the toxin genes are expressed at a detectable level from their own lytic-development independent promoters.
Detection of prophage excision by LT-II prophages
Comparison with preliminary sequence data for several LT-IIc loci indicates that the LT-IIa locus has an ≈3 kb deletion following the LT-IIa genes–gene 39 appears to be a deletion/fusion event joining the coding sequences for 30 residues at the start of an endolysin-like protein with the majority (deleted for ∼50 N-terminal residues) of a small terminase subunit. In this region, other similar phages have three highly conserved but hypothetical genes. It is thus possible that this LT-IIa prophage is now defective, and I have been unsuccessful in detecting plaque formation or Rac phage production from SA53. However, spontaneous excision of the prophage can be detected in some type II ETEC isolates by PCR. Outward-directed primers (RacrhsF3-right arm, and racIntF, left arm) from both ends of the prophage detect circularized phage DNA in stationary phase cultures of several LT-IIc isolates, but not SA53 (LT-IIa) (Fig. 4). The defective K12 Rac prophage can also excise (Evans, Seeley and Kuempel 1979), in a process recently found to be regulated by HN-S (Hong, Wang and Wood 2010), by recombination between direct repeats of 5′-TTGTTCAGGTTGTATTGTTCTTTCTT-3′, that flank the K12 locus and define the att site. Thus, the LT-IIc phages are functional for at least a portion of the lytic pathway, but we do not yet know of conditions that may induce the lytic cycle or if these are capable of replicating and forming functional phage particles. LT-IIc prophages must be significantly different from LT-IIa in their right arms, as five outward-directed LT-IIa-specific primers from 4 kb distal to the insertion site, down to only 170 bp 5′ of the racrhsF3 positive primer, fail to give products with uspFR on any LT-IIc isolate DNA template (not shown). For comparison, there are few published studies describing Rac-like phages. The Sakai Rac prophage, Sp10 (Asadulghani et al. 2009) is reported to be competent for all phage functions except for the repressor which appears to be described as defective only because it lacks an SOS-induction domain. That study found Sp10 DNA was packaged into phage particles, and these could transduce marked phage genomes to some E. coli K12 strains, where they integrated specifically at the rac locus. Further work is warranted to understand the life cycle of Rac prophages in greater detail.
Figure 4.
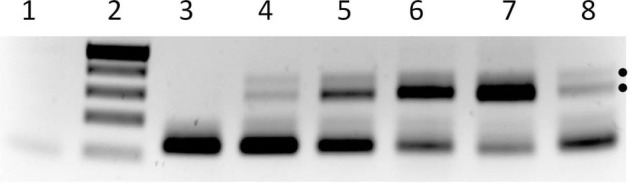
Demonstration of prophage circularization in type II ETEC isolates. Agarose gel separation of PCR products using E. coli cultures as template. Primers read out from the prophage genome (ttcAR at the left end and racrhsF3 at the right end) Lanes—1, K12 strain AB1157 (Racempty, no prophage); 2, size standards – 500, 400, 300, 200 and 75 bp; 3–8, type II ETEC isolates -3, LT-IIa; 4, LT-IIc1; 5, LT-IIc2; 6, LT-IIc3; 7, LT-IIc4; 8, LT-IIc5. Dots mark 325/295 bp products produced only by circular phage DNA. RacrhsF3 has a partial secondary priming site 7 bp 5′ of its complete match in the LT-II prophage genome, and so also produces a minor product 30 bp larger. The smallest band in each lane is the primer-dimer of racrhsF3 and ttcAR primers, which is preferentially produced in the absence of functional template.
Bioinformatic comparison of LT-II Rac prophages to other Rac prophages in sequenced E. coli genomes
There are currently no complete E. coli genomes published that have EplBA/EalAB and LT-II-encoding toxin prophages at the rac locus. Some complete E. coli genomes encode large prophages at the rac locus (inserted within ttcA and distal to uspF), including E. coli W (CP002185), the prototype EHEC strains EDL933 (NC_002655) and Sakai (NC_002695), EAEC O42 (FN554766) and avian pathogenic isolate APEC O1 (NC_008563). Although none of these prophage genomes encode similar enterotoxins, the O42 and O1 Rac prophages have morons following the structural genes that encode an iron transport system (SitABCD) and a cytolethal distending toxin (CdtABC), respectively. Following their lysis cassettes, these two phages encode trkG homologs, also found in the K12 Rac prophage. One Rac prophage in a 0.9 Mb contig from the draft genome for an avian E. coli B088 isolate (GG749133) does encode a different LT-II-like toxin, but no PT-like toxin locus. I will describe the LT-II toxin from this strain in a future publication. The E. coli W Rac prophage is most similar to the LT-IIa Rac prophage with 73% identity over the whole prophage genome. A global comparison shows more than half of these two prophage genomes are ≥90% identical, interspersed with regions of no or low DNA homology, indicative of module swapping (Casjens 2003; Grose and Casjens 2014). These non-homologous regions include portions of the recombination genes, key regulatory genes (dicA/dicC or cI/cro), replication genes and, significantly, the morons encoding EplBA and LT-IIa toxins. Notably, the moron flanking (Q and terminase subunit) and intervening (two holin) genes are well conserved although the third lysis cassette (endolysin) gene appears divergent. This analysis supports the concept of prophage evolution by recombination between homologous modules (Lima-Mendez, Toussaint and Leplae 2011; Menouni et al. 2015).
EplBA loci are present in the majority of LT-II ETEC isolates
Since both the LT-IIa and the LT-IIc2 loci, from SA53 and T442/2, respectively, encode highly related EplBA toxins, I next determined if similar toxins were also encoded by our other LT-II strains, and if so, how different they were. The conserved Q-gene and holin gene-specific primers RacatermF and RacholinR, respectively (Table S1, Supporting Information), used to amplify the LT-IIc2 locus from 442/2, also amplified similar sized (2.5–2.8 kb) products from all bar four of our 50 LT-II isolates (no products were obtained from three LT-IIc1 subtype strains and the lone LT-IIb strain). Variation in the size of the amplimers indicated that, while the sequence and order of the Q-holin genes is conserved in the majority of the LT-II isolates’ prophages, the intervening DNA shows some heterogeneity. I determined the DNA sequence of these PCR products for the other five of the six LT-IIc type strains (LT-IIc1, c3-c6, Jobling and Holmes 2012), and found that LT-IIc1, c2, c3 and c4 encoded highly similar eplBA loci. PCR with the RacatermF primer and a reverse primer internal to eplB (EplBR) was used to determine the frequency that eplBA toxin genes are associated with their LT-II locus for each of the 46 Q-holin PCR-positive LT-II strains in my collection. Thirty six type II ETEC isolates were PCR positive (all producing an ∼1.1 kb product) using RacatermF and EplBR as the reverse primer, showing that the majority of LT-IIc-positive type II ETEC also encode eplBA loci.
Variations within EplB genes are responsible for the diversity within EplBA-encoding loci
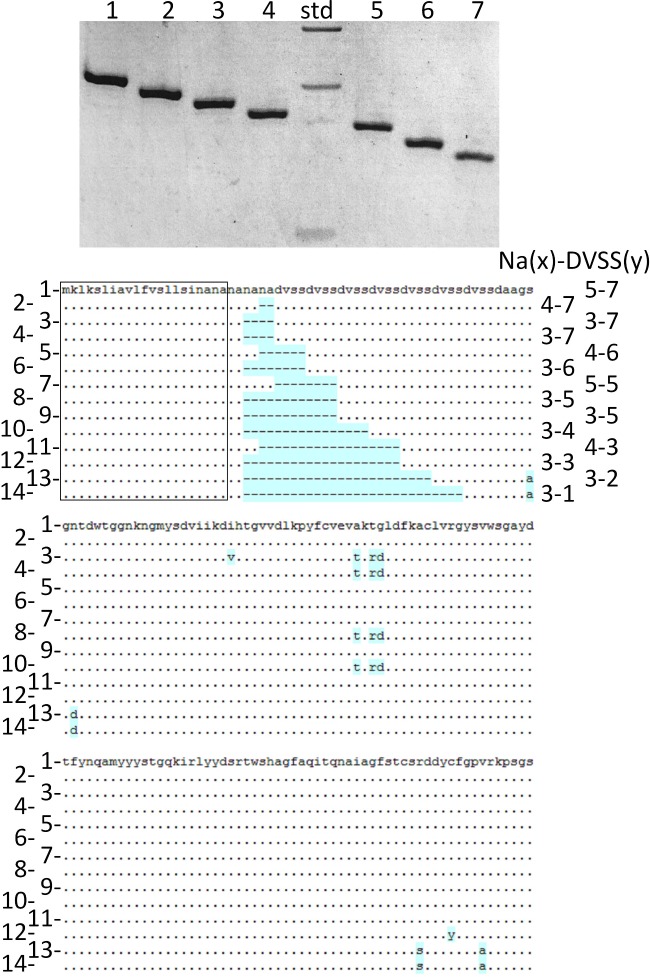
Sequence analysis of the four EplBA-positive LT-IIc subgroup prototype strains revealed that their EplA polypeptides are almost identical, differing at only the last residue, and all differ from EplA at the LT-IIa locus by 6 of their 248 residues. The final residue for each EplA polypeptide for LT-IIa and LT-IIc1-c4 type strains is L, E, V, D and G, respectively. The 4 LT-IIc1-c4 type strain EplB proteins are also very similar and showed at least 98% identity, and 90% identity with the LT-IIa EplB protein. Each gene encodes a polypeptide with a well-defined 21 residue signal sequence ending with canonical cleavage sites (ala-xxx-ala)—NANA-NA predicted to be cleaved after the second alanine; surprisingly, the major differences in the predicted protein sequences is due to a variable number of repeats directly following the predicted signal sequence—comprising the sequence (NA)x(DVSS)y, where, for LT-IIa and LT-IIc1 through LT-IIc4 type strains, x is either 3 or 4 (which includes the signal peptide cleavage site from −4 to −1) and y is 2 for the LT-IIa-encoded EplB, and 6 or 7 for the LT-IIc1-c4-encoded EplB polypeptides. The number of repeats is responsible for the majority of differences between the loci, and excluding the repeats individual mature EplB proteins differ by at most 8 residues in 9 of 125 positions (Fig. 5). The mature EplB subunits from LT-II prophages thus form two major groups that differ at 3 of 4 residues (having either AKTG or—TKRD signatures) in the center of the protein sequence, but this does not correlate with any specific LT-II type. The two LT-IIa EplB subunits are variants of the AKTG signature group.
Figure 5.
Variation in the coding sequences for EplB subunits from selected type II ETEC isolates. (A) High-resolution agarose gel of PCR amplimers from E. coli cells using EplBmatF and EplBR primers. From left to right, isolates tested are (LT-II type and number of DVSS repeats given in parentheses) lane 1, 442/2 (IIc2, 7); 2, 357900 (IIc1, 6); 3, W517 (IIc4, 5); 4, F150 (IIc3, 4); size markers—300, 200 and 75 bp, noted on left side of gel; 5, F250 (IIc4, 3); 6, SA53(IIa, 2) and 7, KH655 (IIa, 1). (B) Aligned amino acid sequences of EplB subunits from representative isolates show variability in the number or repeats of NA (asn-ala) and DVSS (asp-val-ser-ser) and identifies three groups of EplB families. Arrowheads above amino acids denote approximate position of primers in the coding region used to produce panel A. Unique EplB subunits are numbered on the left, and the numbers of NA and DVSS repeats given on the right in the form x–y. EplB variants are shown for the following isolates (LT-II type is in parentheses) 1, T5 (IIc2); 2, 33548 (IIc4); 3, E21845 (IIc2); 4, 442/2 (IIc2); 5, D217 (IIc3); 6, 357900 (IIc1); 7, NK87 (IIc2); 8, T1 (IIc3); 9, W517 (IIc4); 10, F150 (IIc3); 11, 35227-2 (IIc4); 12, F250 (IIc4); 13, SA53 (IIa); 14, KH655 (IIa). The EplB amino acid sequence for isolate T5 is given in row 1, and dots show identical residues in other EplB subunits; differences from T5 are shown with a blue background, gaps in the protein sequences are shown as dashes. Signal sequences are boxed. Outside of the repeat region, EplB subunits divide into two main groups with either AKTG or TKRD signatures; two single residue variants are also identified, one in each group, and the two LT-IIa-specific EplB alleles are further variants of the AKTG signature.
To analyze further the variation in the number of N-terminal repeats, I performed PCR on each of the remaining RacatermF-EplBR PCR-positive type II ETEC isolates, with a primer pair that closely flank the variable region (EplBmatF, priming at the end of the signal peptide coding sequence and EplBR) and using high-resolution borate-based agarose gels (Brody et al. 2004) each isolate could be put into one of seven groups based on the size of the amplimer (Fig. 5), from 0.12 to 0.2 kb. By sequencing the complete EplB gene of representative isolates from each group, EplB variants were found with between three and five NA repeats, and DVSS repeats from one up to a maximum of seven, with variants with all intervening number of DVSS repeats found (Fig. 5). EplB subunits from two LT-IIa prophages are identical to each other and have the lowest number (one or two) of repeats, while LT-IIc loci generally have five or more repeats (only five LT-IIc loci encode three repeats and two encode four); the number of LT-IIc loci encoding five, six or seven repeats are eight, five and 13, respectively.
Most EplBA-negative LT-II isolates encode alleles of a second novel PT-like toxin family
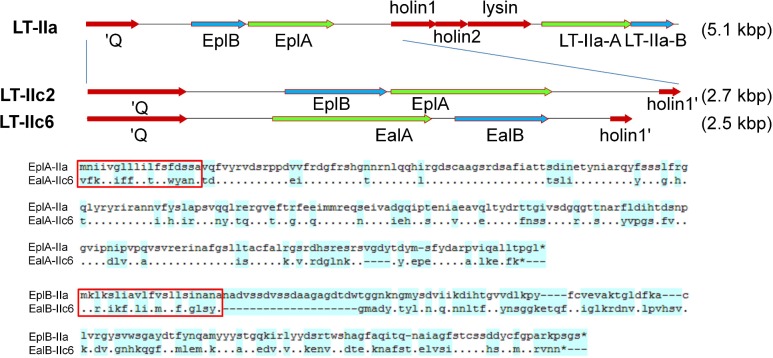
While the type strains for LT-IIc5 and LT-IIc6 did not produce PCR products with RacatermF and an eplB-specific reverse primer, they did produce slightly smaller (2.5 versus 2.8 kb) amplicons with RactermF and RacholinR primers compared to eplBA-positive isolates, indicating that the intervening DNA sequences differed. DNA sequencing of these PCR products showed, surprisingly, that both LT-IIc isolates did not encode EplBA toxins but instead encoded unique alleles of a second different, but still related, two-gene ADP-ribosylating toxin operon that has a more typical A-B gene order (Fig. 6). Comparing Eal to Epl polypeptides, the A polypeptides show 64% identity, while the B polypeptides show only 28% identity (Fig. 6). I designated the genes encoded in these LT-IIc isolates ealAB, for E. coli artAB-like AB toxin, as both predicted A and B polypeptides are most closely related to the ArtAB toxin subunits from S. enterica serovar Typhimurium DT104 (Saitoh et al. 2005) (67% –69% identity; Figs S5 and 6, Supporting Information). The ArtAB-encoding genes of Salmonella are present in the same location (between Q and holin genes) within a functional lambdoid prophage (Saitoh et al. 2005), and have been proposed as contributing to the enhanced virulence characteristics of this phage type (Uchida et al. 2009). EalA polypeptides from LT-IIc5 and LT-IIc6 are almost identical (99%), differing at only 2/242 residues, while their EalB polypeptides are more divergent (84% identity) differing at 22/142 residues (Fig. S2, Supporting Information).
Figure 6.
Identification of LT-II loci encoding alternative PT-like toxin (EalAB) genes and comparison of the predicted polypeptides with EplBA. A, A graphical map of the Q-EplBA-lysis-LT-IIa region of SA53 (top) is compared to the Q-holin PCR products using RacatermF and RacholinR primers in two LT-IIc isolates (expanded, lower). Filled blue arrows denote toxin B (binding) subunits, green arrows show toxin A (catalytic) subunits and red arrows show flanking prophage genes. Actual sizes of PCR products are 2789 bp (LT-IIc2) and 2488 bp (LT-IIc6). The complete LT-IIa toxin locus is 5.1 kb, and the equivalent PCR product on LT-II is 2731 bp. B and C, Amino-acid comparison of the Epl and Eal toxin A subunits (B) and B subunits (C). Signal sequences, matches and mismatches are identified and colored as in Fig. 1.
Determining the DNA sequences of the other eight RacatermF-holinR PCR-positive but eplBA-negative loci confirmed that each of these loci also encoded EalAB toxins. EalAB-encoding loci were found in two more LT-IIc5 isolates, all five LT-IIc1v isolates and one LT-IIa isolate. The IIc1v EalAB polypeptides were all identical and showed 99%–100% amino acid identity to EalAB from LT-IIc5, respectively, while the LT-IIa EalAB locus was unique, and the encoded polypeptides differed from all the others (73% identity for EalB; Fig. S2, Supporting Information). The major differences between the EalAB-encoding loci occur in the intergenic region between the end of the EalB gene and the start of the first holin gene; the coding sequences are highly conserved (red boxed regions; Fig. S3, Supporting Information), but the intervening region varies greatly in length and sequence, from 320 bp for IIc1v/IIc5 variants, 284 bp for IIc6 variant down to only 57 bp for the IIa variant locus. The significance of this is unclear. By comparison, all EplA and holin genes are separated by 494 bp, 99%–100% identical for the LT-IIc loci and 87% identical to the LT-IIa loci.
Non-random association of EplBA/EalAB and LT-II loci
Table 2 shows that the particular BA/AB toxin type also correlates closely with LT-II type and origin of isolate. Strikingly, four subgroups of LT-IIc (c1, c2, c3 and c4) isolates are only associated with eplBA genes and constitute two-thirds (33/50) of all LT-II isolates. All LT-IIc1v and the single c6 isolates encode EalAB toxins. While 90% (9/10) of the EalAB-encoding loci are found in isolates of animal origin, 90% (9/10) of human type II ETEC isolates encode EplBA toxins. This tight and non-random linkage between EalAB/EplBA and LT-II type raises the questions as to how the various combinations might have arisen, and what the selective forces are that produce the tight linkage these two toxins. Both toxin loci fit the definitions of morons, as beneficial elements that usually have their own promoters and terminators (Cumby et al. 2012b). Here, they are inserted between highly conserved Q and terminase genes, and these are potential sites where homologous recombination can occur. The most likely way to generate recombinants is by superinfection, of a type II ETEC isolate with a resident rac prophage, by an active toxin-encoding phage with homologous Q, holin and terminase genes but different EplBA, EalAB or LT-II loci. Relaxed recombination specificity of phage recombination enzymes such as λredαβ or Rac recE/recT gene products (De Paepe et al. 2014) drives assortment of phage modules, and can occur even when the resident prophage is defective. Bobay, Touchon and Rocha (2013) finds that ‘phages encoding their own recombination machinery have more mosaic genomes resulting from recent recombination events and have more diverse gene repertoires’. Superinfection of stx lysogens by other stx phages has been demonstrated (Fogg et al. 2011), and in a comparative genomics study of stx phages, Smith concludes that ‘recombination events between the host, phages and their remnants within the same infected bacterial cell will continue to drive the evolution of Stx phage variants’ (Smith et al. 2012). These events have apparently occurred frequently enough in type II ETEC to generate the many combinations of LT-II toxin and Epl/Eal toxin types we find here, and suggest that toxin-encoding Rac phages are or have recently been able to produce infectious particles. The tight linkage of a PT-like toxin with an LT-II toxin within the same prophage indicates that the ability to produce both toxins provides the cell with a currently undetermined selective advantage in nature.
Table 2.
Distribution of ealBA and ealAB loci among 50 LT-II ETEC isolates.
| LT-II toxin type; number of isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| c1 | c2 | c3 | c4 | c5 | c6 | a | b | ||
| Source | eplBA or ealAB type | 9a | 15 | 6 | 10 | 5 | 1 | 3 | 1 |
| Human | BA | 1 | 6 | 1 | 1 | - | - | - | - |
| AB | - | - | - | - | - | - | 1 | - | |
| Animal or food | BA | - | 9 | 5 | 9 | 2 | - | 2 | - |
| AB | 5b | - | - | - | 3 | 1 | - | - | |
Three isolates were negative by PCR with Q-holin primers.
All were LT-IIc1v subtype.
LT-IIb is encoded by a different highly defective prophage remnant and does not encode a PT-like toxin
From a collection of 50 type II ETEC isolates, only four produced no PCR product using the Q and holin gene-specific primers. Three of these were LT-IIc1 isolates, one from Brazil (raw beef), one from Sri Lanka and one from Belgium (both bovine); these could simply be primer mismatches or they may be more divergent; they were not studied further. The single LT-IIb isolate also gave no product, and neither the isolate, strain 41, nor TE1 carrying the cosmid clone, D27, showed clustering activity on CHO cells, indicating that the LT-IIb strain does not produce a PT-like toxin. It does, however encode a lysin gene 5′ of the LT-IIb locus (Jobling and Holmes 2012), indicating LT-IIb may also be encoded by a prophage. To verify this, I determined the prophage-encoding sequence of the single available LT-IIb cosmid clone D27; this cosmid appears chimeric (more than one contiguous genomic Sau3AI fragment inserts), and has a 10 kb Sau3AI fragment encoding the LT-IIb locus, joined to ∼26 kb of unlinked chromosomal DNA (shown graphically in Fig. S4, Supporting Information). The 10 kb DNA fragment encompasses 5 kb of sequence 5′ of the LT-IIb toxin genes, the LT-IIb genes and the remainder of the right arm, including the chromosomal integration site of the prophage. This shows that the LT-IIb locus is encoded within a prophage-like element, although the LT-IIb prophage is not found at the rac locus but is instead inserted into the serU tRNA gene, adjacent to the K12 yodB locus at 2.04 Mb in MG1655. Additionally, the LT-IIb prophage appears severely defective, missing the majority of the phage structural genes. It has only four genes (one hypothetical, a small terminase subunit and two phage tail fiber genes) following the LT-IIb gene before the predicted chromosomal insert site is found. Preceding the LT-IIb genes are homologs of the three gene lysis cassette, but no PT-like toxin locus. Instead, this region appears to have undergone rearrangement and/or duplication, having a second copy of a holin gene flanked by multiple tRNA genes. Phage-related sequences in the LT-IIb cosmid end with a partial gene showing homology to phage-encoded methylases. This gene is joined at a Sau3AI site to what appears to be an otherwise unlinked chromosomal DNA fragment, the ends of which encode ′xdhA and ′araE homologous sequences (Fig. S4, Supporting Information)that are separated by 26 kb in the K12 genome. Thus, strain 41, whose LT-IIb genes are nevertheless syntenic with their homolog and 5′ flanking genes in the LT-IIa/c prophages, encodes LT-IIb (but no PT-like toxin) within a non-Rac prophage. This SerU-prophage is unique, degenerate and divergent from the Rac prophages that encode all other LT-II loci.
Comparison of the A subunits between EplBA, EalAB and other PT-like families
I have identified two new families of PT-like toxins, EplBA most closely related to PltBA, and EalAB most closely related to ArtAB, predicted to encode an enzymatic (ADP-ribosyltransferase) A subunit and a ligand-binding B subunit. While the gene order is reversed between the eplBA and ealAB families (as it is for pltBA and artAB), their corresponding A- and B polypeptides are clearly structurally and functionally related. The A subunits of both novel toxins show 66% overall identity with each other, having more conserved amino-terminal regions and more variable carboxyl-terminal halves (Fig. 6). Together with the A subunits of the other two-gene toxins from Salmonella (ArtAB and PltBA), they share extensive amino acid homology with the enzymatic S1 subunit of PT Fig. S5, Supporting Information). The more divergent carboxyl termini are likely a function of the need to preserve interaction with their similarly related but quite different B subunits. Crystal structures are available for PltBA (Song, Gao and Galan 2013) (in conjunction with its second enzymatic A subunit, CdtB) and PT (Stein et al. 1994b). These show that the carboxyl domains of the enzymatic subunits extend into and interact significantly with the pore of the B pentamer. The amino-termini of these catalytic subunits show very similar folds and key catalytic residues are conserved with PT-S1 for all EplA and EalA alleles (Fig. S5, Supporting Information). Variants of both EplBA and EalAB with glu-to-ala substitutions for the predicted catalytic glutamate residues (E113A and E115A, respectively) in their A polypeptides assemble into holotoxin normally, but these do not intoxicate host cells (data not shown), indicating that their enzymatic function is required to induce the morphological response. In contrast, comparison of each of these A subunit sequences with PltA shows that two enzymatically critical residues (homologs of PT-S1 R13 and H35), that are also conserved in all other A/S1 subunits, are variant in PltA (substituted by T and F, respectively; identified in Fig. 1 and Fig. S5, Supporting Information). These are likely to affect the catalytic activity of PltA, and it is possible that PltA has devolved into a mainly structural and not catalytic role in Typhi toxin. The critical function of PltA may then be coupling the CdtB toxin protein to the PltB subunit (ligand-binding component) in its unique tripartite (A2-B5) toxin complex (Song, Gao and Galan 2013). All other toxins with S1-like subunits are therefore likely to be functional ADP-ribosyltransferases, and like PT, modify and inactivate inhibitory G-proteins; this has been proven for ArtAB (Uchida et al. 2009). Further work is necessary to identify specific subtrates for EplBA and EalAB toxins.
Comparison of the B subunits between EplBA, EalAB and other PT-like families
While their A subunits are highly related, both to each other and PltA and ArtA, the predicted mature EplB and EalB subunits are the most divergent polypeptides in the two-gene PT-like toxin group. EplB polypeptides are unique in possessing a variable number of amino-terminal DVSS-repeats, of unknown function, but with the majority of EplB subunits having higher numbers of repeats, the repeat may have biological significance. Comparing EplB (from LT-IIc2) and EalB (from LT-IIc5), they show only 23% identity, matching at only 38 of 156/121 residues. Excluding the repeat region of EplB, this rises to 28% identity. EplB shows progressively decreasing percent identity compared to SubB, PltB and ArtB (40%, 33% and 25%, respectively) while the percent identity increases for EalB (26%, 29% and 68%, respectively) (Fig. S6, Supporting Information). All five B subunits show weak homology to PT-S2/S4 subunits. Features conserved in all B subunits include cysteine pairs that in the structures of PltB, SubB and PT-S2/S4 form disulfide bridges, and critical ligand-binding residues also identified in Fig. S6 (Supporting Information). These data indicate that these novel EplBA and EalAB toxins are structurally and functionally related to the PltBA and ArtAB toxins and may bind similar ligands. To permit further characterization of these novel toxins, I proceeded to engineer toxin clones with carboxyl-terminal his-tagged B subunits with which to over-expression and purify the holotoxins.
Cloning and expression of EplBA and EalAB toxins
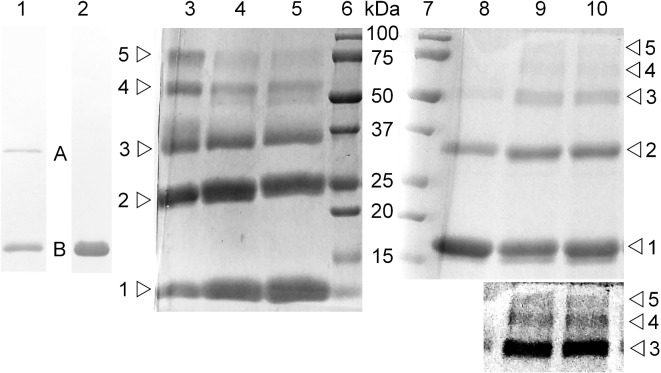
I cloned the complete eplBA locus from the original cosmid clone A38 (Pickett et al. 1986) as a 1.5 kb BglII-EcoRI fragment (see Fig. 1) into BamHI-EcoRI cut pBluescript SKII- (Alting-Mees and Short 1989); cell extracts of TE1 carrying this clone (pAEplBA-IIa) showed PT-like only activity on CHO cells (Fig. 7); extracts of cells carrying only the cloned eplB (a 1.5 kb ApoI deletion, removing the 3′ end of eplA making pAEplB-IIa) or eplA genes (a subclone of a PstI-KpnI fragment into pSKII-, deleting the 5′ end of eplB, making pAEplA-IIa) alone show no activity, demonstrating that both eplB and eplA genes are necessary and sufficient to express this novel toxin activity (Fig. 7). I then appended a 6-his tag with a (gly-pro)2-encoding linker to the carboxyl terminus of EplB, and expressed this gene as an operon with native eplA (see methods). Essentially pure assembled EplBA holotoxin was affinity purified from cell lysates of TE1 [pAEplBgpgpH6EplA] by metal-chelate (Talon) chromatography followed by anion exchange chromatography. Holotoxin (A + B subunits) eluted first, and consisted of two bands by SDS-PAGE (Fig. 8 lane 1), consistent with assembly into a multimeric complex of mature EplA (26 kDa) with tagged EplB-h6 (16 kDa). A second peak consisted of free B subunits (Fig. 8 lane 2). Intramolecular cross-linking of the purified EplBgpgpH6 B subunit only complex, followed by SDS-PAGE, showed a ladder of five bands consistent with the holotoxin consisting of a single A subunit associated with a homopentamer of B subunits (Fig. 8). For EplB, cross-linkers with larger spacers (DMP and DMS) more efficiently cross-linked all subunits of the pentamer, while for the control CT-B subunits, the cross-linker with the shortest spacer (DMA) was more efficient. This makes EplBA a bona fide member of the AB5 family of toxins. Assembly of the single EplB subunit into a pentamer is in agreement with the crystal structure of the tripartite A2B5 Typhi toxin (Song, Gao and Galan 2013) where the CdtB toxin is covalently coupled to PltA (via a disulfide bond), and PltA is non-covalently coupled to a pentamer of PltB subunits. Using a similar strategy, I cloned, expressed and purified his-tagged EalAB holotoxin (see methods). Both tagged variants of EplBA and EalAB holotoxin formed stable multimeric complexes that produced the characteristic clustering phenotype when added to CHO cells, indicating that the his-tag did not interfere with their ability to bind to and intoxicate target cells.
Figure 7.
Functional EplBA toxin requires both EplB and EplA subunits. A, Graphical maps of the wt eplBA clone (pAEplBA-IIa) expressed from the vector lacUV5 promoter, and the two deletion subclones pAEplA-IIa (PstI-KpnI subclone) and pAEplB-IIa (ApoI deletion). B, CHO cell clustering titer of periplasmic extracts of overnight cultures of plasmid constructs in E. coli TE1. Dashed line shows the limit of detection (25 μL per well, a titer of 40 U mL−1). Actual titers are show above the bars. Extracts from strains producing either polypetide alone do not show any detectable clustering activity.
Figure 8.
Purification and cross-linking of EplBA holotoxin subunits. Peaks eluting with NaCl gradient from Talon-purified EplBgpgpH6-EplA bound to HS10 anion exchange column. Lane 1, holotoxin; lane 2, EplBgpgpH6-only subunits; lanes 3, 4, 5 control cross-linking of CTB; lanes 6 and 7, molecular size standards (in kDa); lanes 8, 9 and 10, cross-linked EplBgpgpH6. Open triangles show the number of subunits in each band. Cross-linkers-DMA—lanes 3, 8; DMP—lanes 4, 9; DMS—lanes 5, 10. Inset box under lanes 9 and 10 shows a contrast-enhanced region of upper portion of the same lanes in order to more readily identify the upper bands. Proven pentameric CTB and EplB both show the monomers and four cross-linked forms consistent with a pentameric assembly of monomers.
Detection by glycan array of ligands bound by EplBA and EalAB toxin B subunits
The ligand-binding function for PT resides in its B subunits (Rossjohn et al. 1997) and is required for intoxication of host cells. PT binds to glycan moieties of host cell surface glycoproteins (Witvliet et al. 1989; Stein et al. 1994a), preferentially sialoglycoproteins (el Bayâ et al. 1995). Similarly, an AB5 toxin with a related B subunit (SubAB) binds to glycoprotein ligands (Beddoe et al. 2010), preferentially an α(2-3)-linked N-glycolylneuraminic acid (Neu5Gc). Although this sugar does not occur naturally in humans, it is incorporated into and sensitizes human intestinal cells by absorption from dietary foods (Byres et al. 2008). Recently, the preferential glycan recognized by the PltB subunit was determined (Song, Gao and Galan 2013) to be terminal galactose with α(2-3)-linked N-acetylneuraminic acid (Neu5Ac). The structures of SubB (Byres et al. 2008), SubAB (Le Nours et al. 2013) and Typhi toxin (PltBACdtB) (Song, Gao and Galan 2013) have all recently been solved and their B subunits have nearly identical backbone folds (PltB and SubB have and a root mean square deviation (rmsd) of <0.5 Å for 97 Cα atoms with 50% identity), and the more divergent PltB and PT-S2 B subunits’ sugar-binding domains have an rmsd of <1.8 Å over 80 Cα atoms (Song, Gao and Galan 2013).
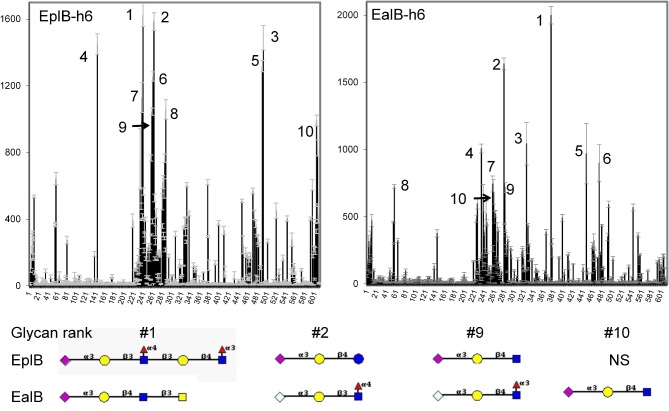
The EplBA and EalAB toxins also appear to use glycoproteins as ligands, as the glycan-deficient CHO cell line CHO-15B (Brennan et al. 1988) is between 333- and 390-fold less susceptible to both toxins than the parent CHO cell line (identical within the limits of the assay) to these cell lines’ response to PT (400-fold less susceptible), as determined by morphological assay (Fig. 9). CHO-15B cells show cell clustering by EplBA, EalAB or PT only when treated with 50–62 ng or greater, whereas CHO cells are susceptible to 125–160 pg amounts of these toxins. I have determined the preferred glycans for the his-tagged EplB and EalB subunits using the version 5.1 glycan array at the Consortium for Functional Glycomics (see www.functionalglycomics.org for complete data). This array has 610 unique mammalian glycans and the data show that these two toxins binds different but overlapping sets of glycans, with terminal sialated galactose sugars in common. While both EplB and EalB bind best to glycans with terminal gal-α(2-3)-Neu5Ac, EalAB also binds to glycans with terminal gal-α(2-3)-Neu5Gc. Figure 10 (top panels) shows the signals obtained for all glycans in the array for each toxin B subunit (left, EplB and right, EalB), and Fig. 10 (lower panel) shows the structures of representative top ten glycans bound by each toxin. The top ten ranking glycans bound by EplB and EalB are shown in Tables 3 and 4, respectively, along with the corresponding ranking for that glycan's binding by the other toxin.
Figure 9.

Sialyated glycoproteins are functional ligands for EplBA and EalAB toxins. Tissue culture clustering assays of serial dilutions of purified his-tagged or pertussis toxin on wt CHO cells or glycosylation-deficient CHO-15B cells; toxin titers are expressed as log10 of the clustering units per μg. Above each bar is given the minimal amount of toxin required to observe clustering on the respective cell line. The almost three log decrease in toxin titer on CHO-15B cells indicates that sialated glycoproteins are the major functional ligands for all these toxins.
Figure 10.
Glycan array (PA_v51) analysis of EplBgpgpHis6 and EalBgpgpH6 toxin binding. Profiles for EplB (left panel) and EalB (right panel) binding to the consortium for functional glycomics printed array v5.1, which has 609 mammalian glycans per slide (numbered on the X-axis). Y-axis values represent the average relative fluorescence unit signal of bound toxin (200 μg mL−1) detected with Alexa488-labeled anti-His antibody. The schematic structure of the most relevant glycans bound (top two glycans for each subunit, first shared glycan—no. 9 for EplB and no. 10 for EalB, and the second Neu5Gc-containing glycan for EalB) for each subunit is shown beneath the panels, along with the specific linkages. Diamonds, Neu5Ac (magenta) or Neu5Gc (pale blue); circles, Gal (yellow) or Glc (blue); squares, GlcNac (yellow) or GalNac (blue); red triangle, Fuc; sialic acids are linked α1-3 to Gal, all other linkages are α1 or β1-3 or -4 as shown. NS, not shown. Terminal sugar is on the left, glycan linkage to the right.
Table 3.
Selected glycans bound by EplB.
| EplB | EalB | Compound | EplB | Fraction | |
|---|---|---|---|---|---|
| rank | rank | no. | signal | of max | Compound (top ten and others selected from 75 high-affinity glycans) |
| 1 | 30 | 240 | 1625 | 1.00 | Neu5Acα2-3Galβ1-3(Fucα1-4)GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ-Sp0 |
| 2 | 15 | 263 | 1586 | 0.98 | Neu5Acα2-3Galβ1-4Glcβ-Sp0 |
| 3 | 13 | 493 | 1488 | 0.92 | Galβ1-4(Fucα1-3)GlcNAcβ1-6(Neu5Acα2-6(Neu5Acα2-3Galβ1-3)GlcNAcβ1-3)Galβ1-4Glc-Sp21 |
| 4 | 32 | 144 | 1451 | 0.89 | Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glcβ-Sp0 |
| 5 | 33 | 491 | 1319 | 0.81 | Neu5Acα2-3Galβ1-3GlcNAcβ1-6GalNAcα-Sp14 |
| 6 | 17 | 261 | 1256 | 0.77 | Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ-Sp0 |
| 7 | 11 | 238 | 1129 | 0.69 | Neu5Acα2-3Galβ1-3(6S)GlcNAc-Sp8 |
| 8 | 35 | 288 | 1059 | 0.65 | Neu5Acα2-3Galβ1-4GlcNAcβ1-6(Galβ1-3)GalNAcα-Sp14 |
| 9 | 10 | 259 | 1045 | 0.64 | Neu5Acα2-3Galβ1-4GlcNAcβ-Sp0 |
| 10 | 62 | 605 | 992 | 0.61 | [X]a β1-3Galβ1-4GlcNAcβ1-6([X]a β1-3Galβ1-4GlcNAcβ1-3)GalNAcα-Sp14 |
| 16 | 9 | 282 | 622 | 0.38 | Neu5Gcα2-3Galβ1-4(Fucα1-3)GlcNAcβ-Sp0 |
| 20 | 22 | 11 | 537 | 0.33 | Neu5Acβ-Sp8 |
| 48 | 23 | 281 | 359 | 0.22 | Neu5Gcα2-3Galβ1-3GlcNAcβ-Sp0 |
[X] = Neu5Acα2-6Galβ1-4GlcNAc.
Neu5Ac, N-acetyl-neuraminic acid; Neu5Gc, N-glycolyl-neuraminic acid; Gal, galactose; GalNAc, N-aceytl-galacotosamine; Glc, glucose; GlcNAc, N-acetyl-glucosamine; Fuc, Fucose; Man, Mannose, Sp, spacer (number indicates specific type).
Table 4.
Selected glycans bound by EalB.
| EalB | EplB | Compound | EalB | Fraction | |
|---|---|---|---|---|---|
| Rank | rank | no. | signal | of max | Compound (top ten and others selected from 35 high-affinity glycans) |
| 1 | 17 | 376 | 1999 | 1.00 | Neu5Acα2-3Galβ1-4GlcNAcβ1-3GalNAc-Sp14 |
| 2 | 26 | 280 | 1638 | 0.82 | Neu5Gcα2-3Galβ1-3(Fucα1-4)GlcNAcβ-Sp0 |
| 3 | 49 | 326 | 1043 | 0.52 | Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-6([X]a β1-2Manα1-3)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 |
| 4 | 14 | 234 | 1008 | 0.50 | Neu5Acα2-3Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glcβ-Sp0 |
| 5 | 25 | 448 | 970 | 0.48 | Galβ1-4GlcNAcβ1-2Manα-Sp0 |
| 6 | 33 | 474 | 901 | 0.45 | Neu5Acα2-3Galβ1-3GlcNAcβ1-6(Neu5Acα2-3Galβ1-3GlcNAcβ1-2)[Z]b β-Sp19 |
| 7 | 12 | 257 | 744 | 0.37 | Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4GlcNAcβ-Sp8 |
| 8 | 15 | 57 | 725 | 0.36 | [X]a β1-2Manα1-6([X]a β1-2Manα1-3)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp24 |
| 9 | 16 | 282 | 716 | 0.36 | Neu5Gcα2-3Galβ1-4(Fucα1-3)GlcNAcβ-Sp0 |
| 10 | 9 | 259 | 684 | 0.34 | Neu5Acα2-3Galβ1-4GlcNAcβ-Sp0 |
| 22 | 20 | 11 | 458 | 0.23 | Neu5Acβ-Sp8 |
| 23 | 48 | 281 | 448 | 0.22 | Neu5Gcα2-3Galβ1-3GlcNAcβ-Sp0 |
| 27 | 55 | 283 | 391 | 0.20 | Neu5Gcα2-3Galβ1-4GlcNAcβ-Sp0 |
[X] = Neu5Acα2-6Galβ1-4GlcNAc;
[Z] = Manα1-6(Neu5Acα2-3Galβ1-3GlcNAcβ1-2Manα1-3)Manβ1-4GlcNAcβ1-4GlcNAc.
Remarkably, EplB and EalB bind very similar glycans despite having only 23% identity in their mature subunits. Both toxins bind sialated galactose residues, as do PltB (Song, Gao and Galan 2013) and SubB (Byres et al. 2008). Overall, although they bind unique sets of glycans, half of the top ten glycans for one toxin are also in the top 20 glycans bound by the other toxin. Common among the top glycans for both toxins are terminal Neu5Acα2-3Galβ1-4GlcNAc units, although Galβ1-3GlcNAc linkages appear equally frequently; glycans with fucosylation of the GlcNAc residue are bound with increased affinity for both toxins (compare EplB glycans no. 1 and 5; EalB glycans ranked no. 9 and 27). Both EplB and EalB bind to Neu5Ac alone, ranked no. 20 and 23, respectively. The increased ability of EalB to bind Neu5Gc-containing glycans (that humans do not produce) correlates with the animal origin of the majority of the EalAB-positive type II ETEC isolates, although the ability of dietary Neu5Gc glycans to become functional receptors for SubAB on human intestinal cells (Byres et al. 2008) suggests that this may also happen with EalAB. Importantly, residues involved in ligand binding by the mature SubB subunit (S12, Y78) are conserved in both EplB and EalB. Additionally, EalB (but not EplB) has D8 in common with SubB that is critical for the SubB pentamer ability to bind Neu5Gc (Byres et al. 2008). However, EplB can still bind Neu5Gc glycans but with apparently lower affinity—four of its top 30 glycans contain Neu5Gc although they rank towards the lower end of this group, whereas EalB has 3 Neu5Gc-containing glycans in its top 15. EplB and EalB appear to be more promiscuous towards the sialic acid group than both PltB and SubB, which are much more specific for Neu5Ac and Neu5Gc, respectively. Differences between toxins in their preferred ligands may result in altered patterns of target cell intoxication and likely lead to differences in host response to each of these toxins. This could include differences in immunomodulatory properties with consequences for the infection process or their potential use a novel adjuvants. Further work in this area is warranted.
CONCLUDING REMARKS
I showed that the genes encoding the heat-labile enterotoxins of type II ETEC are present within a lambdoid prophage inserted at the rac locus, that in E. coli K12 strains encodes a defective prophage. Although I have yet to detect production of plaque-forming phage, enterotoxigenicity of type II ETEC can be added to the growing list of virulence factors of pathogenic bacteria that are phage-encoded and may be acquired by lysogenic conversion (Casas and Maloy 2011). Almost all (46/50) of these LT-II toxin prophages in my collection also encoded one of two novel, two-gene PT-like toxins (EplBA and EalAB) that have homologs in S. enterica prophages found in the Typhi serovar (PltBA, encoded by a degenerate and defective remnant prophage) and in the Typhimurium serovar DT104 phage type lineage (ArtAB, encoded within a functional prophage). The location within the prophage of these dual toxin operons (EplBA or EalAB and LT-IIa/c), like the Shiga toxin genes of EHEC prophages, indicates that their expression is likely to be increased during lytic induction of the prophage, being the first genes to be expressed by Q-directed antitermination that drives synthesis of the late structural and lysis genes of the phage particle. This is the first description of a pathogen producing two ADP-ribosylating toxins that are predicted to target both arms of host cell control over adenylate cyclase, by modifying and constitutively activating the stimulatory G-protein Gsα (CT/LT-I/LT-II action) and additionally removing inhibitory control by modifying and inactivating Giα (PT-like action). The extremely tight linkage of LT-II toxins with a PT-like toxin in the vast majority of LT-II isolates strongly implies that this combination is biologically advantageous to the host strain. Simultaneous production of both toxins thus has the potential to synergistically increase cAMP production by intoxicated cells. In addition to affecting cAMP levels in intoxicated cells, both toxins, through their modification of both stimulatory and inhibitory host cell Gα subunits, are likely, like cholera and PT, to have strong immunomodulatory effects on the host cell mucosal tissue, including both cytokine production by target enterocytes (Soriani, Bailey and Hirst 2002; Wang et al. 2013) and immune responses by the gut-associated lymphoid tissue (Elson and Ealding 1984; Queen and Satchell 2013; Mattsson et al. 2014).
Supplementary Material
Acknowledgments
I thank Lisa Gotow for technical assistance in protein purification and cross-linking experiments and Randall Holmes for support and access to strains.
SUPPLEMENTARY DATA
FUNDING
This work was supported in part by National Institutes of Health grants AI-31940 and AI-14107. Glycan array screenings were performed by the Consortium for Functional Glycomics supported in part by National Institutes of Health Grants GM62116 and GM098791.
Conflict of interest. None declared.
REFERENCES
- Allen KP, Randolph MM, Fleckenstein JM. Importance of heat-labile enterotoxin in colonization of the adult mouse small intestine by human enterotoxigenic Escherichia coli strains. Infect Immun. 2006;74:869–75. doi: 10.1128/IAI.74.2.869-875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alting-Mees MA, Short JM. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989;17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadulghani M, Ogura Y, Ooka T, et al. The defective prophage pool of Escherichia coli O157: prophage–prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 2009;5:e1000408. doi: 10.1371/journal.ppat.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddoe T, Paton AW, Le Nours J, et al. Structure, biological functions and applications of the AB5 toxins. Trends Biochem Sci. 2010;35:411–8. doi: 10.1016/j.tibs.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay L-M, Touchon M, Rocha EPC. Manipulating or superseding host recombination functions: a dilemma that shapes phage evolvability. PLoS Genet. 2013;9:e1003825. doi: 10.1371/journal.pgen.1003825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan MJ, David JL, Kenimer JG, et al. Lectin-like binding of pertussis toxin to a 165-kilodalton Chinese hamster ovary cell glycoprotein. J Biol Chem. 1988;263:4895–9. [PubMed] [Google Scholar]
- Brody JR, Calhoun ES, Gallmeier E, et al. Ultra-fast high-resolution agarose electrophoresis of DNA and RNA using low-molarity conductive media. Biotechniques. 2004;37:598, 600, 602. doi: 10.2144/04374ST04. [DOI] [PubMed] [Google Scholar]
- Byres E, Paton AW, Paton JC, et al. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature. 2008;456:648–52. doi: 10.1038/nature07428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonetti NH. Contribution of pertussis toxin to the pathogenesis of pertussis disease. Pathog Dis. 2015;73 doi: 10.1093/femspd/ftv073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonetti NH, Artamonova GV, Van Rooijen N, et al. Pertussis toxin targets airway macrophages to promote bordetella pertussis infection of the respiratory tract. Infect Immun. 2007;75:1713–20. doi: 10.1128/IAI.01578-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas V, Maloy S. Role of bacteriophage-encoded exotoxins in the evolution of bacterial pathogens. Future Microbiol. 2011;6:1461–73. doi: 10.2217/fmb.11.124. [DOI] [PubMed] [Google Scholar]
- Casey TA, Connell TD, Holmes RK, et al. Evaluation of heat-labile enterotoxins type IIa and type IIb in the pathogenicity of enterotoxigenic Escherichia coli for neonatal pigs. Vet Microbiol. 2012;159:83–9. doi: 10.1016/j.vetmic.2012.03.018. [DOI] [PubMed] [Google Scholar]
- Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Hendrix RW. Bacteriophage lambda: early pioneer and still relevant. Virology. 2015;479-480:310–30. doi: 10.1016/j.virol.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly CE, Sun Y, Carbonetti NH. Pertussis toxin exacerbates and prolongs airway inflammatory responses during Bordetella pertussis infection. Infect Immun. 2012;80:4317–32. doi: 10.1128/IAI.00808-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conter A, Bouche JP, Dassain M. Identification of a new inhibitor of essential division gene ftsZ as the kil gene of defective prophage Rac. J Bacteriol. 1996;178:5100–4. doi: 10.1128/jb.178.17.5100-5104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumby N, Davidson AR, Maxwell KL. The moron comes of age. Bacteriophage. 2012a;2:225–8. doi: 10.4161/bact.23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumby N, Edwards AM, Davidson AR, et al. The bacteriophage HK97 gp15 moron element encodes a novel superinfection exclusion protein. J Bacteriol. 2012b;194:5012–9. doi: 10.1128/JB.00843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe M, Hutinet G, Son O, et al. Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: the role of Rad52-like recombinases. PLoS Genet. 2014;10:e1004181. doi: 10.1371/journal.pgen.1004181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Bayâ A, Linnemann R, von Olleschik-Elbheim L, et al. Identification of binding proteins for pertussis toxin on pancreatic β cell-derived insulin-secreting cells. Microb Pathog. 1995;18:173–85. doi: 10.1016/s0882-4010(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Elson CO, Ealding W. Generalized systemic and mucosal immunity in mice after mucosal stimulation with cholera toxin. J Immunol. 1984;132:2736–41. [PubMed] [Google Scholar]
- Evans R, Seeley NR, Kuempel PL. Loss of rac locus DNA in merozygotes of Escherichia coli K12. Mol Gen Genet. 1979;175:245–50. doi: 10.1007/BF00397223. [DOI] [PubMed] [Google Scholar]
- Fogg PCM, Rigden DJ, Saunders JR, et al. Characterization of the relationship between integrase, excisionase and antirepressor activities associated with a superinfecting Shiga toxin encoding bacteriophage. Nucleic Acids Res. 2011;39:2116–29. doi: 10.1093/nar/gkq923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco BD, Gomes TA, Jakabi M, et al. Use of probes to detect virulence factor DNA sequences in Escherichia coli strains isolated from foods. Int J Food Microbiol. 1991;12:333–8. doi: 10.1016/0168-1605(91)90147-h. [DOI] [PubMed] [Google Scholar]
- Fullner Satchell KJ. Activation and suppression of the proinflammatory immune response by Vibrio cholerae toxins. Microbes Infect. 2003;5:1241–7. doi: 10.1016/j.micinf.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Glenn GM, Francis DH, Danielsen EM. Toxin-mediated effects on the innate mucosal defenses: implications for enteric vaccines. Infect Immun. 2009;77:5206–15. doi: 10.1128/IAI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb C, Baenziger J, Kornfeld S. Deficient uridine diphosphate-N-acetylglucosamine:glycoprotein N-acetylglucosaminyltransferase activity in a clone of Chinese hamster ovary cells with altered surface glycoproteins. J Biol Chem. 1975;250:3303–9. [PubMed] [Google Scholar]
- Gottlieb C, Skinner AM, Kornfeld S. Isolation of a clone of Chinese hamster ovary cells deficient in plant lectin-binding sites. P Natl Acad Sci USA. 1974;71:1078–82. doi: 10.1073/pnas.71.4.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BA, Neill RJ, Ruyechan WT, et al. Evidence that a new enterotoxin of Escherichia coli which activates adenylate cyclase in eucaryotic target cells is not plasmid mediated. Infect Immun. 1983;41:383–90. doi: 10.1128/iai.41.1.383-390.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grose JH, Casjens SR. Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology. 2014;468–470:421–43. doi: 10.1016/j.virol.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrant RL, Brunton LL, Schnaitman TC, et al. Cyclic adenosine monophosphate and alteration of Chinese hamster ovary cell morphology: a rapid, sensitive in vitro assay for the enterotoxins of Vibrio cholerae and Escherichia coli. Infect Immun. 1974;10:320–7. doi: 10.1128/iai.10.2.320-327.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth BE, Pickett CL, Twiddy EM, et al. Production of type II heat-labile enterotoxin by Escherichia coli isolated from food and human feces. Infect Immun. 1986;54:587–9. doi: 10.1128/iai.54.2.587-589.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW, Lawrence JG, Hatfull GF, et al. The origins and ongoing evolution of viruses. Trends Microbiol. 2000;8:504–8. doi: 10.1016/s0966-842x(00)01863-1. [DOI] [PubMed] [Google Scholar]
- Hewlett EL, Myers GA, Sauer KT, et al. Alteration of Chinese hamster ovary (CHO) cell growth and cyclic amp metabolism by pertussis toxin (PT) Clin Res. 1982;30:368A. [Google Scholar]
- Hewlett EL, Sauer KT, Myers GA, et al. Induction of a novel morphological response in Chinese hamster ovary cells by pertussis toxin. Infect Immun. 1983;40:1198–203. doi: 10.1128/iai.40.3.1198-1203.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodak H, Galan JE. A Salmonella Typhi homologue of bacteriophage muramidases controls typhoid toxin secretion. EMBO Rep. 2013;14:95–102. doi: 10.1038/embor.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn B, Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980;11:291–8. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Holmes RK, Jobling MG, Connell TD. Cholera toxin and related enterotoxins of Gram-negative bacteria. In: Moss J, Iglewski B, Vaughan M, et al., editors. Bacterial Toxins and Virulence Factors in Disease. Vol. 8. New York: Marcel Dekker, Inc.; 1995. pp. 225–56. [Google Scholar]
- Hong SH, Wang X, Wood TK. Controlling biofilm formation, prophage excision and cell death by rewiring global regulator H-NS of Escherichia coli. Microb Biotechnol. 2010;3:344–56. doi: 10.1111/j.1751-7915.2010.00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Cai Z, Ho SN, et al. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–35. [PubMed] [Google Scholar]
- Hsia JA, Moss J, Hewlett EL, et al. Requirement for both choleragen and pertussis toxin to obtain maximal activation of adenylate cyclase in cultured cells. Biochem Bioph Res Co. 1984;119:1068–74. doi: 10.1016/0006-291x(84)90883-0. [DOI] [PubMed] [Google Scholar]
- Ichihara Y, Kurosawa Y. Construction of new T vectors for direct cloning of PCR products. Gene. 1993;130:153–4. doi: 10.1016/0378-1119(93)90361-6. [DOI] [PubMed] [Google Scholar]
- Jobling MG, Holmes RK. Identification of motifs in cholera toxin A1 polypeptide that are required for its interaction with human ADP-ribosylation factor 6 in a bacterial two-hybrid system. P Natl Acad Sci USA. 2000;97:14662–7. doi: 10.1073/pnas.011442598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MG, Holmes RK. Type II heat-labile enterotoxins from 50 diverse Escherichia coli isolates belong almost exclusively to the LT-IIc family and may be prophage encoded. PLoS One. 2012;7:e29898. doi: 10.1371/journal.pone.0029898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MG, Palmer LM, Erbe JL, et al. Construction and characterization of versatile cloning vectors for efficient delivery of native foreign proteins to the periplasm of Escherichia coli. Plasmid. 1997;38:158–73. doi: 10.1006/plas.1997.1309. [DOI] [PubMed] [Google Scholar]
- Juhala RJ, Ford ME, Duda RL, et al. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- Kaiser K, Murray NE. Physical characterisation of the “Rac prophage” in E. coli K12. Mol Gen Genet. 1979;175:159–74. doi: 10.1007/BF00425532. [DOI] [PubMed] [Google Scholar]
- Kaiser K, Murray NE. On the nature of sbcA mutations in E. coli K 12. Mol Gen Genet. 1980;179:555–63. doi: 10.1007/BF00271745. [DOI] [PubMed] [Google Scholar]
- Katada T. The inhibitory G protein Gi identified as pertussis toxin-catalyzed ADP-ribosylation. Biol Pharm Bull. 2012;35:2103–11. doi: 10.1248/bpb.b212024. [DOI] [PubMed] [Google Scholar]
- Le Nours J, Paton AW, Byres E, et al. Structural basis of subtilase cytotoxin SubAB assembly. J Biol Chem. 2013;288:27505–16. doi: 10.1074/jbc.M113.462622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Hajishengallis G. Heat-labile enterotoxins as adjuvants or anti-inflammatory agents. Immunol Invest. 2010;39:449–67. doi: 10.3109/08820130903563998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Mendez G, Toussaint A, Leplae R. A modular view of the bacteriophage genomic space: identification of host and lifestyle marker modules. Res Microbiol. 2011;162:737–46. doi: 10.1016/j.resmic.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Low B. Restoration by the rac locus of recombinant forming ability in recB - and recC - merozygotes of Escherichia coli K-12. Mol Gen Genet. 1973;122:119–30. doi: 10.1007/BF00435185. [DOI] [PubMed] [Google Scholar]
- Lu C, Erickson HP. Expression in Escherichia coli of the thermostable DNA polymerase from Pyrococcus furiosus. Protein Expres Purif. 1997;11:179–84. doi: 10.1006/prep.1997.0775. [DOI] [PubMed] [Google Scholar]
- Matsushiro A, Sato K, Miyamoto H, et al. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with Norfloxacin. J Bacteriol. 1999;181:2257–60. doi: 10.1128/jb.181.7.2257-2260.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J, Schon K, Ekman L, et al. Cholera toxin adjuvant promotes a balanced Th1/Th2/Th17 response independently of IL-12 and IL-17 by acting on Gsα in CD11b+ DCs. Mucosal Immunol. 2014;8:815–27. doi: 10.1038/mi.2014.111. [DOI] [PubMed] [Google Scholar]
- Melsom CB, Ørstavik Ø, Osnes J-B, et al. Gi Proteins regulate adenylyl cyclase activity independent of receptor activation. PLoS ONE. 2014;9:e106608. doi: 10.1371/journal.pone.0106608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menouni R, Hutinet G, Petit M-A, et al. Bacterial genome remodeling through bacteriophage recombination. FEMS Microbiol Lett. 2015;362:1–10. doi: 10.1093/femsle/fnu022. [DOI] [PubMed] [Google Scholar]
- Nardi ARM, Salvadori MR, Coswig LT, et al. Type 2 heat-labile enterotoxin (LT-II)-producing Escherichia coli isolated from ostriches with diarrhea. Vet Microbiol. 2005;105:245–9. doi: 10.1016/j.vetmic.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Nawar HF, King-Lyons ND, Hu JC, et al. LT-IIc, a new member of the type II heat-labile enterotoxin family encoded by an Escherichia coli strain obtained from a nonmammalian host. Infect Immun. 2010;78:4705–13. doi: 10.1128/IAI.00730-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely MN, Friedman DI. Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a Shiga-like toxin. Gene. 1998a;223:105–13. doi: 10.1016/s0378-1119(98)00236-4. [DOI] [PubMed] [Google Scholar]
- Neely MN, Friedman DI. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol Microbiol. 1998b;28:1255–67. doi: 10.1046/j.1365-2958.1998.00890.x. [DOI] [PubMed] [Google Scholar]
- Oppenheim AB, Kobiler O, Stavans J, et al. Switches in bacteriophage lambda development. Annu Rev Genet. 2005;39:409–29. doi: 10.1146/annurev.genet.39.073003.113656. [DOI] [PubMed] [Google Scholar]
- Paton AW, Srimanote P, Talbot UM, et al. A new family of potent AB5 cytotoxins produced by Shiga toxigenic Escherichia coli. J Exp Med. 2004;200:35–46. doi: 10.1084/jem.20040392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Grishin NV. COG3926 and COG5526: a tale of two new lysozyme-like protein families. Protein Sci. 2005;14:2574–81. doi: 10.1110/ps.051656805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett CL, Twiddy EM, Belisle BW, et al. Cloning of genes that encode a new heat-labile enterotoxin of Escherichia coli. J Bacteriol. 1986;165:348–52. doi: 10.1128/jb.165.2.348-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett CL, Twiddy EM, Coker C, et al. Cloning, nucleotide sequence, and hybridization studies of the type IIb heat-labile enterotoxin gene of Escherichia coli. J Bacteriol. 1989;171:4945–52. doi: 10.1128/jb.171.9.4945-4952.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett CL, Weinstein DL, Holmes RK. Genetics of type IIa heat-labile enterotoxin of Escherichia coli: operon fusions, nucleotide sequence and hybridization studies. J Bacteriol. 1987;169:5180–7. doi: 10.1128/jb.169.11.5180-5187.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce NF, Kaper JB, Mekalanos JJ, et al. Role of cholera toxin in enteric colonization by Vibrio cholerae O1 in rabbits. Infect Immun. 1985;50:813–6. doi: 10.1128/iai.50.3.813-816.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl P, Lintermans P, Mainil J, et al. ETEC-like strains from cattle. Vet Rec. 1989;125:382. doi: 10.1136/vr.125.14.382. [DOI] [PubMed] [Google Scholar]
- Queen J, Satchell KJF. Promotion of colonization and virulence by cholera toxin is dependent on neutrophils. Infect Immun. 2013;81:3338–45. doi: 10.1128/IAI.00422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossjohn J, Buckley JT, Hazes B, et al. Aerolysin and pertussis toxin share a common receptor-binding domain. EMBO J. 1997;16:3426–34. doi: 10.1093/emboj/16.12.3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack R. The discovery of cholera - like enterotoxins produced by Escherichia coli causing secretory diarrhoea in humans. Indian J Med Res. 2011;133:171–8. [PMC free article] [PubMed] [Google Scholar]
- Saitoh M, Tanaka K, Nishimori K, et al. The artAB genes encode a putative ADP-ribosyltransferase toxin homologue associated with Salmonella enterica serovar Typhimurium DT104. Microbiology. 2005;151:3089–96. doi: 10.1099/mic.0.27933-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schneider OD, Weiss AA, Miller WE. Pertussis toxin utilizes proximal components of the T-cell receptor complex to initiate signal transduction events in T cells. Infect Immun. 2007;75:4040–9. doi: 10.1128/IAI.00414-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears CL, Kaper JB. Enteric bacterial toxins: mechanisms of action and linkage to intestinal secretion. Microbiol Rev. 1996;60:167–215. doi: 10.1128/mr.60.1.167-215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seriwatana J, Echeverria P, Taylor DN, et al. Type II heat-labile enterotoxin-producing Escherichia coli isolated from animals and humans. Infect Immun. 1988;56:1158–61. doi: 10.1128/iai.56.5.1158-1161.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöling A, Wiklund G, Savarino SJ, et al. Comparative analyses of phenotypic and genotypic methods for detection of enterotoxigenic Escherichia coli toxins and colonization factors. J Clin Microbiol. 2007;45:3295–301. doi: 10.1128/JCM.00471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Rooks D, Fogg P, et al. Comparative genomics of Shiga toxin encoding bacteriophages. BMC Genomics. 2012;13:311. doi: 10.1186/1471-2164-13-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Gao X, Galan JE. Structure and function of the Salmonella Typhi chimaeric A2B5 typhoid toxin. Nature. 2013;499:350–4. doi: 10.1038/nature12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriani M, Bailey L, Hirst TR. Contribution of the ADP-ribosylating and receptor-binding properties of cholera-like enterotoxins in modulating cytokine secretion by human intestinal epithelial cells. Microbiology. 2002;148:667–76. doi: 10.1099/00221287-148-3-667. [DOI] [PubMed] [Google Scholar]
- Spano S, Ugalde JE, Galan JE. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host & Microbe. 2008;3:30–8. doi: 10.1016/j.chom.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Stein PE, Boodhoo A, Armstrong GD, et al. Structure of a pertussis toxin-sugar complex as a model for receptor binding. Nat Struct Biol. 1994a;1:591–6. doi: 10.1038/nsb0994-591. [DOI] [PubMed] [Google Scholar]
- Stein PE, Boodhoo A, Armstrong GD, et al. The crystal structure of pertussis toxin. Structure. 1994b;2:45–57. doi: 10.1016/s0969-2126(00)00007-1. [DOI] [PubMed] [Google Scholar]
- Stojković EA, Rothman-Denes LB. Coliphage N4 N-Acetylmuramidase defines a new family of murein hydrolases. J Mol Biol. 2007;366:406–19. doi: 10.1016/j.jmb.2006.11.028. [DOI] [PubMed] [Google Scholar]
- Tripet B, Howard MW, Jobling M, et al. Structural characterization of the SARS-coronavirus spike S fusion protein core. J Biol Chem. 2004;279:20836–49. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida I, Ishihara R, Tanaka K, et al. Salmonella enterica serotype Typhimurium DT104 ArtA-dependent modification of pertussis toxin-sensitive G proteins in the presence of [32P]NAD. Microbiology. 2009;155:3710–8. doi: 10.1099/mic.0.028399-0. [DOI] [PubMed] [Google Scholar]
- Wang H, Paton JC, Herdman BP, et al. The B subunit of an AB5 toxin produced by Salmonella enterica serovar typhi up-regulates chemokines, cytokines and adhesion molecules in human macrophage, colonic epithelial and brain microvascular endothelial cell lines. Infect Immun. 2013;81:673–768. doi: 10.1128/IAI.01043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AD, Robinson A, Irons L, et al. Cholera and pertussis toxins, but not forskolin or LT-B, adjuvant IgA antibody responses to orally administered antigen. Adv Exp Med Biol. 1995;371B(2):1523–6. [PubMed] [Google Scholar]
- Witvliet MH, Burns DL, Brennan MJ, et al. Binding of pertussis toxin to eucaryotic cells and glycoproteins. Infect Immun. 1989;57:3324–30. doi: 10.1128/iai.57.11.3324-3330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CS, Jelacic S, Habeeb RL, et al. The risk of the hemolytic–uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N Engl J Med. 2000;342:1930–6. doi: 10.1056/NEJM200006293422601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Future directions for research on enterotoxigenic Escherichia coli vaccines for developing countries. Wkly Epidemiol Rec. 2006;81:97–104. [PubMed] [Google Scholar]
- Yang X, Goliger JA, Roberts JW. Specificity and mechanism of antitermination by Q proteins of bacteriophages λ and 82. J Mol Biol. 1989;210:453–60. doi: 10.1016/0022-2836(89)90122-8. [DOI] [PubMed] [Google Scholar]
- Zhang X, McDaniel AD, Wolf LE, et al. Quinolone antibiotics induce shiga toxin-encoding bacteriophages, toxin production, and death in mice. J Infect Dis. 2000;181:664–70. doi: 10.1086/315239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.