Abstract
Background
Corynebacterium xerosis is a commensal organism found in skin and mucous membranes of humans. It is considered an unusual pathogen, and it is rarely found in human and animal clinical samples. Here we describe the isolation of C. xerosis from a 4-months-old Pelifolk lamb located in Tesistán, central western Mexico. This microorganism should be considered for differential diagnosis in cutaneous abscessed lesions in sheep, as it represents a zoonotic risk factor for human infection in sheep farms.
Case presentation
The animal exhibited a hard-consistency, 5 cm diameter abscess, without drainage, in the neck. The presumptive clinical diagnosis was caseous lymphadenitis, caused by Corynebacterium pseudotuberculosis. Samples were obtained by puncture and cultured in 8 % sheep blood agar under microaerophilic conditions. Colonies were non-haemolytic, brown-yellowish and showed microscopic and biochemical features similar to C. pseudotuberculosis, except for the urea test. A multiplex-PCR for the amplification of partial sequences of the pld, rpoB and intergenic fragment from 16S to 23S genes suggested that isolate could be C. xerosis, which was later confirmed by sequencing analysis of the rpoB gene.
Conclusions
This study shows for the first time isolation and molecular characterization of C. xerosis from a clinical sample of an ovine cutaneous abscess in Mexico. This finding highlights the need for differential diagnosis of this pathogen in ovine skin abscesses, as well as epidemiological and control studies of this pathogen in sheep farms.
Keywords: Corynebacterium xerosis, Corynebacterium pseudotuberculosis, Skin abscess, Sheep, Pld gene, RpoB gene, Case report
Background
Corynebacterium xerosis is a commensal organism normally present in skin and mucous membranes of humans and animals [1]. It is considered an unusual pathogen but it is able to cause endocarditis, skin infections and other illnesess [1–6]. Furthermore, it has been shown that human clinical isolates originally identified as C. xerosis sometimes correspond to other species of Corynebacterium (C. amycolatum, C. freneyi and C. hansenii). This misidentification is due to their close phylogenetic relationship and similar phenotypic characteristics [7–10]. C. xerosis grows in colonies of 0.2–1.0 mm in diameter, brown-yellowish in colour, slightly dry in appearance and non-haemolytic when cultured in blood agar. Microscopically, C. xerosis is irregularly stained, pleomorphic Gram positive rods presenting club-like ends. Biochemically, they are catalase-positive, and are able to ferment glucose, sucrose and maltose [1, 11–13]. Vela et al. [1] described for the first time the identification of eight isolates of C. xerosis from animal clinical samples, using classical microbiological methods as well as molecular genetics. More recently, Palacios et al. [14] showed a more thorough analysis of several C. xerosis isolates from animal clinical samples, mostly lesions from swine, including subcutaneous abscesses.
Case presentation
The present case report is derived from a routine veterinary visit to a 155 animals farm, with two sheep breeds (Pelifolk and Blackbelly), located in Tesistán, municipality of Zapopan, state of Jalisco, Mexico. Facilities from this farm were adapted from previous pig farm, and are still surrounded by pig and cattle farms in the near neighbourhood. The flock was kept on elevated floors to facilitate cleansing and handling, however many roasted sharp objects (such as wire, broken fences, nails, etc.) were present in the pens and cleansing was not being performed properly in the farm at the time of inspection. Feeding was based on locally available feedstuffs, such as; low-quality roughage, swine manure-silage and high-protein commercial feed concentrate. This farm called the veterinary services because of frequent cutaneous abscesses found in the animals, no other complaints related to skin problems were reported. Abscesses do not seemed to affect productivity of the animals, however the presence of such lesions negatively affects animal marketing, and therefore, the owner was interested in finding out about the etiological agent and the recommended treatment to eliminate the disease from the farm. Content of the abscesses was sampled from 31 animals (29 ewes, 1 ram and 1 lamb) by a certified veterinarian and sent to the laboratory for bacteriological diagnosis, in order to find the etiological agents that were causing the problem. Based on appearance of the abscesses, presumptive diagnosis of infection in the farm was Caseous lymphadenitis for most animals [4]. However, bacteriological analysis demonstrated that 13 animals were infected with C. pseudotuberculosis, 1 with Corynebacterium spp., 2 with Proteus spp., 2 with Streptococcus spp., and in the remaining 13 cases the pathogen could not be identified. The categorization for the Corynebacterium spp. isolate was unclear because the results of the bacteriological and biochemical tests were not concluding.
This paper describes the isolation of C. xerosis from a 4-months-old Pelifolk (3/4 Pelibuey, 1/4 Suffolk) lamb in good body condition. To the best of our knowledge this is the first report for C. xerosis producing a clinical cutaneous abscess in sheep. Informed consent was obtained from the animal’s owner for the publication of the results of the clinical study, including photographic material. The animal exhibited a hard-consistency abscess, without drainage, on the left side of the neck (Fig. 1a). Initial clinical diagnosis suggested caseous lymphadenitis, possibly caused by C.pseudotuberculosis. A sample for bacteriological analysis was obtained by puncturing the abscess with a sterile, 5 ml syringe, with a 20 gauge hypodermic needle after cleaning and disinfecting the surface of the abscess. The type of exudate recovered was serous-like and white in appearance. The biological sample was kept at 4 °C until biological characterization at the Centro de Investigación y Estudios Avanzados en Salud Animal (CIESA, km 15.5 Toluca-Atlacomulco road, Toluca, Mexico, z.c. 50200). The time between sampling and processing did not exceed 72 h. Sample was cultured in duplicate, in 8 % sheep blood agar [3] and incubated 24–48 h at 37 °C in aerobic and microaerophilic conditions [1, 14]. Colonies 1.0 mm in diameter, non-haemolytic, yellowish-brown in colour and slightly dry in appearance were observed (Fig. 2a). These morphological characteristics did not correspond to those previously described for C. pseudotuberculosis, but they rather seemed colonies from C. xerosis [1, 13]. Microscopically (10× magnification; Fig. 2c) bacteria were observed as club-like ended, pleomorphic, irregularly stained Gram-positive rods. Biochemical tests showed the following results: Triple sugar Iron (TSI,−), Lysine Iron agar (LIA,−), Citrate test (CIT,−), Sulphide Indole-Motility (SIM,−), Motility, Indole, Ornitine (MIO,−), Oxidative-fermentative (OF,−) Methyl red (−), Peptone Water (−), Voger Pascaguer (−), Urea (−), Nitrate Broth (+), Trehalose (+), Sucrose (+), Maltose (+) and Glucose fermentation (positive at 37 °C and negative at 42 °C). These results could correspond to a C. pseudotuberculosis profile, with the exception of the urea test, which was negative instead of positive, and therefore better coincident with a C. xerosis profile [13]. Diagnostic results were not conclusive, therefore, even if the API system is not specific to identify C. xerosis [14, 15], we decided to study the isolate with this system, to see if this isolate could be identified as other Corynebacterium species. Furthermore, in order to find out if this isolate belongs to other species such as C. freneyi and C. amycolatum which can rarely be found in sheep, we tested if the colony was able to grow at 20 °C and if it was able to ferment glucose at 42 °C. The isolate grew fine at 20 °C and did not fermented glucose at 42 °C, both characteristics associated to C. xerosis and C. hansenii [14], therefore we decided to perform a molecular analysis to find out if we could molecularly identify the isolate as C. xerosis. We analysed three loci to increase our diagnostic accuracy. We tested for a gene normally absent in C. xerosis but present in C. pseudotuberculosis, the pld gene [1, 16] where we expected to find no amplicon after PCR test if the isolate was C. xerosis in opposition to C. pseudotuberculosis which will amplify a 203 bp band; a second locus targeted the amplification of the intergenic spacer region of the 16S–23S rRNA genes (16S–23S) using primers designed for C. pseudotuberculosis, which have been reported not to work for C. xerosis [16], and we would also expect no amplification band for C. xerosis and a 816 bp band for C. pseudotuberculosis. Finally we performed PCR amplification and sequencing of the rpoB gene (446 bp), which has previously been reported to differentiate species of Corynebacterium [14, 16]. DNA extraction was carried out using a commercial kit (KAPA Express Extract) following the manufacturer’s protocol. A Multiplex-PCR technique for amplification of partial sequences of the pld, 16S–23S and rpoB genes was performed using the protocol published by Pacheco in 2007 [16]. Reactions were carried out using a commercial multiplex PCR kit (QIAGEN Multiplex PCR) following the manufacturer’s specifications. The PCR analysis included the following samples: The isolate to be characterized, the putatively C. xerosis isolate, and two C. pseudotuberculosis (one local isolate previously characterized as biovar ovis and, a reference strain, ATCC 43924, biovar equi) used as controls. As expected for the putative C. xerosis, a single PCR 446 bp amplicon band, corresponding to the rpoB gene fragment was amplified, and the intergenic 16S–23S gene and pld gene fragments were not amplified. Also as expected, both C. pseudotuberculosis strains showed three bands of 203, 446 and 816 bp, corresponding to genes pld, rpoB and 16S, respectively (Fig. 3). RpoB gene amplicons of all the three samples were purified using the Promega purification kit (Wizard® SV Gel and PCR Clean-Up System) and were sent for automatic sequencing by Macrogen (Rockville, MD, USA). Multiple sequence alignments obtained from a BLAST (NCBI) were analysed along with our isolate sequences using Clustal W analysis from Mega 6.0.6 (Fig. 4). Phylogenetic analysis was performed using the neighbour-joining method (MEGA software 6.0.6). Bootstrap values were obtained generating 1000 random trees. Phylogenetic analysis also included the sequences from the rpoB gene from C. xerosis (GenBank AY492233.1), C. pseudotuberculosis biovar ovis (GenBank CP002924.1) and C. pseudotuberculosis biovar equi (GenBank CP003540.2). It was possible to observe different phylogenetic groups that corresponded to particular species of Corynebacterium. These results, confirm that this study’s isolate (rpoB C53) is C. xerosis (Fig. 4).
Fig. 1.
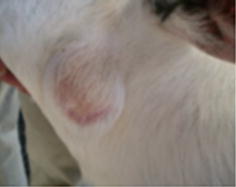
Studied abscess. A hard-consistency abscess without drainage was reported in the neck region of a 4-months-old lamb
Fig. 2.
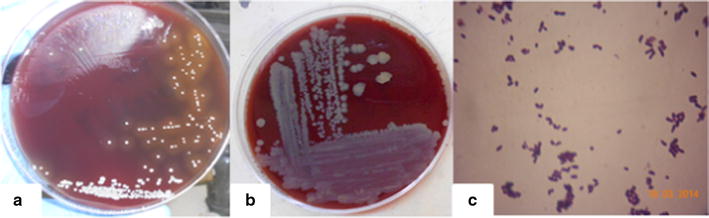
In-vitro Bacteriological culture of Corynebacterium xerosis and Corynebacterium pseudotuberculosis. Bacteria were cultured in 8 % sheep blood agar. a Corynebacterium pseudotuberculosis (ATCC 43924) showed whitish colonies with beta haemolysis. b Corynebacterium xerosis isolate, grew as small yellowish-brown colonies without haemolysis. c Gram-stained smear preparation of Corynebacterium xerosis showing characteristic pleomorphic Gram-positive rods, with club-like ends
Fig. 3.
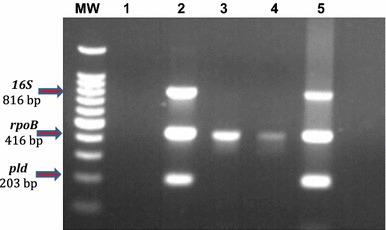
Multiplex PCR. Amplification of partial sequences of 16S rRNA, rpoB and pld genes. MW lane: molecular weight marker of 1 Kb Plus DNA Ladder™ (Invitrogen). Lane 1: negative control (reaction without template DNA). Lane 2: Corynebacterium pseudotuberculosis biovar equi. Lanes 3–4: Corynebacterium xerosis isolate (10–0.001 ng of DNA, respectively). Lane 5: Corynebacterium pseudotuberculosis biovar ovis
Fig. 4.
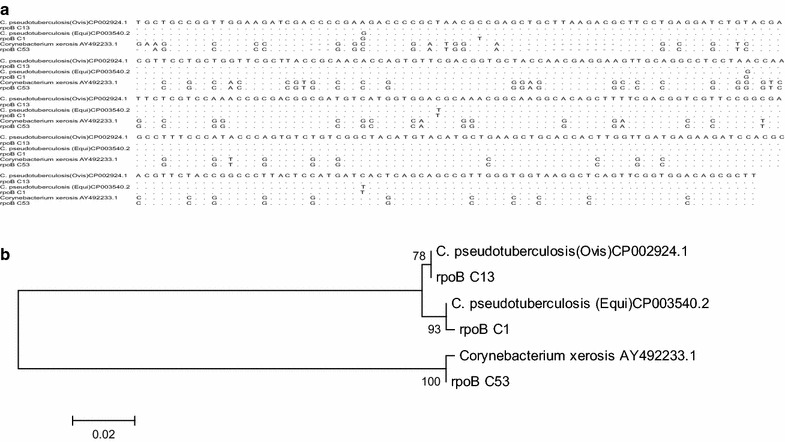
Bioinformatic analysis. a Shows the sequence alignment from the sequences used to construct the Phylogenetic tree in (b). Three sequences were downloaded from GenBank: C. pseudotuberculosis biovar ovis (CP002924.1), C. pseudotuberculosis biovar equi (CP003540.2), and Corynebacterium xerosis (AY492233.1). The other three were specimens sent for sequencing (Macrogen, Rockville, MD, USA): rpoB13 (local isolate characterized as C. pseudotuberculosis, biovar ovis), rpoB C1 (C. pseudotuberculosis, biovar equi, reference strain ATCC43924) and rpoB C53 (local isolate, characterized as C. xerosis). b The tree shows the genetic relationship of Corynebacterium pseudotuberculosis and Corynebcaterium xerosis for the rpoB gene. The tree was built from the alignment of partial sequence of the gene. Bootstrap values were obtained generating 1000 random trees, and the strength of each branch is indicated in the respective node
Genotyping techniques such as PCR as well as sequence analysis of the rpoB C53 isolate in this study contributed greatly towards a correct species identification, which was originally misleading when using phenotypic microscopic and biochemical features. Corynebacterium xerosis has been previously reported in clinical samples of human cases in lesions in endocarditis, pneumonitis, osteomyelitis and skin infections, especially in immunocompromised patients [3, 12]. It has also been found in animal clinical samples, for instance, in goat liver lesions (suspected pseudotuberculosis), and cow milk, from animals presenting mastitis. In swine, C. xerosis has been isolated from lesions in different tissues such as liver, kidney, lung, spleen, joints, as well as from subcutaneous abscess. Additionally, C. xerosis has been isolated from ovine clinical samples of uterus, in a case of abortion, and from lung tissue, from animals presenting respiratory problems [1, 14]. These isolates were identified and characterized by amplification of the rRNA 16S–23S gene using PCR–RFLP. However, in those studies, it was not possible to obtain a band pattern clear enough to differentiate C. xerosis from other species of Corynebacterium. The authors also conducted an analysis and sequence comparison of genes 16S and rpoB, which allowed them to differentiate C. xerosis from other species, genetically similar to Corynebacterium [1, 14].
As mentioned above, this study was conducted based on the presence of the rpoB gene, which has been reported by other authors as the gene of choice for phylogenetic analysis of the genus Corynebacterium, as it presents high polymorphism, even greater than the intergenic spacer region of the 16S–23S rRNA genes [17, 18]. It is known that cutaneous abscesses in sheep (caseous lymphadenitis) is caused by C. pseudotuberculosis and its main factor of virulence and pathogenicity is the exotoxin phospholipase D, encoded by the pld gene and expressed in the cell membrane of the bacterium [19]. This exotoxin is a permeability factor that promotes hydrolysis of the ester bonds in sphingomyelin in mammalian cell membranes, possibly contributing to the spread of bacteria from the initial site of infection to secondary sites through the lymphatic system to regional ganglia, and it seems to be involved in the reduction of macrophage viability following infection [19]. The exotoxin also causes dermonecrotic lesions [20, 21]. However, this exotoxin has not been reported in C. xerosis as a pathogenic toxin that could contribute to the development of abscesses. This highlights the importance of carrying out further research regarding the mechanisms of infection present in this Corynebacterium species.
A major point of discussion of epidemiological nature, include the fact that the ovine production system, where the sample was obtained from, was previously used for swine, a species where C. xerosis has been reported as a common pathogen [14]. Moreover, one of the main ingredients of sheep diet includes pig by-products (swine manure-silage), which could eventually have active pathogens, maybe including Corynebacterium spp. In addition, sharp objects such as metallic rods, nails and wires, are widespread all over the facilities, which increase the chance of injury of the animals, opening a way of entrance of Corynebacterium spp. to the organism including C. xerosis. All these factors may have contributed to the presence of C. xerosis inside the facility. No further studies were conducted, however recommendations for the owner was to debride and disinfect abscesses from all animals, in a infection containing area (to avoid spread of the pathogens), reinforce pens cleansing and remove all sharp edges and objects from the fences, feeders and floors. Since C. xerosis, as well as C. pseudotuberculosis, are potentially zoonotic microorganisms, recommendations were made to the farmer to extreme precautions during handling of animals and waste produced by the farm to prevent human and animal infection and the possible widespread of the microorganism to other farms.
Conclusions
Here we report the isolation and molecular characterization of C. xerosis from a clinical sample from a sheep cutaneous abscess for the first time in Mexico. These results encourage further exploration of the factors responsible for C. xerosis causing cutaneous abscesses.
Authors’ contributions
FH participated in animal sampling, molecular studies and article scripting, JA participated in the phenotypic studies and designing of the molecular structure for the first bacterial isolates and article scripting, JV participated in designing molecular studies and article scripting and revision, PF participated as general advisor for the study, RM participated in the first bacterial isolated and phenotypic studies, project coordination and manuscript finalization. All authors agreed on the final manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors wish to thank the University of the State of Mexico (projects 3249/2012CHT and 3806/2014/CIA) for the financial support to the study and to CONACYT for the scholarship awarded to Fernando Hernandez Leon.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Ethics approval is “Not applicable”, since this was not an experimental research project, but written informed consent was obtained from the animal’s owner for publication of this Case report and any accompanying images.
Abbreviations
- C. pseudotuberculosis
Corynebacterium pseudotuberculosis
- C. xerosis
Corynebacterium xerosis
- C. amycolatum
Corynebacterium amycolatum
- C. freneyi
Corynebacterium freneyi
- C. hansenii
Corynebacterium hansenii
- C. ulcerans
Corynebacterium ulcerans
- CIESA
Centro de Investigación y Estudios Avanzados en Salud Animal
- TSI
triple sugar iron
- LIA
lysine iron agar
- CIT
citrate test
- SIM
sulfide indole-motility
- MIO
motility, indole, ornitine
- OF
oxidative-fermentative
Contributor Information
Fernando Hernández-León, Email: erlich_511@hotmail.com.
Jorge Acosta-Dibarrat, Email: jpacosta00@hotmail.com.
Juan Carlos Vázquez-Chagoyán, Email: jcvch@yahoo.com.
Pomposo Fernandez Rosas, Email: hap_5@yahoo.com.mx.
Roberto Montes de Oca-Jiménez, Email: romojimenez@yahoo.com.
References
- 1.Vela AI, Gracía E, Fernández A, Domínguez L, Fernández-Garayzábal JF. Isolation of Corynebacterium xerosis from animal clinical specimens. J Clin Microbiol. 2006;2006(44):2242–2243. doi: 10.1128/JCM.02473-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coyle MB, Lipsky BA. Coryneform bacteria in infectious diseases: clinical and laboratory aspects. Clin Microbiol Rev. 1990;3:227–246. doi: 10.1128/CMR.3.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funke G, Lawson PA, Bernard KA, Collins MD. Most Corynebacterium xerosis strains identified in the routine clinical laboratory correspond to Corynebacterium amycolatum. J Clin Microbiol. 1996;1996(34):1124–1128. doi: 10.1128/jcm.34.5.1124-1128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riegel P, Ruimy R, Christen R, Monteil H. Species identities and antimicrobial susceptibilities of corynebacteria isolated from various clinical sources. Eur J Clin Microbiol Infect Dis. 1996;15:657–662. doi: 10.1007/BF01691153. [DOI] [PubMed] [Google Scholar]
- 5.Funke G, von Graevenitz A, Clarridge JE, Bernard KA. Clinical microbiology of coryneform bacteria. Clin Microbiol Rev. 1997;10:125–159. doi: 10.1128/cmr.10.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wauters G, Van Bosterhaut B, Janssens M, Verhaegen J. Identification of Corynebacterium amycolatum and other nonlipophilic fermentative corynebacteria of human origin. J Clin Microbiol. 1998;36:1430–1432. doi: 10.1128/jcm.36.5.1430-1432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renaud FNR, Aubel D, Riegel P, Meugnier H, Bollet C. Corynebacterium freneyi sp. nov.,-glucosidase-positive strains related to Corynebacterium xerosis. Int J Syst Evol Microbiol. 2001;51:1723–1728. doi: 10.1099/00207713-51-5-1723. [DOI] [PubMed] [Google Scholar]
- 8.Auzias A, Bollet C, Ayari R, Drancourt M, Raoult D. Corynebacterium freneyi bacteremia. J Clin Microbiol. 2003;41:2777–2778. doi: 10.1128/JCM.41.6.2777-2778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renaud FNR, Le Coustumier A, Wilhem N, Aubel D, Riegel P, Bollet C, Freney J. Corynebacterium hansenii sp. nov., an-glucosidase- negative bacterium related to Corynebacterium xerosis. Int J Syst Evol Microbiol. 2007;57:1113–1116. doi: 10.1099/ijs.0.64665-0. [DOI] [PubMed] [Google Scholar]
- 10.Funke G, Frodl R. Comprehensive study of Corynebacterium freneyi strains and extended and emended description of Corynebacterium freneyi. J Clin Microbiol. 2008;46:638–643. doi: 10.1128/JCM.01491-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollis DG, Weaver RE. Gram-positive organism: A guide to identification. Atlanta: Special Bacteriology Laboratory, Centers for Disease Control; 1981. pp. 1–10. [Google Scholar]
- 12.Krish G, Beaver W, Sarubbi F, Verghese A. Corynebacterium xerosis as a cause of vertebral osteomyelitis. J Clin Microbiol. 1989;1989(27):2869–2870. doi: 10.1128/jcm.27.12.2869-2870.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coyle MB, Leonard RB, Nowowiejski DJ, Malekniazi A, Finn DJ. Evidence of multiple taxa within commercially available reference strains of Corynebacterium xerosis. J Clin Microbiol. 1993;31:1788–1793. doi: 10.1128/jcm.31.7.1788-1793.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palacios L, Vela AI, Molin K, Fernández A, Latre MV, Chacón G, Falsen E, Fernández-Garayzábal JF. Characterization of some bacterial strains isolated from animal clinical materials and identified as Corynebacterium xerosis by molecular biological techniques. J Clin Microbiol. 2010;2010(48):3138–3145. doi: 10.1128/JCM.02373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Funke G, Renaud FNR, Freney J, Riegel P. Multicenter evaluation of the updated and extended API (RAPID) Coryne database 2.0. J Clin Microbiol. 1997;35:3122–3126. doi: 10.1128/jcm.35.12.3122-3126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pacheco LG, Pena RR, Castro TLP, Dorella FA, Bahia RC, Carminati R, Frota MN, Oliveira SC, Meyer R, Alves FS, Miyoshi A, Azevedo V. Multiplex PCR assay for identification of Corynebacterium pseudotuberculosis from pure cultures and for rapid detection of this pathogen in clinical samples. J Med Microbiol. 2007;56:480–486. doi: 10.1099/jmm.0.46997-0. [DOI] [PubMed] [Google Scholar]
- 17.Khamis A, Raoult D, La Scola B. Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. J Clin Microbiol. 2005;43:1934–1936. doi: 10.1128/JCM.43.4.1934-1936.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Retamal P, Ríos M, Cheuquepán F, Abalos P, Pizarro-Lucero J, Borie C, Gutiérrez J. Host associated polymorphisms in the Corynebacterium pseudotuberculosisrpoB gene sequence. Vet Microbiol. 2011;151:400–403. doi: 10.1016/j.vetmic.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 19.McKean SC, Davies JK, Moore RJ. Expression of phospholipase D, the major virulence factor of Corynebacterium pseudotuberculosis, is regulated by multiple environmental factors and plays a role in macrophage death. Microbiology. 2007;2007(153):2203–2211. doi: 10.1099/mic.0.2007/005926-0. [DOI] [PubMed] [Google Scholar]
- 20.McNamara PJ, Cuevas WA, Songer JG. Toxic phospholipases D of Corynebacterium pseudotuberculosis, C. ulcerans and Arcanobacterium haemolyticum: cloning and sequence homology. Gene. 1995;156:113–118. doi: 10.1016/0378-1119(95)00002-N. [DOI] [PubMed] [Google Scholar]
- 21.Tachedjian M, Krywult J, Moore RJ, Hodgson AL. Caseous lymphadenitis vaccine development: site-specific inactivation of the Corynebacterium pseudotuberculosis phospholipase D gene. Vaccine. 1995;13:1785–1792. doi: 10.1016/0264-410X(95)00144-P. [DOI] [PubMed] [Google Scholar]


