Abstract
Shape-persistent macrocycles are attractive functional targets for synthesis, molecular recognition, and hierarchical self-assembly. Such macrocycles are non-collapsible and geometrically well-defined, and they are traditionally characterized by having repeat units and low conformational flexibility. Here, we find it necessary to refine these ideas in the face of a highly flexible yet shape-persistent macrocycles. A molecule is shape-persistent if it has a small change in shape when perturbed by external stimuli (e.g., heat, light, and redox chemistry). In support of this idea we provide the first examination of the relationships between a macrocycle’s shape persistence, its conformational space, and the resulting functions. We do this with a star-shaped macrocycle called cyanostar that is flexible as well as being shape-persistent. We employed molecular dynamics (MD), density functional theory (DFT), and NMR experiments. Considering a thermal bath as a stimulus we found a single macrocycle has 332 accessible conformers with olefins undergoing rapid interconversion by up-down and in-out motions on short time scales (0.2 ns). These many interconverting conformations classify single cyanostars as flexible. To determine and confirm that cyanostars are shape-persistent, we show that it has a high 87% shape similarity across these conformations. To further test the idea, we use the binding of diglyme to the single macrocycle as guest-induced stimulation. This guest has almost no effect on the conformational space. However, formation of a 2:1 sandwich complex involving two macrocycles enhances rigidity and dramatically shifts the conformer distribution towards perfect bowls. Overall, the present study expands the scope of shape-persistent macrocycles to include flexible macrocycles if and only if their conformers have similar shapes.
Graphical abstract

INTRODUCTION
Shape-persistent macrocycles are traditionally defined as cyclic compounds with low conformational flexibility to ensure they are non-collapsible and geometrically well-defined.1 Shape persistence is advantageous for many applications: synthesis of macrocycles,2 preorganization,3 in silico design,4 catalysis,5 configurationally stable stereochemistry,6 and hierarchical self-assembly7,8,9 of multi-functional architectures,3a,8a,10 To design shape-persistent macrocycles, Moore has suggested that they have limited degrees of conformational freedom, i.e., they are rigid.11 Yet, through the results obtained in the present study, we were confronted with an apparent contradiction involving cyanostar macrocycles (Figure 1a).12 Cyanostars appear to be flexible based on their many conformations and yet geometrically well-defined based on their pre-organization towards anionic guests.
Figure 1.
(a) Chemical structures of cyanostar (CS) and diglyme. (b) Bowl chirality of cyanostars and their cartoon representations. (c) Crystal structure of 2:1 cyanostar-diglyme complex (represented by a P-M dimer). tert-Butyl groups and hydrogen atoms are omitted for clarity. The tilting angle of CN groups is related to the bowl shape. The molecular electrostatic potential of cyanostar is calculated with M06-2X/6-311++G(3df,2p) (−130 to 175 kJ / mol, red to blue).
To resolve the contradiction, we propose a more robust definition of shape persistence: A molecule is shape-persistent if it undergoes small change in shape when perturbed by external stimuli (e.g., heat, light, redox chemistry, etc.). This definition retains Moore’s original ideas while providing a broader context by introducing the ability to assess the extent of shape changes under any type of perturbation. This refined definition subsumes the concept of preorganization in which guest binding is considered as just one type of perturbation from among many. Here, we present a close examination of the shape persistence of flexible cyanostar macrocycles, and how the shape is largely unchanged with temperature and guest association. We show that, with the new definition, flexible molecules can be shape-persistent if the differences in shape between their conformers are quantified to be small.
The conformational space of several macrocycles had been studied in the past13-17 but, to the best of our knowledge, their shape persistence had not been assessed. Considering Moore’s phenylacetylene macrocycles, they were shown13 to have very few conformations, with only one unique conformation in the planar hexamer and two in the octamer (boat and chair). Similarly, prior work with triazolophane macrocycles showed only four low-lying, semi-planar conformations.14 These macrocycles clearly follow the structural definition of shape-persistence by displaying little conformational freedom. In these cases, shape persistence is self-evident. Other examples of macrocycles include the classic calixarenes,15 cyclic peptides,16 and crown ethers17 of host-guest chemistry. The last, crown ethers, are seen to collapse upon themselves thus not conforming to being shape-persistent, despite being considered pre-organized18 relative to their linear counterparts, podands. For the case of cyanostars, their shape persistence has yet to be formally analyzed. The potential for conformational freedom in the cyanostar is suggested by the crystal structure that shows accommodation of a shallow, bowl-like shape (Figure 1c). Cyanostar also possesses bowl chirality (Figure 1b)12,19 and, like other bowl-shaped compounds,20 is therefore expected to undergo bowl inversions21 that offer access to multiple conformations.
Bowl-shaped molecules can be shape-persistent. For example, they have been conformationally preorganized as chiral hosts by imposing extremely high inversion barriers as a means to achieve enantioselective recognition and asymmetric catalysis.5,19-20 In contrast, rapidly-inverting bowls that exhibit high conformational freedom have started to find applications such as in supramolecular polymerization by cofacial association.22 Aida and coworkers designed the first living and stereoselective supramolecular polymerization by using corannulene derivatives.22b In Aida’s polymerization scheme, shape persistence of the monomers is critical for propagating the chirality in the growing self-assembled polymers; whereas flexibility in the initiator bowls allows production of either enantiomeric form of the monomer. Unlike corannulenes, however, cyanostar has a cavity in its center, a feature that provides a unique model for testing the shape persistence of cyanostars upon guest binding.
Herein, we report that conformationally rich flexibility can coexist with shape persistence in the cyanostar macrocycle. This assertion is based on the results from density functional theory (DFT), molecular dynamics (MD),11,23 and NMR experiments. Although as many as 332 conformations are thermally accessible by single cyanostars, the Boltzmann-weighted variation in atomic positions relative to the most stable conformer leads to ~87% retention of the molecule’s size,24 indicative of high shape persistence. Unlike the crystal of the 2:1 complex, the single macrocycle has a ruffled rim with two olefins rotated up and three down. The ensemble-averaged bowl shape of cyanostar is further verified by through-space 1H-1H NMR measurements based on nuclear Overhauser effect (NOE), which closely matches with theory to within ±2%. We also test the use of a diglyme guest and cofacial stacking of a second macrocycle as perturbations. We see the shape is retained based on the conformational analysis. By evaluating the conformational landscape, we are able to show that the steric hindrance between the two cofacial macrocycles, rather than the interactions with the weakly bound diglyme, is responsible for the shift in conformer population to enhance shape persistence. The correlations between theory and experiment substantiate the refined definition of shape persistence to enable inclusion of macrocycles that are both conformationally flexible and geometrically well-defined.
RESULTS AND DISCUSSION
A. Delineating Cyanostar’s Conformational Landscape
To investigate the conformational landscape and examine the shape persistence of a single cyanostar macrocycle, MD simulations and DFT were used (details of the two methods can be found in Supporting Information). To date, cyanostars have only been observed as guest-bound dimers,12, 25 and the strong tendency of cyanostar to form such 2:1 complexes has precluded our ability to examine single cyanostars experimentally.
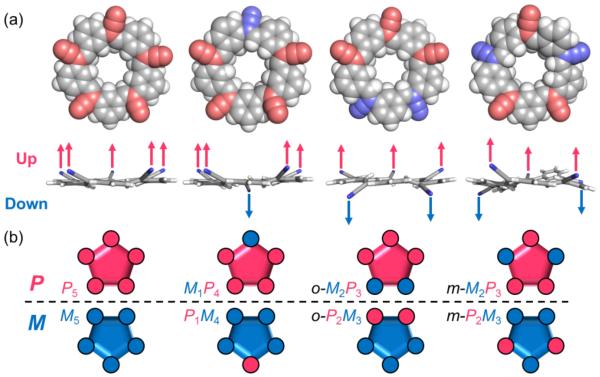
The shapes of the cyanostar’s conformations were analyzed geometrically. To facilitate data mining of the many MD-generated geometries, dihedral angles between the olefins and their neighbouring phenyl groups were measured to characterize local tilting (ΦCN, Figures 1c and S4) and the overall shape of the bowl. Using these dihedrals two isomerization phenomena were found. First, up-and-down motions of cyano-olefins change the bowl chirality of cyanostar (Figure 2a). Second, in-and-out motions change the orientation of cyano groups relative to the cavity but leave the bowl chirality unchanged (Figure 3a).
Figure 2.
All-out conformers of cyanostars. (a) Top and side views of the DFT-optimized conformations with global P chirality. The cyano groups are colored in pink for P chirality (up) and blue for M (down). (b) Cartoon representations of bowl chirality at the local (circles) and global (pentagon) levels. (Geometries: M06-2X/6-31+G(d,p), IEFPCM implicit solvation of CH2Cl2)
Figure 3.
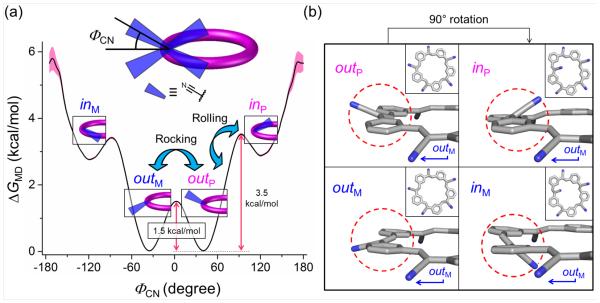
(a) Free energy landscape of olefin rotation. The rotation energy landscape was computed based on MD simulations with a bin size of 5°; errors are presented as pink areas (see Section 4.3 of Supporting Information). Rocking and rolling motions are indicated. (b) The four rotamers obtained from a DFT relaxed scan for the m-M2P3 conformer (Insets: top views).
Up-and-down Conformations of Cyanostar Determine Local and Global Bowl Chirality
The up-and-down motions are closely associated with changes in bowl chirality of the cyanostar. Therefore, use of local chirality to augment the traditional definition of bowl chirality provides a means to describe the conformational diversity of cyanostars. Bowl chirality is usually defined globally20 by looking from the top into the bottom of the bowl (Figure 1b) and then using priority rules to define when symmetry is first broken when moving away from the benzene in either a clockwise (P) or counter-clockwise (M) direction. Here, we identified the P face to be the specific face of a single cyanostar in which a clockwise direction is observed. For simplicity, all top views of cyanostar are shown with P faces. Thus, cyano groups oriented towards the readers will have P local chirality and are oriented “up”, and others with local M chirality are oriented “down” (Figure 2a).
To begin the conformation count, there are four pairs of up-and-down combinations of olefins that are possible (Figure 2b). They can be named using local chiralities: P5, M1P4, o-M2P3, m-M2P3; where o and m refer to the ortho and meta relationship of the two olefins with M chirality. Global chirality can also be interpreted from this nomenclature by the majority rule: monomers that have more P local chirality will be P in the globally.
The thermal accessibility of the four types of all-out conformations (Figure 2) was confirmed by MD simulations and independently by DFT geometry optimization (Figure 2). Furthermore, the root-mean-square deviation (RMSD) of atomic positions for the DFT-optimized P5 conformer matches with the X-ray solid-state structures12 to within 0.1 Å, reflecting the accuracy of DFT methods.
In-and-out Conformations of Cyanostar Are Connected by Olefin Rock and Roll Rotations
The rotation energy landscape of a single olefin was computed (Figure 3a) by utilizing the MD-generated distribution of dihedral angles. Four rotamers were identified (see minima in Figure 3a and representative images in Figure 3b) along with two types of barriers (rocking and rolling). The two global minima are out rotamers (ΦCN~ ±30°) and the high-energy minima are in rotamers (ΦCN~ ±120°). These rotamers can either be up (P) or down (M). The free energy difference between the two types of rotamers is sufficiently low (2.8 kcal / mol) that the in rotamers are thermally accessible.
The same energy profile was computed using DFT and the same four rotamers were found (Figure 3b). Interestingly, DFT shows that a complete 360º rotation is possible. With a barrier of 6.5 kcal / mol (Figures S2 and S3), this motion would be expected on a 10-ns time scale (assuming a typical 1013 pre-exponential factor for unimolecular reactions18). However, this motion was never observed in the MD simulation that covered a 400-ns time period. We attribute this apparent contradiction to differences in the calculation methods: In DFT, all of the internal coordinates are fully relaxed other than the rotating olefin; whereas in the MD simulations – and in reality – all internal modes are thermally excited. Thus, the rotations of nearby olefins in the MD simulation hinder the 360° rotation of any individual olefin.
A Landscape of 332 Thermally Accessible Conformers
Based on the up-down and in-out orientations of cyano groups, there will be 1024 mathematically possible conformations for single cyanostars. However, many combinations are not energetically accessible, except for a group of 332 conformations that are predicted to be thermally accessible (Supporting Information). These 332 conformations create a free energy landscape of equal populations of P and M chirality at both local and goal levels (Figure 4a and b). When the degeneracies are factored out, the landscape of 332 states can be simplified into 34 unique geometries, or 17 pairs of enantiomers, that exist as local minima (Figure S1).26 Overall, theory predicts a very rich conformational landscape for the single cyanostar macrocycle.
Figure 4.
Calculated results showing the distribution of local and global chiralities for a single cyanostar macrocycle using (a) DFT and (b) MD. (c) Snapshots of P conformers during the MD simulation. For simplicity, DFT energies (M06-2X/6-311++G(3df,2p), IEFPCM implicit solvation of CH2Cl2) are Boltzmann-weighted for the listed conformers, e.g., ΔGDFT(M2P3) is a Boltzmann average of all ortho, meta, and in-out combinations.
At equilibrium, the single macrocycle is not a perfect bowl but instead is ruffled with two out of the five cyano groups on opposite sides. DFT predicts that 82% of the total population will be P2M3 and M2P3 (Figure 4a), while MD predicts 66%. The relative stabilities of the various conformers can be understood by assessing dipole-dipole interactions between cyano-olefins, which carry a strong 5.3 D dipole moment. This value is comparable to the 5.4 D dipole moment of an analogous benzene-triazole-benzene triad that has been widely used in CH-based anion binding.4,14,27 Repulsions between local dipoles favor alternating orientations of the cyano groups. With five olefins, m-M2P3 or m-P2M3 conformation has the best up-down alternation and is thus the global minimum. The perfect bowl conformation present in the crystal structure of the dimers,12 P5 or M5, is the least stable with all dipoles pointing in the same direction. However, this perfect bowl provides a better environment for guest binding with all CH hydrogen bond donors focused towards the center of the dimer.
Rapidly Inverting Bowl Chirality in Flexible Cyanostars
The 332 thermally accessible conformers undergo rapid isomerization and racemization (Figures 4b, S6, and S8). Although most bowl-shaped compounds have one global inversion,20a the simultaneous inversion of all five cyano groups in the cyanostar was never observed in MD simulations. Rather, local inversions involving movements of a single olefin contribute 94% of all stereoisomerizations, the two-olefin inversion pathways are limited (5%), and all others are rare. Consequently, the energetic profile for rocking single olefins between different all-out conformers was explicitly characterized by DFT (Figure 5).
Figure 5.
Rocking pathways between all-out conformers (pink P) and their enantiomers (blue M), are established through corresponding transition states (yellow). The free energies were calculated by DFT with implicit solvation (see Section 5 of Supporting Information).
The rocking motions are rapid. DFT-derived barriers (Figure 5) range from 0.7 to 1.7 kcal / mol, and are in good agreement with the MD ensemble-averaged rocking barrier of 1.5 kcal / mol (Figure 3). The very low barriers highlight again the flexibility of cyanostars at 298 K. It has been suggested11,28 that at 298 K, a barrier of 24 kcal / mol is a practical limit to distinguish between rigid and flexible conformers. Others consider slow exchange on the NMR time scale (16 kcal / mol) to demarcate rapid versus slow dynamics.29 With either standard, cyanostar rocking motions are much faster.
Rapid interconversions were confirmed using lifetime correlation functions that were calculated from the MD data (see Section 8.1 of Supporting Information). The expedited loss of coherence between local chiral centers was reflected by a short relaxation time of 35 ps. Beyond 200 ps, the correlation coefficient dropped to zero, suggesting that the olefins have completely lost the memory of their past status of chirality.
B. Shape persistence of Single Cyanostars
Cyanostar macrocycles are flexible: They rapidly convert between multiple conformations. Now we have to determine if they are shape-persistent.
Quantitative Definition of Shape Persistence for Flexible Cyanostar Monomers
The shape persistence of single cyanostars was determined from the coordinates of all 332 cyanostar conformers. Specifically, the Boltzmann-weighted RMSD relative to the global minimum provides a temperature-dependent determination of the shape deformation. To calibrate the relative significance of the shape changes, we refer the RMSD to the mean molecular radius (assuming the volume of the molecule is distributed as a sphere). Accordingly, a normalized shape-change index, Δσ, can be expressed:
Where p(i) is the relative population of conformer i, and VCPK is the CPK volume of the molecule. The basic logic of the shape-change index is consistent with the concepts of shape similarity,24 and of conformational heterogeneity.30 Both concepts have been widely used in biology31 and in developing algorithms for conformational searches.32
Cyanostar has a small shape-change index, Δσ. Using DFT-optimized geometries, the typical RMSD from m-P2M3 to other out conformers of a single cyanostar is 0.4 Å (Table S2). The Boltzmann-weighted RMSD across all conformers was found to be 0.7 Å. Considering the mean molecular radius of 5.5 Å of cyanostar, the normalized shape change index (Δσ) is only 0.13 for single cyanostars in solution at room temperature. In other words, the flexibility of cyanostar produces, on average, only 13% variation in its radius. Thus, there is a corresponding 87% shape persistence that originates in the relatively small RMSD values. The modest variations in shape can also be seen from the MD simulations (see Movies S1 and S2). When the intensity of the stimulus is lowered by reducing the temperature in the thermal bath to 218 and 178 K, the shape persistence increases to 91% and 93%, respectively. These theoretical findings unambiguously show cyanostars are shape-persistent as well as being flexible.
Interestingly, the noticeable dynamic motions of the single cyanostar macrocycle originate entirely from the olefinic units while the aromatic units remain quite static (see Movie S1). Thus it can be understood that the aromatic component of the backbone is contributing to the shape persistence of cyanostar and the rocking-and-rolling olefins are responsible for the flexibility. This observation may be the reason why cyanostar macrocycles are more flexible than other aromatic-based receptors.13,14
Quantitative NOE Verifies the Bowl Shape of Single Cyanostars as Predicted by Simulations
The accuracy of the theoretical calculations were verified experimentally using quantitative NOE experiments.33 NMR spectra obtained at 212 K are consistent with fast exchange between conformers on the NMR time scale (see Section 11 of Supporting Information). The relatively static aromatic components provide a reference point to determine intramolecular proton-proton distances from olefinic proton, Hc, to the aromatic ones, Ha and Hd. These enable calculation of the ensemble-averaged distances and thus the bowl shape of single cyanostars. Specifically, a ratio of two proton-proton distances, n, was measured from a 1H-1H 2D NOESY experiment (Figure 7). This distance ratio (n) was shown from DFT (Figure 7b) to decrease steadily with increasing dihedral angle. Thus, the average depth of the bowl and the shape of single cyanostars can be determined.
Figure 7.
Structural correlations between proton distances and the cyanostar’s bowl shape. (a) Aromatic region of the 1H-1H NOESY spectrum of cyanostar (298 K, 500 MHz, CD2Cl2, mixing time 600 ms). (b) Plot of the ratio of proton-proton distances (n) as a function of dihedral angle (ΦCN). Open circles were obtained from DFT. The value of n = 1.6 obtained from NOE experiments and theory are marked.
The quantitative NOESY experiment indicates that the conformational ensembles generated by the DFT and MD approaches are quite accurate (Table S22). The range of possible distance ratio (n) can be related to a perfectly planar cyanostar (n = 1.9) and a conformation with the olefin making a ~60 angle (n ~ 1.4). The data analysis on NOE experiments yielded n values of 1.6, clearly indicating that cyanostars on average have the ruffled out conformation in solution. The distance ratio obtained from the NOE experiment is smaller than the crystal structures (P5, n = 1.7) , which is consistent with the existence of multiple conformations, including in rotamers with a distance ratios of n ~ 1.3. To our satisfaction, distance ratios predicted by theory deviate only by 0.03 from experimental values (2% error), with MD slightly overestimating planarity (1.63) and DFT slightly underestimating it (1.57).
C. Shape Persistence under the Perturbation of Diglyme Binding and Molecular Self-association
With the shape persistence of flexible cyanostars confirmed for the single macrocycle at various temperatures, we made the same assessment as a result of guest binding.
The Conformational Landscape changes to Favor Perfect Bowls in Dimers of Cyanostars stabilized by Complexation with Diglyme
The shallow free energy surface of single cyanostars suggests that even modest external forces are sufficient to perturb the conformer distribution, shape persistence and flexibility of cyanostars. To test this idea, the impact of guest binding was examined using the cyanostar-diglyme model system seen in the 2:1 binding stoichiometry in the crystal.12 Specifically, we focus here on the meso-dimer formed by the rim-sharing arrangement of one bowl with P chirality and the other with M chirality.34 The weak perturbation introduced by diglyme binding is manifested by the modest 2:1 binding constant, β2, of only 30 M−2 (2 kcal / mol; see Section 10 of Supporting Information). MD simulation was the primary modeling tool for the 2:1 complex, as its size of ~300 atoms prevents DFT from efficiently characterizing the entire conformation space.
MD simulations of the 2:1 cyanostar-diglyme complex revealed substantial motion of hosts and guest within the complex. The bound diglyme was observed to gyrate within the cavity, and occasionally displayed escape and rebinding dynamics (Figure 8a, e, and i). Upon cooling to 218 K, the dissociation is diminished enough to resemble the corresponding crystal structures (Figure 8a). Cyanostars in the dimers showed partial rotations, although restricted by the steric gearing of tert-butyl groups between the pair of stacked macrocycles (Figure 8b, f, and j).
Figure 8.
MD simulations of 2:1 cyanostar-diglyme complexes (P-M) at various temperatures: 178, 218, and 298 K. Images in (a, e, i) show overlays of diglyme recorded over 400 ns within a cyanostar dimer. (b, f, j) Top views and (c, g, k) side views of structural overlays of cyanostar dimers in the 2:1 complexes but with the diglymes omitted for clarity. (d, h, l) Histogram from the MD simulation of the equilibrium distribution of states for the P cyanostars in the 2:1 complex (dashed boxes). (Oxygen atoms are colored in red, nitrogen in blue, and carbon in cyan; hydrogen atoms are omitted. Overlays in (a) include an additional, faded data set generated with an 800-ns simulation. The corresponding average displacements between diglyme and cyanostar dimers are noted.)
Consistent with our hypothesis, binding of diglyme to the dimers dramatically alters the conformational landscape of cyanostars. Compared to the flexible free macrocycle (Figure 4), the population of conformers in the 2:1 complex becomes highly concentrated in the newly emerged global minimum of P5 or M5 with a shape of perfect bowls (Figure 8d, h, and l), predominating 85% of the time. The free energy differences between out and in conformers upon diglyme binding are raised from 2.7 to 3.9 kcal / mol; consequently, the overall population of in-rotamers dropped from 30% for the single macrocycle to 4% in the 2:1 complex. Rocking motions were infrequent and resulted in negligible bowl inversions: at 298 K, only 0.1% of all cyanostars in the dimer had an inversion of global chirality from their P (or M) starting point. Only 3% were inverted locally (Figure 8d, h, and l). The rigidification of cyanostar in the 2:1 complex upon diglyme binding was accompanied by suppressed olefin rotations and an extended chirality lifetime beyond 800 ns (Figure 6, vide supra). The steady state correlation coefficient remained high, indicating that >96% of the original chirality configuration was preserved (Figure S34). The number of thermally accessible conformers is reduced upon formation of the 2:1 complex around diglyme from 332 to 20 (P5, M1P4, o-M2P3, and m-M2P3 with their five-fold degeneracy).35
Figure 6.

Average lifetime correlation functions of local bowl chirality for cyanostars as 2:1 cyanostar-diglyme complexes (black) and single macrocycles (red).
The shape persistence of the cyanostars in the 2:1 complex increased over the single macrocycle. Using the MD-simulated population and DFT-optimized geometries of single cyanostars with the corresponding chirality, the shape change index (Δσ) associated with the thermal energy present at room temperature was estimated to be 2%. This finding indicates that the shape persistence of cyanostar was greatly enhanced in the 2:1 complex up to 98%.
The change in conformational landscape for the sandwich complex can be attributed to either one or two structural features (Figure 9). First, steric hindrance between tert-butyl groups on one macrocycle and cyano groups from the other macrocycle could play a role (Figure 9). Rocking any olefins from the top macrocycle towards the other macrocycle generates more steric contacts. Second, hydrogen bonding between cyanostar and diglyme could also be a factor. The perfect bowls maximize guest binding when all of the olefinic CH donors are pointed towards the center between the two macrocycles (Figure 9, “better binding” label). Rotating any P olefins into M olefins would redirect the hydrogen bond donor away from the bound guest.
Figure 9.

Side view of a DFT-modeled meso 2:1 cyanostar-diglyme complex. (Diglyme and parts of cyanostars are removed for clarity, M06-2X/6-31G(d), CH2Cl2).
Steric Hindrance, Not Binding, Rigidifies Cyanostar in the 2:1 Complex with Diglyme
MD simulations show that steric hindrance from cofacial stacking and not hydrogen bonding from guest binding alters the conformational landscape. This finding emerges from an examination of a putative 1:1 complex, in which the second cyanostar partner was absent. The equilibrium distribution of conformers for the 1:1 complex is found to be almost identical to the distributions for the case of the free cyanostar (Figure 10b). Even the lifetime correlations and relaxation of local inversions are indistinguishable from those in the case of a single macrocycle (see Section 8 of Supporting Information). The only noticeable change is a slightly increased free energy differences between the in and out rotamers by 0.2 kcal / mol, reflected by the reduced number of in rotamers in the 1:1 complex (Figure 10b). The negligible changes between the free macrocycle and the 1:1 cyanostar-diglyme complex indicate that steric hindrance between sandwiched macrocycles play a decisive role36 in reducing the flexibility of olefinic units and thus enhancing the overall shape persistence in the 2:1 complex.
Figure 10.
(a) Structural overlays of diglyme inside a macrocycle, generated from a 400 ns simulation trajectory. (b) Top and side views of structural overlays of cyanostar with diglyme omitted. (Oxygen atoms are colored in red, nitrogen in blue, and carbon in cyan; hydrogen atoms are omitted.)
CONCLUSIONS
In conclusion, we have shown that flexibility can coexist with shape persistence in cyanostar macrocycles. This coexistence appeared to be a contradiction until we defined shape-persistent molecules as having highly similar shape in response to an external perturbation. When placed in a thermal bath, the conformers were calculated to have similar shapes and resulted in 87% shape persistence. The ensemble averaged geometries matched experimental values determined by NOE experiments and corresponded to an overall puckered bowl for the single cyanostar. The shape persistence and flexibility is unperturbed in response to binding a diglyme guest molecule. Yet, cofacial stacking of a second cyanostar macrocycle shows enhanced shape persistence of 98% and leads to perfect chiral bowls being the preferred ensemble averaged structure. These findings show that macrocycles need not be completely rigid to enjoy the same advantages of traditional shape-persistent macrocycles.
Supplementary Material
ACKNOWLEDGMENTS
AHF and YL acknowledge project support from the NSF (CHE-1412401). AS, CGM, and KS acknowledge support from the National Institutes of Health (NIH, 9P41GM104601). AS acknowledges a Beckman Postdoctoral Fellowship. KR acknowledges support from the NSF (CHE-1266154). DFT calculations were conducted on Indiana University’s supercomputer system, Big Red II, which was supported in part by Lilly Endowment, Inc., through its support for the Indiana University Pervasive Technology Institute, and in part by the Indiana METACyt Initiative. The Indiana METACyt Initiative at IU is also supported in part by Lilly Endowment, Inc. AHF and YL thank B. Qiao at Indiana University for a resourceful discussion on processing MD-simulated data. AHF and YL also thank J. R. Dobscha at Indiana University for a discussion on shape persistence in host-guest chemistry.
Footnotes
Supporting Information
Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.XXXXXXX.
Coordinates for the 34 energy minima and the 9 rocking transition states (MOL)
Movies for representative trajectories of MD simulations (MPG)
DFT calculations, raw data of MD simulations, additional data analysis on dihedral angles, bowl chirality and diglyme displacements, derivation of statistical tests, lifetime correlation functions, and NMR spectra (PDF)
Notes
The authors declare no competing financial interest.
REFERENCES
- 1.Zhang J, Pesak DJ, Ludwick JL, Moore JS. J. Am. Chem. Soc. 1994;116:4227. [Google Scholar]
- 2.(a) Zhang W, Moore JS. Angew. Chem. Int. Ed. 2006;45:4416. doi: 10.1002/anie.200503988. [DOI] [PubMed] [Google Scholar]; (b) Feng W, Yamato K, Yang L, Ferguson J, Zhong L, Zou S, Yuan L, Zeng X, Gong B. J. Am. Chem. Soc. 2009;131:2629. doi: 10.1021/ja807935y. [DOI] [PubMed] [Google Scholar]; (c) Jin Y, Voss BA, Noble RD, Zhang W. Angew. Chem. Int. Ed. 2010;49:6348. doi: 10.1002/anie.201001517. [DOI] [PubMed] [Google Scholar]; (d) Zhang C, Wang Q, Long H, Zhang W. J. Am. Chem. Soc. 2011;133:20995. doi: 10.1021/ja210418t. [DOI] [PubMed] [Google Scholar]; (e) Gross DE, Zang L, Moore JS. Pure Appl. Chem. 2012;84:869. [Google Scholar]; (f) Zhang C, Yu C, Long H, Denman RJ, Jin Y, Zhang W. Chem. Eur. J. 2015;21:16935. doi: 10.1002/chem.201502848. [DOI] [PubMed] [Google Scholar]; (g) Ren C, Shen J, Zeng H. Org. Lett. 2015;17:5946. doi: 10.1021/acs.orglett.5b02780. [DOI] [PubMed] [Google Scholar]
- 3.(a) Zhao D, Moore JS. Chem. Commun. 2003;39:807. doi: 10.1039/b207442g. [DOI] [PubMed] [Google Scholar]; (b) Choi K, Hamilton AD. J. Am. Chem. Soc. 2003;125:10241. doi: 10.1021/ja034563x. [DOI] [PubMed] [Google Scholar]; (c) Chang K-J, Moon D, Lah MS, Jeong K-S. Angew. Chem. Int. Ed. 2005;44:7926. doi: 10.1002/anie.200503121. [DOI] [PubMed] [Google Scholar]; (d) Rekharsky MV, Mori T, Yang C, Ko YH, Selvapalam N, Kim H, Sobransingh D, Kaifer AE, Liu S, Isaacs L, Chen W, Moghaddam S, Gilson MK, Kim K, Inoue Y. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20737. doi: 10.1073/pnas.0706407105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Tahara K, Lei S, Mamdouh W, Yamaguchi Y, Ichikawa T, Uji IH, Sonoda M, Hirose K, De Schryver FC, De Feyter S, Tobe Y. J. Am. Chem. Soc. 2008;130:6666. doi: 10.1021/ja711422h. [DOI] [PubMed] [Google Scholar]; (f) Trabolsi A, Khashab N, Fahrenbach AC, Friedman DC, Colvin MT, Coti KK, Benítez D, Tkatchouk E, Olsen J-C, Belowich ME, Carmielli R, Khatib HA, Goddard WA, Wasielewski MR, Stoddart JF. Nat. Chem. 2010;2:42. doi: 10.1038/nchem.479. [DOI] [PubMed] [Google Scholar]; (g) Biedermann F, Uzunova VD, Scherman OA, Nau WM, De Simone A. J. Am. Chem. Soc. 2012;134:15318. doi: 10.1021/ja303309e. [DOI] [PubMed] [Google Scholar]; (h) Barnes JC, Juricek M, Strutt NL, Frasconi M, Sampath S, Giesener MA, McGrier PL, Bruns CJ, Stern CL, Sarjeant AA, Stoddart JF. J. Am. Chem. Soc. 2013;135:183. doi: 10.1021/ja307360n. [DOI] [PubMed] [Google Scholar]; (i) Biedermann F, Vendruscolo M, Scherman OA, De Simone A, Nau WM. J. Am. Chem. Soc. 2013;135:14879. doi: 10.1021/ja407951x. [DOI] [PubMed] [Google Scholar]; (j) Cao L, Sekutor M, Zavalij PY, Mlinaric-Majerski K, Glaser R, Isaacs L. Angew. Chem. Int. Ed. 2014;53:988. doi: 10.1002/anie.201309635. [DOI] [PubMed] [Google Scholar]; (k) Biedermann F, Nau WM, Schneider HJ. Angew. Chem. Int. Ed. 2014;53:11158. doi: 10.1002/anie.201310958. [DOI] [PubMed] [Google Scholar]; (l) Popov I, Chen T-H, Belyakov S, Daugulis O, Wheeler SE, Miljanic OS. Chem. Eur. J. 2015;21:2750. doi: 10.1002/chem.201406073. [DOI] [PubMed] [Google Scholar]; (m) Liu Y, Shen J, Sun C, Ren C, Zeng H. J. Am. Chem. Soc. 2015;137:12055. doi: 10.1021/jacs.5b07123. [DOI] [PubMed] [Google Scholar]; (n) Assaf KI, Ural MS, Pan F, Georgiev T, Simova S, Rissanen K, Gabel D, Nau WM. Angew. Chem. Int. Ed. 2015;54:6852. doi: 10.1002/anie.201412485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramabhadran RO, Liu Y, Hua Y, Ciardi M, Flood AH, Raghavachari K. J. Am. Chem. Soc. 2014;136:5078. doi: 10.1021/ja500125r. [DOI] [PubMed] [Google Scholar]
- 5.(a) Zheng Y-S, Luo J. J. Incl. Phenom. Macrocycl. Chem. 2011;71:35. [Google Scholar]; (b) Li SY, Xu YW, Liu JM, Su CY. Int. J. Mol. Sci. 2011;12:429. doi: 10.3390/ijms12010429. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Adriaenssens L, Ballester P. Chem. Soc. Rev. 2013;42:3261. doi: 10.1039/c2cs35461f. [DOI] [PubMed] [Google Scholar]
- 6.(a) Araki K, Inada K, Shinkai S. Angew. Chem. Int. Ed. 1996;35:72. [Google Scholar]; (b) Odermatt S, Alonso-Gomez JL, Seiler P, Cid MM, Diederich F. Angew. Chem. Int. Ed. 2005;44:5074. doi: 10.1002/anie.200501621. [DOI] [PubMed] [Google Scholar]; (c) Yang Y, Xue M, Xiang JF, Chen CF. J. Am. Chem. Soc. 2009;131:12657. doi: 10.1021/ja9029335. [DOI] [PubMed] [Google Scholar]; (d) Xu Y, Smith MD, Geer MF, Pellechia PJ, Brown JC, Wibowo AC, Shimizu LS. J. Am. Chem. Soc. 2010;132:5334. doi: 10.1021/ja9107066. [DOI] [PubMed] [Google Scholar]; (e) Ogoshi T, Akutsu T, Yamafuji D, Aoki T, Yamagishi T. Angew. Chem. Int. Ed. 2013;52:8111. doi: 10.1002/anie.201302675. [DOI] [PubMed] [Google Scholar]
- 7.(a) Gong HY, Rambo BM, Karnas E, Lynch VM, Sessler JL. Nat. Chem. 2010;2:406. doi: 10.1038/nchem.597. Selected exmaples for 1D self-assemblies: [DOI] [PubMed] [Google Scholar]; (b) Ren C, Maurizot V, Zhao H, Shen J, Zhou F, Ong W, Du Z, Zhang K, Su H, Zeng H. J. Am. Chem. Soc. 2011;133:13930. doi: 10.1021/ja206457b. [DOI] [PubMed] [Google Scholar]; (c) Yang Y, Feng W, Hu J, Zou S, Gao R, Yamato K, Kline M, Cai Z, Gao Y, Wang Y, Li Y, Yang Y, Yuan L, Zeng X, Gong B. J. Am. Chem. Soc. 2011;133:18590. doi: 10.1021/ja208548b. [DOI] [PubMed] [Google Scholar]; (d) Wittenberg JB, Zavalij PY, Isaacs L. Angew. Chem. Int. Ed. 2013;52:3690. doi: 10.1002/anie.201300404. [DOI] [PubMed] [Google Scholar]; (e) Jin GX, Zhang YY, Zhang L, Lin YJ. Chem. Eur. J. 2015;21:14893. doi: 10.1002/chem.201502194. [DOI] [PubMed] [Google Scholar]
- 8.(a) Shetty AS, Fischer PR, Stork KF, Bohn PW, Moore JS. J. Am. Chem. Soc. 1996;118:9409. Selected exmaples for 2D self-assemblies: [Google Scholar]; (b) Pan GB, Cheng XH, Hoger S, Freyland W. J. Am. Chem. Soc. 2006;128:4218. doi: 10.1021/ja060469f. [DOI] [PubMed] [Google Scholar]; (c) Chen T, Pan GB, Wettach H, Fritzsche M, Hoger S, Wan LJ, Yang HB, Northrop BH, Stang PJ. J. Am. Chem. Soc. 2010;132:1328. doi: 10.1021/ja907220f. [DOI] [PubMed] [Google Scholar]; (d) Tahara K, Balandina T, Furukawa S, De Feyter S, Tobe Y. CrystEngComm. 2011;13:5551. [Google Scholar]; (e) Cui K, Schlutter F, Ivasenko O, Kivala M, Schwab MG, Lee SL, Mertens SF, Tahara K, Tobe Y, Mullen K, Mali KS, De Feyter S. Chem. Eur. J. 2015;21:1652. doi: 10.1002/chem.201405305. [DOI] [PubMed] [Google Scholar]
- 9.(a) Balakrishnan K, Datar A, Zhang W, Yang X, Naddo T, Huang J, Zuo J, Yen M, Moore JS, Zang L. J. Am. Chem. Soc. 2006;128:6576. doi: 10.1021/ja0618550. Selected exmaples for 3D self-assemblies: [DOI] [PubMed] [Google Scholar]; (b) Finke AD, Gross DE, Han A, Moore JS. J. Am. Chem. Soc. 2011;133:14063. doi: 10.1021/ja204795q. [DOI] [PubMed] [Google Scholar]; (c) Li M, Schluter AD, Sakamoto J. J. Am. Chem. Soc. 2012;134:11721. doi: 10.1021/ja3038905. [DOI] [PubMed] [Google Scholar]; (d) He Y, Xu M, Gao R, Li X, Li F, Wu X, Xu D, Zeng H, Yuan L. Angew. Chem. Int. Ed. 2014;53:11834. doi: 10.1002/anie.201407092. [DOI] [PubMed] [Google Scholar]; (e) Li X, Li B, Chen L, Hu J, Wen C, Zheng Q, Wu L, Zeng H, Gong B, Yuan L. Angew. Chem. Int. Ed. 2015;54:11147. doi: 10.1002/anie.201505278. [DOI] [PubMed] [Google Scholar]; (f) Xue J, Izumi T, Yoshii A, Ikemoto K, Koretsune T, Akashi R, Arita R, Taka H, Kita H, Sato S, Isobe H. Chem. Sci. 2016;7:896. doi: 10.1039/c5sc03807c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Moore JS. Acc. Chem. Res. 1997;30:402. [Google Scholar]; (b) Bieri M, Nguyen MT, Groning O, Cai J, Treier M, Ait-Mansour K, Ruffieux P, Pignedoli CA, Passerone D, Kastler M, Mullen K, Fasel R. J. Am. Chem. Soc. 2010;132:16669. doi: 10.1021/ja107947z. [DOI] [PubMed] [Google Scholar]; (c) McDonald KP, Hua Y, Lee S, Flood AH. Chem. Commun. 2012;48:5065. doi: 10.1039/c2cc17459f. [DOI] [PubMed] [Google Scholar]; (d) Rambo BM, Gong HY, Oh M, Sessler JL. Acc. Chem. Res. 2012;45:1390. doi: 10.1021/ar300076b. [DOI] [PubMed] [Google Scholar]; (e) Schlutter F, Rossel F, Kivala M, Enkelmann V, Gisselbrecht JP, Ruffieux P, Fasel R, Mullen K. J. Am. Chem. Soc. 2013;135:4550. doi: 10.1021/ja400857g. [DOI] [PubMed] [Google Scholar]; (f) Gong B, Shao Z. Acc. Chem. Res. 2013;46:2856. doi: 10.1021/ar400030e. [DOI] [PubMed] [Google Scholar]; (g) Isaacs L. Acc. Chem. Res. 2014;47:2052. doi: 10.1021/ar500075g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Shimizu LS, Salpage SR, Korous AA. Acc. Chem. Res. 2014;47:2116. doi: 10.1021/ar500106f. [DOI] [PubMed] [Google Scholar]; (i) Barnes JC, Dale EJ, Prokofjevs A, Narayanan A, Gibbs-Hall IC, Juricek M, Stern CL, Sarjeant AA, Botros YY, Stupp SI, Stoddart JF. J. Am. Chem. Soc. 2015;137:2392. doi: 10.1021/ja512959g. [DOI] [PubMed] [Google Scholar]; (j) Wu Y, Nalluri SK, Young RM, Krzyaniak MD, Margulies EA, Stoddart JF, Wasielewski MR. Angew. Chem. Int. Ed. 2015;54:11971. doi: 10.1002/anie.201504576. [DOI] [PubMed] [Google Scholar]; (k) Chen D, Avestro AJ, Chen Z, Sun J, Wang S, Xiao M, Erno Z, Algaradah MM, Nassar MS, Amine K, Meng Y, Stoddart JF. Adv. Mater. 2015;27:2907. doi: 10.1002/adma.201405416. [DOI] [PubMed] [Google Scholar]; (l) Schweez C, Shushkov P, Grimme S, Hoger S. Angew. Chem. Int. Ed. 2016;55:3328. doi: 10.1002/anie.201509702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belostotskii AM. Conformational Concept for Synthetic Chemists’ Use: Principles and in Lab Exploitation. World Scientific; Singapore: 2008. p. 166. [Google Scholar]
- 12.Lee S, Chen CH, Flood AH. Nat. Chem. 2013;5:704. doi: 10.1038/nchem.1668. [DOI] [PubMed] [Google Scholar]
- 13.(a) Li Y, Zhao J, Yin X, Yin G. ChemPhysChem. 2006;7:2593. doi: 10.1002/cphc.200600416. [DOI] [PubMed] [Google Scholar]; (b) Ali MA, Krishnan MS. J. Org. Chem. 2010;75:5797. doi: 10.1021/jo100603a. [DOI] [PubMed] [Google Scholar]
- 14.Hua Y, Ramabhadran RO, Uduehi EO, Karty JA, Raghavachari K, Flood AH. Chem. Eur. J. 2011;17:312. doi: 10.1002/chem.201002340. [DOI] [PubMed] [Google Scholar]
- 15.(a) den Otter WK, Briels WJ. J. Chem. Phys. 1997;107:4968. [Google Scholar]; (b) den Otter WK, Briels WJ. J. Am. Chem. Soc. 1998;120:13167. [Google Scholar]; (c) Aleman C, Zanuy D, Casanovas J. J. Org. Chem. 2006;71:6952. doi: 10.1021/jo061062w. [DOI] [PubMed] [Google Scholar]; (d) Pochorovski I, Knehans T, Nettels D, Muller AM, Schweizer WB, Caflisch A, Schuler B, Diederich F. J. Am. Chem. Soc. 2014;136:2441. doi: 10.1021/ja4104292. [DOI] [PubMed] [Google Scholar]
- 16.(a) Ranghino G, Romans S, Lehn JM, Wipff G. J. Am. Chem. Soc. 1985;107:7873. [Google Scholar]; (b) Billeter M, Howard AE, Kuntz ID, Kollman PA. J. Am. Chem. Soc. 1988;110:8385. [Google Scholar]; (c) Grootenhuis PDJ, Kollman PA. J. Am. Chem. Soc. 1989;111:2152. [Google Scholar]; (d) Grootenhuis PDJ, Kollman PA. J. Am. Chem. Soc. 1989;111:4046. [Google Scholar]; (e) Troxler L, Wipff G. J. Am. Chem. Soc. 1994;116:1468. [Google Scholar]; (f) Hosokai T, Horie M, Aoki T, Nagamatsu S, Kera S, Okudaira KK, Ueno N. J. Phys. Chem. C. 2008;112:4643. [Google Scholar]
- 17.(a) Bonnet P, Agrafiotis DK, Zhu F, Martin E. J. Chem. Inf. Model. 2009;49:2242. doi: 10.1021/ci900238a. [DOI] [PubMed] [Google Scholar]; (b) Oakley MT, Johnston RL. J. Chem. Theory Comput. 2013;9:650. doi: 10.1021/ct3005084. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Oakley MT, Johnston RL. J. Chem. Theory Comput. 2014;10:1810. doi: 10.1021/ct500004k. [DOI] [PubMed] [Google Scholar]; (d) Watts KS, Dalal P, Tebben AJ, Cheney DL, Shelley JC. J. Chem. Inf. Model. 2014;54:2680. doi: 10.1021/ci5001696. [DOI] [PubMed] [Google Scholar]
- 18.Anslyn EV, Dougherty DA. Modern Physical Organic Chemistry. University Science Books; Sausalito: 2006. Strain and Stability. [Google Scholar]
- 19.Wu YT, Siegel JS. Chem. Rev. 2006;106:4843. doi: 10.1021/cr050554q. [DOI] [PubMed] [Google Scholar]
- 20.Szumna A. Chem. Soc. Rev. 2010;39:4274. doi: 10.1039/b919527k. [DOI] [PubMed] [Google Scholar]
- 21.(a) Scott LT, Hashemi MM, Bratcher MS. J. Am. Chem. Soc. 1992;114:1920. [Google Scholar]; (b) Biedermann PU, Pogodin S, Agranat I. J. Org. Chem. 1999;64:3655. doi: 10.1021/jo9900174. [DOI] [PubMed] [Google Scholar]; (c) Seiders TJ, Baldridge KK, Grube GH, Siegel JS. J. Am. Chem. Soc. 2001;123:517. doi: 10.1021/ja0019981. [DOI] [PubMed] [Google Scholar]; (d) Priyakumar UD, Sastry GN. J. Org. Chem. 2001;66:6523. doi: 10.1021/jo015775k. [DOI] [PubMed] [Google Scholar]; (e) Priyakumar UD, Sastry GN. J. Phys. Chem. A. 2001;105:4488. [Google Scholar]; (f) Dinadayalane TC, Sastry GN. Tetrahedron. 2003;59:8347. [Google Scholar]; (g) Amaya T, Sakane H, Muneishi T, Hirao T. Chem. Commun. 2008;44:765. doi: 10.1039/b712839h. [DOI] [PubMed] [Google Scholar]; (h) Nishida S, Morita Y, Ueda A, Kobayashi T, Fukui K, Ogasawara K, Sato K, Takui T, Nakasuji K. J. Am. Chem. Soc. 2008;130:14954. doi: 10.1021/ja806708j. [DOI] [PubMed] [Google Scholar]; (i) Hayama T, Baldridge KK, Wu YT, Linden A, Siegel JS. J. Am. Chem. Soc. 2008;130:1583. doi: 10.1021/ja073052y. [DOI] [PubMed] [Google Scholar]; (k) Amaya T, Hirao T. Pure Appl. Chem. 2012;84:1089. [Google Scholar]; (k) Wu TC, Chen MK, Lee YW, Kuo MY, Wu YT. Angew. Chem. Int. Ed. 2013;52:1289. doi: 10.1002/anie.201208200. [DOI] [PubMed] [Google Scholar]; (l) Jaafar R, Pignedoli CA, Bussi G, Ait-Mansour K, Groening O, Amaya T, Hirao T, Fasel R, Ruffieux P. J. Am. Chem. Soc. 2014;136:13666. doi: 10.1021/ja504126z. [DOI] [PubMed] [Google Scholar]; (m) Juricek M, Strutt NL, Barnes JC, Butterfield AM, Dale EJ, Baldridge KK, Stoddart JF, Siegel JS. Nat. Chem. 2014;6:222. doi: 10.1038/nchem.1842. [DOI] [PubMed] [Google Scholar]
- 22.(a) Sato K, Itoh Y, Aida T. Chem. Sci. 2014;5:136. [Google Scholar]; (b) Kang J, Miyajima D, Mori T, Inoue Y, Itoh Y, Aida T. Science. 2015;347:646. doi: 10.1126/science.aaa4249. [DOI] [PubMed] [Google Scholar]
- 23.(a) Yoon J, Sheu C, Houk KN, Knobler CB, Cram DJ. J. Org. Chem. 1996;61:9323. Selected papers using MD simulations in supramolecular chemistry: [Google Scholar]; (b) Raymo FM, Houk KN, Stoddart JF. J. Am. Chem. Soc. 1998;120:9318. doi: 10.1021/ja010443i. [DOI] [PubMed] [Google Scholar]; (c) Jang SS, Jang YH, Kim YH, Goddard WA, 3rd, Choi JW, Heath JR, Laursen BW, Flood AH, Stoddart JF, Norgaard K, Bjornholm T. J. Am. Chem. Soc. 2005;127:14804. doi: 10.1021/ja0531531. [DOI] [PubMed] [Google Scholar]; (d) Jang SS, Jang YH, Kim YH, Goddard WA, 3rd, Flood AH, Laursen BW, Tseng HR, Stoddart JF, Jeppesen JO, Choi JW, Steuerman DW, Deionno E, Heath JR. J. Am. Chem. Soc. 2005;127:1563. doi: 10.1021/ja044530x. [DOI] [PubMed] [Google Scholar]; (e) Cruz C, Delgado R, Drew MG, Felix V. J. Org. Chem. 2007;72:4023. doi: 10.1021/jo062653p. [DOI] [PubMed] [Google Scholar]; (f) Kim H, Goddard WA, 3rd, Jang SS, Dichtel WR, Heath JR, Stoddart JF. J. Am. Chem. Soc. 2009;113:2136. doi: 10.1021/jp809213m. [DOI] [PubMed] [Google Scholar]; (g) Santos SM, Costa PJ, Lankshear MD, Beer PD, Felix V. J. Phys. Chem. B. 2010;114:11173. doi: 10.1021/jp100724e. [DOI] [PubMed] [Google Scholar]; (h) Altarsha M, Yeguas V, Ingrosso F, Lopez R, Ruiz-Lopez MF. J. Phys. Chem. B. 2013;117:3091. doi: 10.1021/jp311671w. [DOI] [PubMed] [Google Scholar]; (i) Langton MJ, Robinson SW, Marques I, Felix V, Beer PD. Nat. Chem. 2014;6:1039. doi: 10.1038/nchem.2111. [DOI] [PubMed] [Google Scholar]; (j) Gao L, Liu W, Lee O-S, Dmochowski IJ, Saven JG. Chem. Sci. 2015;6:7238. doi: 10.1039/c5sc02401c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Narayan ARH, Jimenez-Oses G, Liu P, Negretti S, Zhao W, Gilbert MM, Ramabhadran RO, Yang Y-F, Furan LR, Li Z, Podust LM, Montgomery J, Houk KN, Sherman DH. Nat. Chem. 2015;7:653. doi: 10.1038/nchem.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Jackson NE, Kohlstedt KL, Savoie BM, de la Cruz MO, Schatz GC, Chen LX, Ratner MA. J. Am. Chem. Soc. 2015;137:6254. doi: 10.1021/jacs.5b00493. [DOI] [PubMed] [Google Scholar]
- 24.(a) Meyer AY, Richards WG. J. Comput. Aided Mol. Des. 1991;5:427. doi: 10.1007/BF00125663. A similar concept of shape similarity was conceived earlier in literature: [DOI] [PubMed] [Google Scholar]; (b) Good AC, Richards WG. J. Chem. Inf. Comput. Sci. 1993;33:112. [Google Scholar]
- 25.(a) Hirsch BE, Lee S, Qiao B, Chen CH, McDonald KP, Tait SL, Flood AH. Chem. Commun. 2014;50:9827. doi: 10.1039/c4cc03725a. [DOI] [PubMed] [Google Scholar]; (b) Singharoy A, Venkatakrishnan B, Liu Y, Mayne CG, Lee S, Chen CH, Zlotnick A, Schulten K, Flood AH. J. Am. Chem. Soc. 2015;137:8810. doi: 10.1021/jacs.5b04407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geometries for the 34 conformations are available as Supplementary Documents.
- 27.(a) Li Y, Flood AH. Angew. Chem. Int. Ed. 2008;47:2649. doi: 10.1002/anie.200704717. [DOI] [PubMed] [Google Scholar]; (b) Li Y, Flood AH. J. Am. Chem. Soc. 2008;130:12111. doi: 10.1021/ja803341y. [DOI] [PubMed] [Google Scholar]; (c) Juwarker H, Lenhardt JM, Pham DM, Craig SL. Angew. Chem. Int. Ed. 2008;47:3740. doi: 10.1002/anie.200800548. [DOI] [PubMed] [Google Scholar]; (d) Li Y, Pink M, Karty JA, Flood AH. J. Am. Chem. Soc. 2008;130:17293. doi: 10.1021/ja8077329. [DOI] [PubMed] [Google Scholar]; (e) Juwarker H, Lenhardt JM, Castillo JC, Zhao E, Krishnamurthy S, Jamiolkowski RM, Kim KH, Craig SL. J. Org. Chem. 2009;74:8924. doi: 10.1021/jo901966f. [DOI] [PubMed] [Google Scholar]; (f) Hua Y, Flood AH. Chem. Soc. Rev. 2010;39:1262. doi: 10.1039/b818033b. [DOI] [PubMed] [Google Scholar]; (g) Wang Y, Xiang J, Jiang H. Chem. Eur. J. 2011;17:613. doi: 10.1002/chem.201001560. [DOI] [PubMed] [Google Scholar]; (h) Hua Y, Liu Y, Chen CH, Flood AH. J. Am. Chem. Soc. 2013;135:14401. doi: 10.1021/ja4074744. [DOI] [PubMed] [Google Scholar]; (i) Qiao B, Sengupta A, Liu Y, McDonald KP, Pink M, Anderson JR, Raghavachari K, Flood AH. J. Am. Chem. Soc. 2015;137:9746. doi: 10.1021/jacs.5b05839. [DOI] [PubMed] [Google Scholar]; (j) Hirsch BE, McDonald KP, Qiao B, Flood AH, Tait SL. ACS Nano. 2014;8:10858. doi: 10.1021/nn504685t. [DOI] [PubMed] [Google Scholar]; (k) Lee S, Hua Y, Park H, Flood AH. Org. Lett. 2010;12:2100. doi: 10.1021/ol1005856. [DOI] [PubMed] [Google Scholar]
- 28.Arteca GA. J. Comput. Chem. 1993;14:718. [Google Scholar]
- 29.Jacobsen NE. NMR Spectroscopy Explained: Simplified Theory, Applications and Examples for Organic Chemistry and Structural Biology. Wiley; New York: 2007. [Google Scholar]
- 30.Lyle N, Das RK, Pappu RV. J. Chem. Phys. 2013;139:121907. doi: 10.1063/1.4812791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(a) Holm L, Sander C. Science. 1996;273:595. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]; (b) Rosen M, Lin SL, Wolfson H, Nussinov R. Protein Eng. 1998;11:263. doi: 10.1093/protein/11.4.263. [DOI] [PubMed] [Google Scholar]; (c) Cramer RD, Poss MA, Hermsmeier MA, Caulfield TJ, Kowala MC, Valentine MT. J. Med. Chem. 1999;42:3919. doi: 10.1021/jm990159q. [DOI] [PubMed] [Google Scholar]; (d) Rush TS, Grant JA, Mosyak L, Nicholls A. J. Med. Chem. 2005;48:1489. doi: 10.1021/jm040163o. [DOI] [PubMed] [Google Scholar]
- 32.(a) Walker PD, Mezey PG. J. Am. Chem. Soc. 1993;115:12423. [Google Scholar]; (b) Bender A, Glen RC. Org. Biomol. Chem. 2004;2:3204. doi: 10.1039/B409813G. [DOI] [PubMed] [Google Scholar]; (c) Ballester PJ, Richards WG. J. Comput. Chem. 2007;28:1711. doi: 10.1002/jcc.20681. [DOI] [PubMed] [Google Scholar]; (d) Rodriguez A, Laio A. Science. 2014;344:1492. doi: 10.1126/science.1242072. [DOI] [PubMed] [Google Scholar]
- 33.(a) Anet FAL, Bourn AJR. J. Am. Chem. Soc. 1965;87:5250. [Google Scholar]; (b) Bell RA, Saunders JK. Can. J. Chem. 1970;48:1114. [Google Scholar]; (c) Rowan R, McCammon JA, Sykes BD. J. Am. Chem. Soc. 1974;96:4773. doi: 10.1021/ja00829a031. [DOI] [PubMed] [Google Scholar]; (d) Mugridge JS, Zahl A, van Eldik R, Bergman RG, Raymond KN. J. Am. Chem. Soc. 2013;135:4299. doi: 10.1021/ja309949q. [DOI] [PubMed] [Google Scholar]
- 34. The other isomer, homo-dimer (P-P or M-M), exhibits a similar behavior in all respects except for its marginally weaker binding strength (see Section 7.4 of Supporting Information)
- 35. The degeneracy of P5 conformer in a sandwich complex is affected by the guest. DFT modeling showed nonsymmetric distribution of diglyme and thus C1 symmetry for P5 or M5 conformer in the 2:1 complex. Overall, these conformers have 5-fold degeneracy relative to none in their monomer states.
- 36. The negligible impact of hydrogen bonding compared to steric hindrance in the 2:1 sandwich complex can be seen from the negligible landscape shifts from an empty dimer to a complexed dimer (see Supporting Information)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.