Abstract
KRAS is frequently mutated in lung cancer. Whereas the mitogen-activated protein kinase (MAPK) is a well-known effector pathway of KRAS, blocking this pathway with clinically-available MAPK inhibitors is relatively ineffective. Here, we report that epithelial-to-mesenchymal transition rewires the expression of receptor tyrosine kinases, leading to differential feedback activation of the MAPK pathway following MEK inhibition. In epithelial-like KRAS mutant lung cancers, this feedback was attributed to ERBB3-mediated activation of MEK and AKT. In contrast, in mesenchymal-like KRAS mutant lung cancers, FGFR1 was dominantly expressed but suppressed by the negative regulator sprouty proteins; MEK inhibition led to de-repression of SPRY4 and subsequent FGFR1-mediated re-activation of MEK and AKT. Therapeutically, the combination of MEK inhibitor and FGFR inhibitor induced cell death in vitro and tumor regressions in vivo. These data establish the rationale and a therapeutic approach to treat mesenchymal-like KRAS mutant lung cancers effectively with clinically available FGFR1 and MAPK inhibitors.
Keywords: KRAS, MEK inhibitor, Epithelial-to-mesenchymal transition, lung cancer
Introduction
Lung cancer is the leading cause of cancer-related mortality, with a 5-year survival of less than 15% across all stages of disease. KRAS is the most frequently mutated oncogene in non-small-cell lung cancer (NSCLC), which is detected in 20 to 25% of Caucasian patients with lung adenocarcinoma (1,2). In the past decade, the discovery of driver oncogenes such as EGFR mutations and ALK rearrangements in lung adenocarcinomas have led to the development and implementation of targeted therapies that block the function of these oncogenes. In contrast, there are no approved targeted therapies for the treatment of KRAS mutant cancers because direct inhibition of mutant KRAS has proven challenging, despite the development of small-molecule inhibitors that interfere with the localization of KRAS or inhibit the activity of mutant KRAS (3).
Recently, targeting KRAS effector pathways has gained traction as a therapeutic alternative (1,2,4). The mitogen-activated protein kinases (MAPK) pathway is now a well-known effector pathway of KRAS, however, targeting this pathway by MEK inhibition results in limited activity in patient with KRAS mutant lung cancer (5,6). The lack of effectiveness may be associated with activation of multiple other effectors by KRAS including phosphoinositide 3-kinase (PI3K)-AKT and nuclear factor-kappa B (NF-kB) pathways (3,4). In addition, MEK inhibition leads to relief of physiologic negative feedback loops and results in activation of several upstream receptor tyrosine kinases (RTKs) (7–9). As such, the combination of MEK inhibitor with IGF-IR inhibitor was shown to enhance cell death in KRAS mutant colorectal cancer (10) and lung cancer (11). Additionally, co-targeting MEK and ERBB3 was shown to be effective in KRAS mutant lung and colorectal cancers (12). The different RTKs that are activated in KRAS mutant cancers reflect significant heterogeneity of these tumors, even within the same tissue type. Furthermore, recent integrative analysis of genomic, transcriptomic, and proteomic data identified three subclasses of KRAS mutant lung adenocarcinoma, supporting the idea of heterogeneity among tumors with same tissue type (13).
The epithelial-to-mesenchymal transition (EMT) is an essential mechanism in the developmental process and repair of tissue (14). EMT also contributes to the progression of cancer by promoting loss of cell-cell adhesion, leading to a shift in cytoskeletal dynamics. Hallmarks of EMT include the loss of E-cadherin expression and concomitant increase of mesenchymal markers such as vimentin (15,16). Emerging evidence suggests that EMT has been shown to associate with primary and acquired drug resistance. For example, EMT predicts sensitivity to EGFR inhibitor in NSCLC (17). In addition, EMT is a cause of resistance to EGFR inhibitors in EGFR mutant lung cancers (18). Tumors with a mesenchymal phenotype are also associated with aggressive disease and poor prognosis (19,20).
Herein, we report that MEK inhibition leads to distinct activation of RTKs in KRAS mutant lung cancers depending on the epithelial or mesenchymal state of the cancer. In epithelial-like KRAS mutant cancer cells, MEK inhibition upregulates ERBB3, which in turn activates PI3K-AKT and MAPK signaling. In contrast, mesenchymal-like KRAS mutant lung cancer cells lack ERBB3 expression. Instead, these cells show higher basal expression of FGFR1 protein, and MEK inhibition relieves activation of FGFR1 signaling by suppressing sprouty expression. Importantly, combinatorial inhibition of FGFR1 and MEK in mesenchymal-like KRAS mutant lung cancers resulted in robust apoptosis in vitro and tumor shrinkage in vivo. These data suggest biomarker-driven combinations of FGFR and MEK inhibitors may be effective for mesenchymal-like KRAS mutant NSCLC.
Results
ERBB3 mediates feedback activation of AKT and ERK signaling in ERBB3 expressed KRAS mutant lung cancers following MEK inhibitor treatment
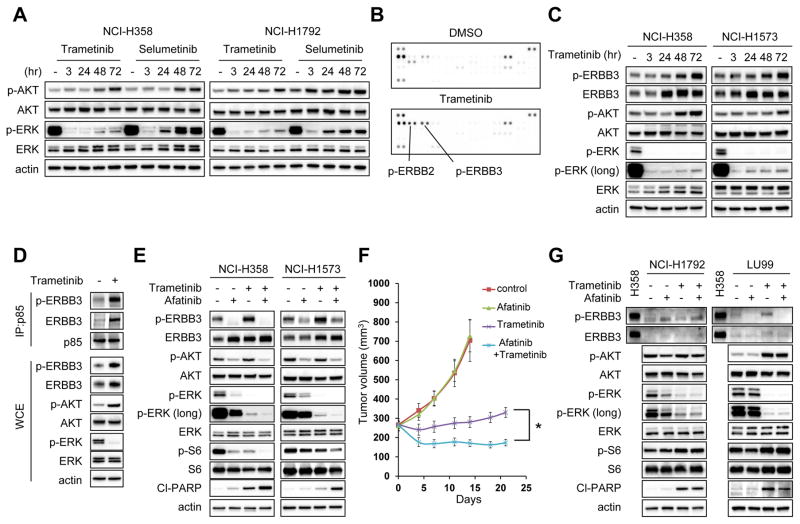
We first evaluated whether MEK inhibitor treatment leads to feedback activation of PI3 kinase and MEK-ERK signaling. Two allosteric MEK inhibitors, trametinib and selumetinib, were employed to determine the lowest drug concentration that maximally inhibited ERK phosphorylation (Supplementary Fig. S1A and S1B). Following 48 hour treatment with MEK inhibitor, rebound activation of ERK signaling occurred with both drugs, as evidenced by phosphorylation of ERK, while the rebound was less significant following treatment with the newer MEK inhibitor trametinib, which also disrupts RAF-MEK complexes (21,22) (Fig. 1A). Treatment with trametinib in a panel of KRAS mutant lung cancer cell lines further demonstrated that long term MEK inhibition induces rebound activation of ERK signaling (Supplementary Fig. S1C). Furthermore, upregulation of AKT phosphorylation was observed in many of the KRAS mutant lung cancer cell lines. Inhibition of PI3K/AKT or MEK is known to relieve feedback suppression of RTKs (8,12,23–25). To determine whether activation of RTK signaling accounts for the rebound activation of ERK and upregulation of AKT signaling following MEK inhibitor treatment, we interrogated global RTK phosphorylation before and after trametinib treatment. The results showed that phosphorylation of ERBB3 and its binding partner ERBB2 were upregulated following trametinib treatment (Fig. 1B); consistently, ERBB3 expressing epithelial-like KRAS mutant NCI-H358 and NCI-H1573 cells showed clear upregulation of ERBB3 phosphorylation following trametinib treatment (Fig. 1C). Furthermore, immunoprecipitation of p85, the regulatory subunit of PI3K, identified the interaction of p85 with ERBB3 was upregulated following trametinib treatment (Fig. 1D). siRNA knockdown or pharmacological inhibition of ERBB3 by a pan-ERBB inhibitor afatinib negated rebound activation of ERK and upregulation of AKT phosphorylation induced by trametinib and resulted in increased apoptosis (Fig. 1E and Supplementary Fig. S1D and S1E). We next determined the effects of the drug combination in vivo. Whereas trametinib monotherapy slowed tumor growth, tumor regression was observed with combination of afatinib with trametinib treatment (Fig. 1F). Collectively, these results showed the feedback ERK and AKT activation induced by ERBB3 mitigates the effect of MEK inhibitor in a subset of KRAS mutant lung cancer cell lines in vitro and in vivo.
Figure 1. ERBB3 mediates feedback activation of AKT and ERK phosphorylation following trametinib treatment in ERBB3 expressing cell lines.
(A) NCI-H358 and NCI-H1792 cells were treated with either 50 nM trametinib or 500 nM selumetinib for indicated times, and lysates were probed with the indicated antibodies. (B) NCI-H358 cells were treated with DMSO or 50 nM trametinib for 72 hours, and cell lysates were analyzed levels of phosphorylated RTKs using phospho-RTK arrays. Key RTKs are indicated. (C) NCI-H358 and NCI-H1573 cells were treated with 50 nM trametinib for the indicated times, and lysates were probed with the indicated antibodies. (D) Cells were treated as in (B) and cell lysates were immunoprecipitated with an antibody to p85. Interaction between p85 and ERBB3 were determined by immunoblotting. In parallel, whole-cell extracts (WCE) were immunoblotted to detect the indicated proteins. (E) Cells were treated with 1uM of the pan-EGFR inhibitor afatinib, 50 nM trametinib, or the combination of these two drugs for 48 hours, and lysates were probed with the indicated antibodies. (F) NCI-H358 xenografts were treated with the vehicle (control), afatinib 7.5 mg/kg, trametinib 0.6 mg/kg, or the combination at the same doses. Drugs were administered once daily by oral gavage. Tumor volumes were plotted over time from the start of treatment (mean ± SEM). *p < 0.05 by Student’s t test. (G) NCI-H1792 and LU99 cells were treated with indicated drug and drug combination as in (E). These cells were absent expression of ERBB3. Lysate from NCI-H358 was used as a positive control for ERBB3 expression. All immunoblots are representative of three independent experiments.
FGFR1 is upregulated in mesenchymal-like KRAS mutant lung cancer cell lines
Since we found that the MAPK pathway was also re-activated in KRAS mutant cells that lacked expression of ERBB3, and its reactivation was not negated by Afatinib treatment (Fig. 1G), we sought to uncover other distinct signaling pathways leading to re-activation of the MAPK pathway in order to develop effective MEK-inhibitor based targeted therapy for these cancers. Previous studies demonstrated that ERBB3 loss is associated with the mesenchymal phenotype in cancers including lung cancer (17,20,26). We therefore hypothesized that it was mesenchymal KRAS mutant lung cancers that re-activated the MAPK pathway following MEK inhibition independent of ERBB3.
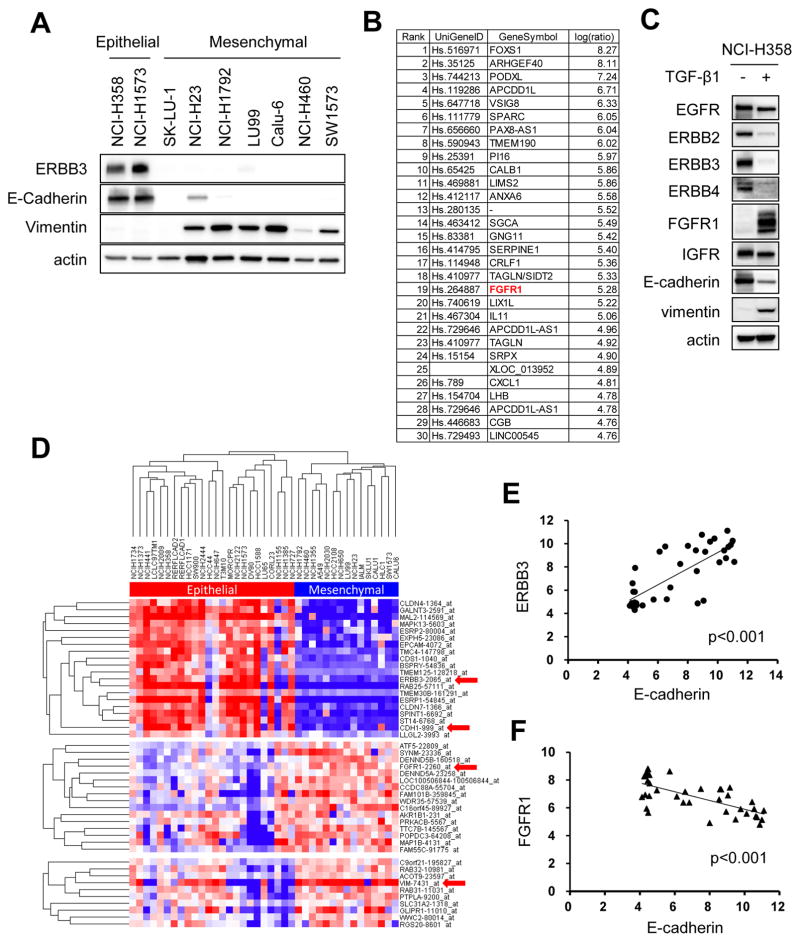
We first performed a Western blot analysis of our KRAS mutant lung cancer cell line panel and determined that our cells that expressed ERBB3 were epithelial, while those null for ERBB3 were mesenchymal (Fig. 2A). To further study this correlation, we induced EMT via chronic TGF-β1 treatment in the epithelial-like NCI-H358 cells, and upregulated and downregulated genes were analyzed (Fig. 2B and Supplementary Table S1). As expected, ERBB3 and CDH1 were strongly suppressed following induction of EMT. Interestingly, we found the receptor tyrosine kinase FGFR1 to be one of the highest upregulated genes in NCI-H358 TGF-β1 treated cells (Fig. 2B). Western blot analysis confirmed that FGFR1 was prominently upregulated following EMT induction while ERBB family proteins EGFR, ERBB2, ERBB3 and ERBB4 had reduced expression (Fig. 2C). We next performed unsupervised hierarchical clustering of 39 KRAS mutant NSCLC cell lines from Cancer Cell Line Encyclopedia (CCLE) (27). A total of 2635 genes, which demonstrated more than a 5-fold expression change among cell lines with median value of 6 or more, were extracted and used for further analyses. These analyses identified two distinct subsets of KRAS mutant cancer cell lines (Fig. 2D). One subtype, including the NCI-H358 and NCI-H1573 cells, typically expressed ERBB3 and E-Cadherin, consistent with the epithelial-like phenotype. The second group had a mesenchymal phenotype with low E-Cadherin, high vimentin, and low ERBB3. Furthermore, these cells had significantly higher FGFR1 expression compared to the epithelial-like cell lines. Linear regression analyses demonstrated positive correlations between ERBB3 and E-Cadherin as well as FGFR1 and vimentin. Conversely, inverse relationships between FGFR1 and E-Cadherin as well as ERBB3 and vimentin were found (Fig. 2E and F and Supplementary Fig. S2). Collectively, these data demonstrate ERBB3 high/FGFR1 low expression pattern in epithelial-like KRAS mutant lung cancers, contrasting with an FGFR1 high/ERBB3 low expression pattern in mesenchymal-like KRAS mutant cancers.
Figure 2. FGFR1 is dominantly expressed in mesenchymal-like KRAS mutant lung cancer cell lines.
(A) Expression of ERBB3, E-Cadherin, and Vimentin protein were analyzed by western blotting of lysates from KRAS mutant lung cancer cell lines. Actin is a loading control. Independent experiments were performed three times, and a representative result is shown. (B, C) NCI-H358 cells were treated with TGF-β1 (4 ng/mL) or PBS for 14 days in order to induce EMT. (B) RNA was extracted from each cells and gene expression profiles were compared. List of 30 genes most upregulated following TGF-β1 treatment was shown. (C) Lysates were extracted from each cells and immunoblotted with antibodies against indicated RTKs and EMT markers. Actin was used as a loading control. Independent experiments were performed three times, and a representative result is shown. (D) Unsupervised hierarchical clustering of 39 KRAS mutant NSCLC cell lines from Cancer Cell Line Encyclopedia (CCLE) database. A total of 2635 genes were analyzed which are showing more than 5-fold change among cell lines with median expression value of 6 or more. (E, F) Scattered plot analysis showing relationship between ERBB3 and E-Cadherin (E) and an inverse relationship between FGFR1 and E-Cadherin (F). p < 0.001, both by linear regression analysis.
Feedback activation of FGFR1-FRS2 pathway following treatment of MEK inhibitor in mesenchymal-like KRAS mutant lung cancer cell lines
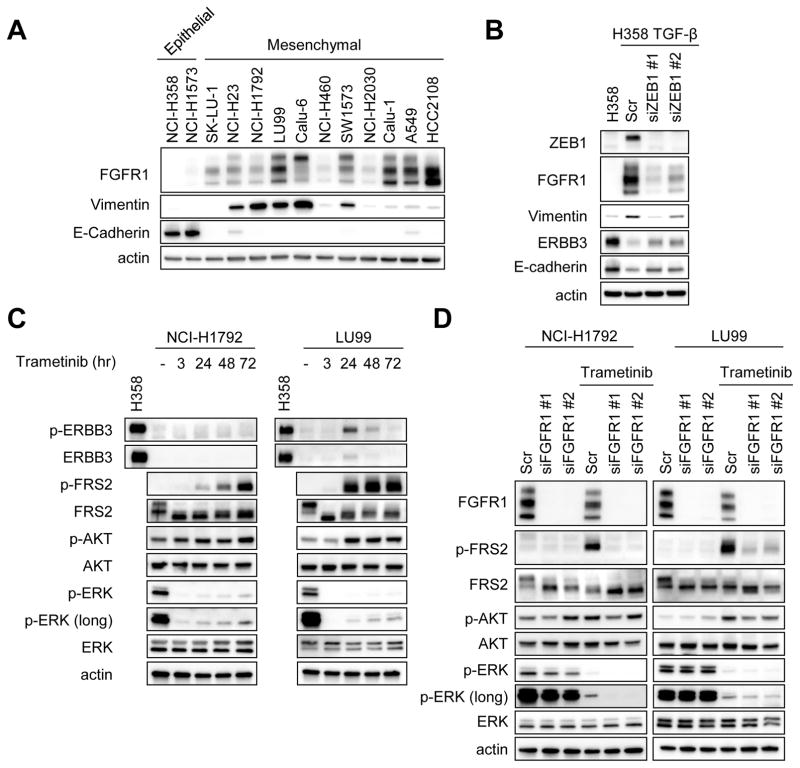
Consistent with the gene expression data from CCLE, Western blot analysis of a panel of KRAS mutant cancer cells identified mesenchymal cells as having higher FGFR1 expression (Fig. 3A). To further determine the relationship between mesenchymal phenotype and FGFR1 expression, we tried to revert EMT to determine how that impacted the expression of FGFR1. Knockdown of the EMT transcriptional repressor ZEB1 partially reversed EMT with upregulation of E-cadherin and downregulation of vimentin demonstrated in the TGF-β1-treated (and thus EMT-induced) NCI-H358 cells (Fig. 3B). Indeed, ZEB1 knockdown resulted in suppression of FGFR1 and induction of ERBB3 expression (Fig. 3B), consistent with our other findings. We next investigated whether FGFR1 expression is related to the feedback activation of ERK signaling following MEK inhibitor treatment in mesenchymal-like KRAS mutant cancers. In NCI-H1792 and LU99 mesenchymal-like KRAS mutant lung cancer cell lines, while MEK inhibitor treatment strongly suppressed ERK phosphorylation, long term MEK inhibition resulted in activation of FRS2, an adaptor protein of FGFR (Fig. 3C). Furthermore, immunoblot analyses identified trametinib did not upregulate phosphorylation of several other RTKs such as EGFR family proteins, AXL, MET, PDGFRα, and IGFR adaptor protein IRS-1, suggesting FRS2 may be the primary signal induced by MEK inhibition (Supplementary Fig. S3). To determine if the upregulation of FRS2 phosphorylation is mediated by FGFR1, we treated the NCI-H1792 and LU99 cells with trametinib after FGFR1 knockdown. Indeed, while either treatment with an FGFR inhibitor NVP-BGJ398 (28) or FGFR1 knockdown modestly downregulated ERK signaling, in the presence of trametinib, FGFR1 knockdown negated trametinib-induced FRS2 phosphorylation and feedback activation of ERK signaling (Fig. 3D and Supplementary Fig. S4). In addition, AKT phosphorylation was upregulated following trametinib treatment, especially in the LU99 cells, which was also negated by FGFR1 knockdown (Fig. 3C and 3D).
Figure 3. Trametinib induces feedback activation of FRS2 phosphorylation via FGFR1.
(A) Expression of FGFR1, vimentin, and E-cadherin protein were analyzed by Western blotting of lysates from KRAS mutant lung cancer cell lines. Actin is the loading control. Independent experiments were performed three times, and a representative result is shown. (B) NCI-H358 cells were treated with TGF-β1 (4 ng/mL) for 14 days in order to induce EMT. Then EMT-induced cells (H358-TGFβ) were transfected with two different siRNAs targeting ZEB1 or scramble siRNA and cultured for 72 hours. Lysates were probed with the indicated antibodies. Independent experiments were performed twice, and a representative result is shown. (C) Long-term MEK inhibition resulted in strong upregulation of FRS2 phosphorylation. NCI-H1792 and LU99 cells were treated with 50 nM trametinib for the indicated times, and lysates were probed with the indicated antibodies. Immunoblots are representative of three independent experiments. (D) FGFR1 mediates FRS2 phosphorylation following trametinib treatment. Cells were transfected with two different siRNAs targeting FGFR1 or scramble siRNA and cultured for 48 hours. Then, media was replaced with or without 50 nM trametinib and cells were treated for an additional 48 hours. Lysates were probed with the indicated antibodies. Independent experiments were performed three times, and a representative result is shown.
SPRY4 suppression relieves FGFR1-FRS2 pathway leading to activation of AKT and ERK signaling in mesenchymal-like KRAS mutant cancer cells
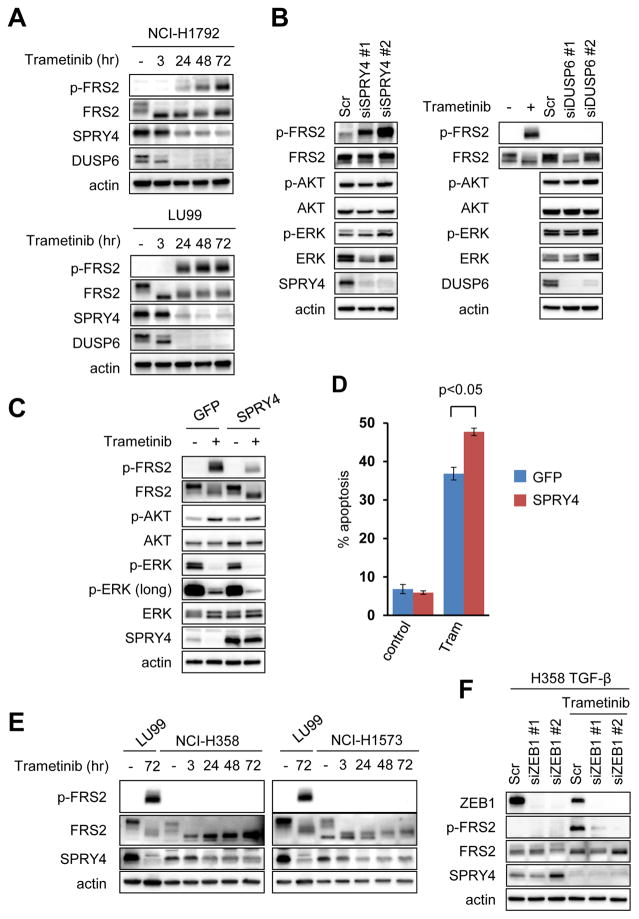
Next, in order to further investigate how MEK inhibition leads to the induction of the FGFR pathway in KRAS mutant mesenchymal lung cancers, we compared gene expression profiles of the mesenchymal-like KRAS mutant lung cancer NCI-H1792 cells before and after treatment with trametinib. Interestingly, we found no significant autocrine upregulation of FGF ligands or receptors following trametinib treatment (Supplementary Table S2). Addition of FGFR1 ligand modestly rescued mesenchymal-like KRAS mutant lung cancer cells from trametinib induced growth inhibition suggesting some, but minor, paracrine contribution from tumor stroma or systemic production. (Supplementary Fig. S5). The MEK/ERK pathway can also be regulated by ligand independent feedback regulation in which downstream proteins transcriptionally regulated by ERK in turn negatively regulate ERK signaling (29). Indeed, trametinib significantly suppressed the expression of inhibitory regulator of MAPK pathway including Sprouty proteins (SPRY1, SPRY2, and SPRY4) and MAPK phosphatases (MPKs and DUSPs) (Supplementary Table S3). Sprouty proteins antagonize FGFR-mediated MAPK activation by competing with FRS2 for binding to GRB2 (growth factor receptor bound protein 2) and the SOS (son of sevenless) complex (30,31). Therefore, we hypothesized that suppression of SPRY or DUSP proteins led to FRS2 activation. We chose to investigate SPRY4 and DUSP6 because these proteins showed the sharpest downregulation following trametinib treatment among proteins in each family (Supplementary Fig. S6). Whereas both SPRY4 and DUSP6 expression were downregulated following trametinib treatment (Fig. 4A), FRS2 phosphorylation was strongly induced by SPRY4 suppression (Fig. 4B). Furthermore, overexpression of SPRY4 protein negated feedback activation of FRS2 following MEK inhibitor treatment (Fig. 4C), resulting in further suppression of ERK phosphorylation and reduction of AKT phosphorylation induced by trametinib and greater induction of apoptosis in LU99 cells (Fig. 4D). Moreover, in low FGFR1 expressing-epithelial-like KRAS mutant lung cancer cells, FRS2 phosphorylation was not induced by trametinib despite SPRY4 downregulation (Fig. 4E). In line with this, ZEB1 knockdown abrogated FRS2 activation following trametinib treatment (Fig. 4F). Collectively, MEK inhibition leads to downregulation of expression of SPRY proteins in KRAS mutant lung cancers, which relieves suppression of basal FGFR-FRS2 function, leading to reactivation of MAPK signaling in the presence of FGFR1.
Figure 4. Downregulation of SPRY4 expression is associated with FGFR1-FRS2 activation.
(A) Trametinib suppresses expression of SPRY4 and DUSP6. NCI-H1792 and LU99 cells were treated with 50 nM trametinib for the indicated times, and lysates were probed with the indicated antibodies. Independent experiments were performed three times, and a representative result is shown. (B) FRS2 phosphorylation is not induced by DUSP6 knockdown, however is induced by SPRY4 knockdown. LU99 cells were transfected with two different siRNAs against SPRY4, DUSP6 or scramble siRNA for 72 hours. Lysates were probed with the indicated antibodies. Lysate from LU99 cells treated with trametinib at 50 nM for 48 hours was used as a positive control for FRS2 activation. Independent experiments were performed three times, and a representative result is shown. (C, D) SPRY4 overexpression negated feedback activation of FGFR signaling. LU99 cells infected with a GFP control or SPRY4 expressing lentiviral plasmid were treated with trametinib for 48 hours. Lysates were probed with indicated antibodies (C) or cells were analyzed by FACS to quantify annexin positive cells (D). The average amount of apoptosis ± SD of 3 independent experiments is shown (p < 0.05 by Student’s t test). (E) Trametinib does not activate FRS2 phosphorylation in epithelial-like KRAS mutant cancer cells with low FGFR1 expression. NCI-H358 and NCI-H1573 cells were treated with 50 nM trametinib for the indicated times, and lysates were probed with the indicated antibodies. Lysate from LU99 was used as a positive control for FRS2 activation following trametinib treatment. Please note: trametinib downregulated SPRY4 expression in both LU99 and epithelial-like cells. Independent experiments were performed three times, and a representative result is shown. (F) H358-TGFβ cells were transfected with two different siRNAs targeting ZEB1 or scramble siRNA and cultured for 48 hours. Then, media was replaced with or without 50 nM trametinib and cells were treated for an additional 48 hours. Lysates were probed with the indicated antibodies. Independent experiments were performed twice, and a representative result is shown.
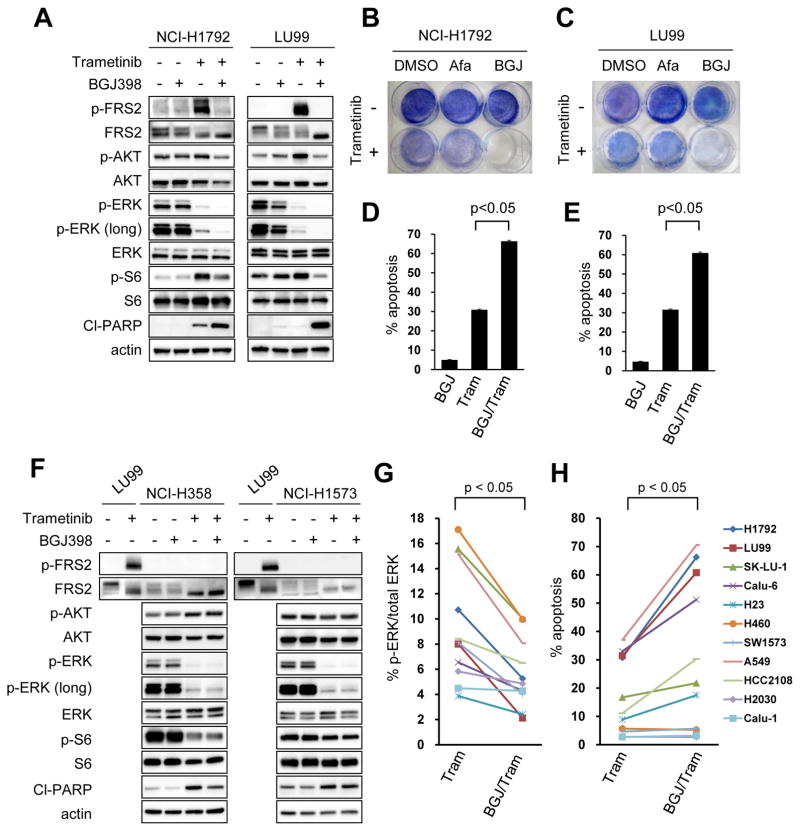
Combination of FGFR inhibition and MEK inhibition suppresses feedback activation of ERK and induces cell death in mesenchymal-like KRAS mutant lung cancer cell lines
To further interrogate the relationship between EMT and FGFR1-mediated ERK re-activation and upregulation of AKT signaling, epithelial and mesenchymal KRAS mutant lung cancer cell lines were treated with the combination of trametinib and NVP-BGJ398. In NCI-H1792 and LU99 mesenchymal-like KRAS mutant lung cancer cell lines, the combination of NVP-BGJ398 and trametinib showed a more complete suppression of ERK phosphorylation and improved suppression of AKT phosphorylation compared to trametinib monotherapy, consistent with a key role of FGFR1 in MEK-inhibitor induced feedback (Fig. 5A). The combination of FGFR and MEK inhibition led to profound inhibition of cell survival compared with single-agent pan-ERBB, FGFR, and MEK inhibitors as well as combined pan-EGFR and MEK inhibition (Fig. 5B and C). This was accompanied by greater induction of apoptosis (Fig. 5D and E). On the contrary, the combination of FGFR and MEK inhibition had no effect on downstream signaling in epithelial-like KRAS mutant cells (Fig. 5F). Accordingly, the combination of FGFR inhibitor with trametinib induced similar levels of apoptosis compared to trametinib monotherapy in the epithelial-like NCI-H358 and NCI-H1573 cells (Fig. 5F). In contrast, in the EMT-induced NCI-H358 cells, the addition of NVP-BGJ398 led to further suppression of ERK phosphorylation compared to trametinib monotherapy and negated AKT activation induced by trametinib (Supplementary Fig. S7A). The combination of trametinib and NVP-BGJ398 or FGFR1 knockdown induced robust apoptosis in these cells (Supplementary Fig. S7B and S7C). Collectively, these data demonstrate MEK inhibition induces distinct RTK activation depending on EMT status, and the activation of the FGFR1-FRS2 pathway is dominantly induced in mesenchymal-like KRAS mutant lung cancer cells.
Figure 5. Combination of trametinib with FGFR inhibition effectively leads to cell death in mesenchymal-like KRAS mutant lung cancer.
(A) Mesenchymal-like KRAS mutant NCI-H1792 and LU99 cells were treated with 1uM pan-FGFR inhibitor NVP-BGJ398, 50 nM trametinib, or the combination of these two drugs for 48 hours, and lysates were probed with the indicated antibodies. (B, C) Cell lines were treated with DMSO, 1uM afatinib, 1uM NVP-BGJ398, with or without 50 nM trametinib, and drug was replenished every 72 hours for 6 days. Plates were then stained with crystal violet and imaged. A representative plate of 2 independent experiments are shown. (D, E) NCI-H1792 (D) or LU99 (E) cells were treated with drug and drug combinations as in (A) for 72 hours and analyzed by FACS to quantify annexin positive cells. The average amount of apoptosis ± SD of 3 independent experiments is shown (p < 0.05 by Student’s t test). (F) Epithelial-like KRAS mutant NCI-H358 and NCI-H1573 cells were treated as in (A). Lysates from LU99 were used as positive control for the induction of FRS2 phosphorylation following trametinib treatment. Independent experiments were performed three times, and a representative result is shown. (G) The levels of phosphorylated ERK after treatment with trametinib or trametinib with NVP-BGJ398 were quantified for eleven mesenchymal-like KRAS mutant cancer cell lines examined (raw data shown in (A) and Supplementary Fig. S8A). A paired Student’s t-test was used for comparisons. (H) Induction of apoptosis by trametinib or the combination of trametinib with NVP-BGJ398 in mesenchymal-like KRAS mutant lung cancer cell lines. Raw data is shown in (D), (E), and Supplementary Fig. S8D. A paired Student’s t-test was used for comparisons.
Encouraged by the activity of the combination of trametinib and NVP-BGJ398, we expanded the number of mesenchymal-like KRAS mutant lung cancer cell lines to further investigate the effects of co-treatment with trametinib and NVP-BGJ398. Immunoblot analysis confirmed that FGFR inhibitor suppressed feedback activation of ERK following trametinib treatment for 48 or 72 hours (Fig. 5A and G, and Supplementary Fig. S8A and S8B). In addition, AKT phosphorylation was upregulated following trametinib treatment in many of the cell lines and the combination showed a trend to induce better suppression of AKT compared to trametinib monotherapy (p=0.08) (Fig. 5A and Supplementary Fig. S8A and S8C). Among eleven mesenchymal-like KRAS mutant lung cancer cell lines, FGFR inhibitor in combination with MEK inhibitor induced robust apoptosis in four, modest apoptosis in three, and minimal apoptosis in four of the cell lines, suggesting heterogeneity of response to the therapy (Fig. 5H and Supplementary Fig. S8D). Moreover, apoptosis induced by NVP-BGJ398 with trametinib was significantly correlated with that of induced by PI3K inhibitor with trametinib (Supplementary Fig. S9A and S9B), suggesting that the FGFR1-FRS2 pathway activated by trametinib plays roles both in PI3K and MEK signaling. To exclude the possibility that residual AKT activity was causing resistance to FGFR and MEK inhibition, SW1573 and NCI-H460 insensitive cell lines were treated with NVP-BGJ398, trametinib and a PI3 kinase inhibitor, GDC-0941 (Pictilisib). Although the combination induced complete suppression of both AKT and ERK signaling (Supplementary Fig. S10A), only modest apoptosis was observed in these cell lines (Supplementary Fig. S10B and S10C). Recent studies have demonstrated that an abnormality in the apoptotic machinery, including Bcl-xL overexpression by Yes-associated protein 1 (YAP1), and decreased ratios of BIM/Bcl-xL and PUMA/Bcl-xL underlie intrinsic resistance to MEK inhibitor with or without PI3K inhibitor (32,33). In line with this, addition of the Bcl-2/Bcl-xL inhibitor ABT-263 (navitoclax) to FGFR inhibitor and MEK inhibitor induced apoptosis in the SW1573 cells, suggesting that deregulation of apoptotic proteins may be a cause of the differential sensitivity to FGFR and MEK inhibition (Supplementary Fig. S10D and S10E).
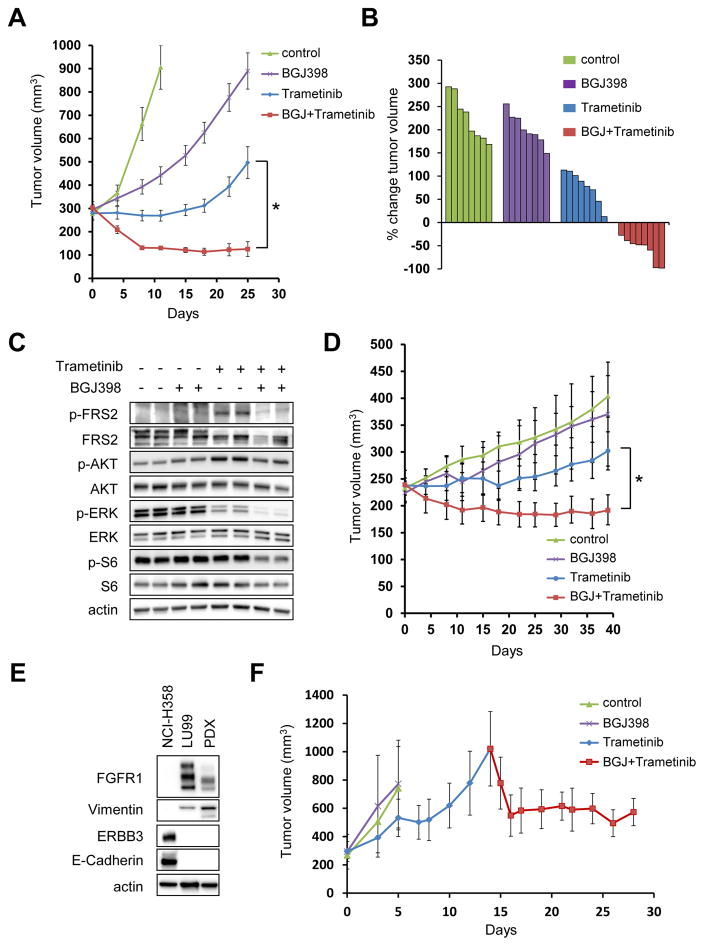
Combination of FGFR and MEK inhibition induces tumor regression in vivo
These findings led us to test the efficacy of combination of FGFR inhibitor with MEK inhibitor in vivo. Whereas either NVP-BGJ398 or trametinib monotherapy had minimal effect of tumor growth in the LU99 tumor xenograft model, tumors regressed when treated with the combination of FGFR and MEK inhibitor (Fig. 6A and B). The drug combination was well tolerated over a four-week treatment period (Supplementary Fig. S11). Pharmacodynamic studies of the drug-treated tumors recapitulated the in vitro results; trametinib partially suppressed ERK phosphorylation, however it induced FRS2 and AKT phosphorylation as well. Addition of FGFR inhibitor suppressed feedback activation of FRS2 leading to greater suppression of ERK and AKT, which resulted in downregulation of S6 phosphorylation (Fig. 6C). Moreover, the efficacy of the combination of FGFR inhibitor with MEK inhibitor was validated in a second NCI-H23 xenograft model (Fig. 6D).
Figure 6. The combination of an FGFR inhibitor and a MEK inhibitor leads to tumor regressions in mesenchymal-like KRAS mutant lung cancer in vivo.
(A) LU99 xenografts were treated with the vehicle (control), NVP-BGJ398 15 mg/kg, trametinib 0.6 mg/kg, or the combination at the same doses. Drugs were administered once daily by oral gavage. Tumor volumes were plotted over time from the start of treatment (mean ± SEM). *p < 0.05 by Student’s t test. (B) Waterfall plot showing the percent change in tumor volume (relative to initial volume) for individual LU99 tumors following 25 days of treatment. Note that data for control group was taken on day 11 due to their growth. (C) LU99 derived xenograft tumors from mice treated as indicated were lysed and immunoblotted with the indicated antibodies. (D) NCI-H23 xenografts were treated same as (A). Tumor volumes were plotted over time from the start of treatment (mean ± SEM). *p < 0.05 by Student’s t test. (E, F) Tumor regression in a patient-derived xenograft (PDX) by FGFR and MEK inhibition. (E) PDX tumors implanted into NSG mice were lysed and immunoblotted with the indicated antibodies. Lysates from NCI-H358 and LU99 were used as control for epithelial and mesenchymal cells, respectively.
(F) PDXs were untreated (control), or treated with NVP-BGJ398 15 mg/kg, or trametinib 0.6 mg/kg (n=3 in each cohort). Once the average of the tumors became more than 1000 mm3 in the trametinib cohort, the combination of trametinib and NVP-BGJ398 was started at the same doses as the single-agent cohorts. Drugs were administered once daily by oral gavage. Tumor volumes were plotted over time from the start of treatment (mean ± SEM).
Lastly, to validate our findings in a clinically relevant setting, we used a patient derived xenograft (PDX) of KRAS mutant lung cancer with a representative mesenchymal phenotype identified by the expression of E-cadherin and vimentin (Fig. 6E). Consistent with the in vitro findings and the cell line xenograft mouse models (Fig. 6A and 6D), the addition of FGFR inhibitor sensitizes these tumors to trametinib (Fig. 6F), supporting the notion that EMT may serve as an informative biomarker to predict the responsiveness of KRAS mutant lung cancers to the combination of FGFR inhibition and MEK inhibition.
Expression of mesenchymal markers is associated with FGFR1 expression in patients with KRAS mutant lung adenocarcinoma
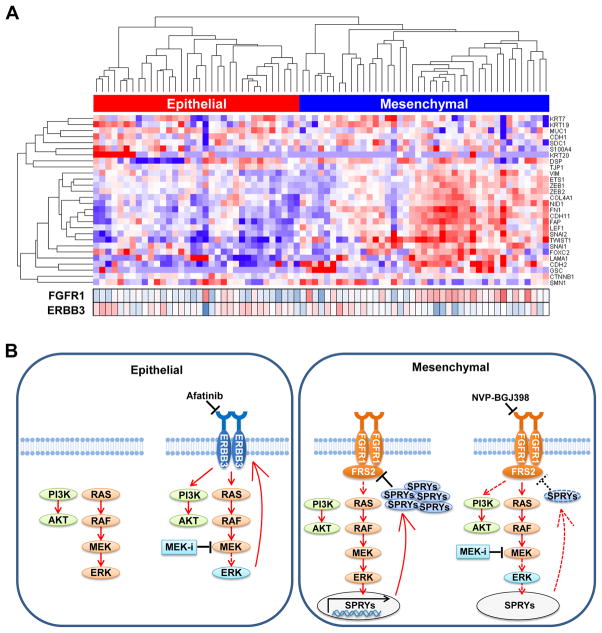
To determine whether FGFR1 expression was also associated with the mesenchymal phenotype in primary KRAS mutant lung cancers as it was in our other cell line studies, we analyzed RNA sequence expression data of 75 KRAS mutant lung adenocarcinomas from The Cancer Genome Atlas (TCGA) (34). Using a set of 28-genes that is highly correlated with the EMT process as reported by Kalluri and Weinberg (35), hierarchical clustering analysis revealed that KRAS mutant cancer could be classified into two groups based on epithelial and mesenchymal markers (Fig. 7A). Importantly, mesenchymal-like KRAS mutant tumors demonstrated significantly higher FGFR1 expression and significantly lower ERBB3 expression compared to epithelial-like KRAS mutant cancer (p < 0.001 for FGFR1 and p = 0.03 for ERBB3, respectively). These data confirm that FGFR1 expression is high specifically in the mesenchymal-like KRAS mutant cancer subgroup.
Figure 7. Expression of mesenchymal marker is associated with FGFR1 expression in patients with KRAS mutant lung adenocarcinoma.
(A) Unsupervised hierarchical clustering of 75 KRAS mutant adenocarcinoma extracted from the TCGA dataset was shown using 28 genes listed as EMT related genes by Kalluri and Weinberg (35). Expression of FGFR1 and ERBB3 in each tumor was also shown in the bottom. FGFR1 expression was significantly higher in mesenchymal-like tumors compared to epithelial-like tumors (p < 0.001 by Student’s t test). (B) Proposed treatment strategies for the treatment of KRAS mutant lung cancer based on EMT status.
Discussion
MAPK is the best characterized downstream pathway of KRAS, however, MEK inhibitor monotherapy demonstrates only modest efficacy in vitro and in vivo (5,6,9). These data are reminiscent of BRAF inhibitors in BRAF mutant colorectal cancer: The subsequent identification and drugging of the EGFR pathway as a feedback activator of ERK signaling has led to clinical trials of combination BRAF and EGFR inhibitors (36,37). In this study, we have shown that resistance to MEK inhibition in KRAS mutant lung cancers is caused by the feedback activation of RTK signaling. While feedback activation is mediated by ERBB3 in epithelial-like KRAS mutant cancer, the FGFR1-FRS2 pathway plays a critical role in the feedback activation of MAPK signaling in mesenchymal-like KRAS mutant cancers. Importantly, the combination of trametinib with FGFR inhibitor induced robust apoptosis in vitro and tumor regressions in vivo in mesenchymal-like KRAS mutant lung cancers, suggesting the biomarker-directed combination of these two drugs would benefit this subset of KRAS mutant lung cancer patients.
Reactivation of ERK signaling has played critical roles in primary and acquired resistance to targeted therapies, even outside of KRAS-driven cancers (36–41). It has been demonstrated that activation of MAPK signaling induces transcription of negative feedback genes including SPRYs and DUSPs (9,42). Interestingly, this pathway is active in benign tumors, presumably acting as a fail-safe negative feedback signaling program enabling the cell to protect itself from malignancy (42). In our study, we found this pathway was also active in the malignant KRAS mutant lung cancers, and relieving this feedback through MEK inhibitor treatment leads to the inhibition of the SPRY proteins. In mesenchymal KRAS mutant cancers, this is sufficient to activate the FGFR1 pathway as FGFR1 appears “primed” in these cancers via high levels of expression. Therefore, in mesenchymal-like KRAS mutant cancer cells, FGFR1 activity is suppressed by the active KRAS-MEK signal, perhaps part of an initial effort by the cell to prevent malignancy; the FGFR1 signal is transduced once negative feedback is relieved by trametinib, and only blocking both the MEK/ERK signal and the FGFR1 signal allows for sustained MEK/ERK inhibition, and subsequent apoptosis and tumor regressions.
In mesenchymal-like KRAS mutant lung cancer cells, SPRY4 plays a primary role in the induction of FRS2 phosphorylation following trametinib treatment. While SPRY proteins have been shown to be involved in ERK dependent feedback inhibition of EGFR family proteins in BRAF mutant melanomas and colon cancers and FGFR family proteins in GISTs (37,38,41), the role of SPRYs and DUSPs is cell type- and context-dependent (43). SPRY4 has been shown to suppress FGF pathways, but it has not been shown to inhibit EGF signaling in contrast to the involvement of other SPRY family proteins (30). Intriguingly, FRS2 phosphorylation was not induced in epithelial-like KRAS mutant cancer cells expressing low FGFR1 protein, despite downregulation of SPRY4 expression by trametinib. These results support the notion that sprouty proteins are downregulated following trametinib treatment in KRAS mutant lung cancers; however, it is only in the high FGFR1-expressing mesenchymal lung cancers does this lead to FGFR1-dependent FRS2 phosphorylation and subsequent re-activation of the MAPK pathway, resulting in resistance to MEK inhibitors (Fig. 7B). SPRY4 expression has been shown to be positively regulated by the ERK pathway previously (43). In line with this, a RAS-responsive element was located between nucleotide positions -69 and -31 in the SPRY4 promoter region and transfection of dominant negative RAS N17 significantly inhibited promoter activity of the gene by up to 70% in A549 cells (44). However, the precise mechanism of SPRY4 regulation remains to be determined.
We identified MEK inhibition led to upregulation of PI3K-AKT signaling in many of the KRAS mutant lung cancer cell lines despite their epithelial or mesenchymal status. In epithelial-like KRAS mutant lung cancer cells, we showed that upregulation of PI3K-AKT signaling was mediated through ERBB3 by upregulation of protein expression and phosphorylation, which led to bound ERBB3:p85, the regulatory subunit of PI3K. Selumetinib was shown to induce degradation of c-MYC, which relieves transcriptional repression of ERBB3 in breast cancer and KRAS mutant lung and colon cancers (8,12). Furthermore, in BRAF mutant cancers, BRAF inhibitor or trametinib upregulates ERBB3 by inducing the expression of the transcription factor FOXD3, or by decreasing the expression of the transcription repressors C-terminal binding protein 1 and 2 (45,46). We also demonstrated that FGFR1 mediated the upregulation of PI3K-AKT signaling in mesenchymal-like cells; PI3 kinase could be activated by FGFR1-FRS2 in which several tyrosine residues of FRS2 serve as docking sites for proteins such as GRB2 and GAB1, allowing recruitment of PI3K. Alternatively, PI3K activation may be mediated by RAS whose activation is upregulated following SPRY4 suppression.
We found ERK suppression-mediated relief of negative feedback of FGFR1 signaling occurred independently of FGF ligands and receptors. Recently, FGFR was also shown to mediate re-activation of the MAPK pathway and attenuate the anti-proliferative effects of Imatinib in gastrointestinal stromal tumor (GIST) (41). In that study, whereas treatment with FGF ligands reduced the antitumor activity of Imatinib in GIST cell lines, no significant alterations in the expression of FGF ligands and receptors were observed after long-term Imatinib treatment. Our results demonstrated only modest induction of trametinib resistance by FGF ligand. These results indicate that relief of ERK dependent feedback may play important roles in the reactivation of ERK in KRAS mutant cancers independently of ligand-mediated activation.
While our study identified subgroups of KRAS mutant lung adenocarcinoma based on EMT status and identified distinct therapeutic approach corresponding to each subgroup, other factors affecting heterogeneity of KRAS mutant lung cancer could contribute to intrinsic resistance. In fact, we observed heterogeneity within each subgroup. For example, the level of ERBB3 and FGFR1 expression was variable among tumors (Fig. 7A). This heterogeneity may be at least partly attributed to co-existing genomic alterations. Recent bioinformatic analysis by Skoulidis et al identified that KRAS mutant lung cancer can be classified into three subgroups by co-occurring genetic alterations in STK11, TP53, and CDKN2A/B (13). In line with this, concurrent loss of LKB1 is a cause of resistance to the combination of selumetinib with docetaxel or PI3 kinase inhibitor in KRAS mutant lung tumor model, likely through activated AKT and/or SRC pathways (47,48). While we noted that p53 or LKB1 status did not affect the level of apoptosis induced by NVP-BGJ398 and trametinib (Supplementary Fig. S12) and that there was no relationship between EMT and subgroups classified by Skoulidis et al (Supplementary Fig. S13) (13), it remains likely that other tumor-specific mutations are impacting drug response.
Another important consideration is the differential activation of effecter pathways by mutant KRAS other than PI3K and MEK in the tumors: for instance, KRAS G12C and G12V mutants harbor increased signaling through Ral effector pathway and decreased signaling through AKT, while KRAS G12D activates both PI3K and MEK signal (49). However, in our study, apoptosis induced by FGFR and MEK inhibitor was not correlated with the site of KRAS amino acid substitutions (Supplementary Fig. S14). Lastly, we noted a marked difference in the amount of apoptosis induced by PI3Ki/MEKi or FGFRi/MEKi in the different models (Fig 5H and Supplementary Fig. S9B). This suggests the apoptotic machinery plays a role in the resistance of KRAS mutant NSCLCs to FGFRi/MEKi (33). In line with this, differences in the balance of pro- and anti-apoptotic BCL-2 family proteins could lead to the insensitivity of the combination in SW1573 cells, and, more generally, may underlie heterogeneity in the apoptotic response to FGFRi/MEKi combination therapy in KRAS mutant NSCLCs. These heterogeneities among KRAS mutant lung cancer indicate the need to identify other targets to combine with MEK inhibitors.
Other studies have identified factors influencing the response of KRAS mutant cancers to targeted inhibition. For instance, YAP1 was recently identified as being able to counteract loss of mutant KRAS in cell lines and mouse models including lung cancer, by activating the FOS transcription factor and inducing expression of mesenchymal genes (50,51). Interestingly, in the same study that identified YAP1 as a survival factor following mutant KRAS suppression (50), members of the FGF family were also identified as genes that rescue loss of viability induced by KRAS knockdown. A second screen also identified FGFR1 as a kinase that was able to rescue KRAS suppression in KRAS-dependent cells (50). In addition, FGFR1 has been demonstrated to induce EMT in embryonic development and prostate cancer models (52,53). These results invoke a link between FGFR1 expression, EMT, and KRAS dependency, and are consistent with some of our findings in this study.
In conclusion, we report that the FGFR1-FRS2 pathway mediates feedback activation of MAPK following MEK inhibition in mesenchymal-like KRAS mutant lung cancer. Our findings support clinical trials with stratification of KRAS mutant lung patients based on EMT status in order to target the mesenchymal tumors with this novel therapy.
Methods
Cell lines and reagents
The lung cancer cell lines A549, NCI-H358, NCI-H1792, NCI-H23, SW900 and SW1573 were purchased from the American Type Culture Collection. SK-LU-1, Calu-1, Calu-6 and NCI-H460 were provided by Takashi Takahashi (Nagoya University, Japan). RERF-LC-AD2, LU-65 and LU-99 cells were obtained from the Japanese Cell Research Bank (Osaka, Japan). HCC2108 cells were obtained from Korean Cell Line Bank (Seoul, South Korea). The NCI-H1573 and NCI-H2030 cells were provided from Massachusetts General Hospital Cancer Center. Cells were cultured in RPMI1640 (Invitrogen) with 10% FBS. Characteristic of cell lines used in this study were summarized in Supplementary Table S4. Cells were obtained between 2012 and 2016. Experiments using A549, SW900, LU-65 and HCC2108 cells were done within 6 months from the acquisition of these cells from cell banks. All other cell lines were tested and authenticated by short tandem repeat (STR) analysis with GenePrint 10 System (Promega) by the Japanese Cell Research Bank at the time of submission. Cells were regularly screened for Mycoplasma using a MycoAlert Mycoplasma Detection Kit (Lonza). NVP-BGJ398, GDC-0941, ABT-263, selumetinib, afatinib, and trametinib were obtained from Active Biochem. Compounds were dissolved in DMSO to a final concentration of 10 mmol/l and stored at −20°C when not in use.
Western blot
Lysates were prepared using Cell Lysis Buffer (Cell Signaling). Procedure for Western blotting was as previously described (10). Antibodies used in this study are Supplementary Table S5.
siRNA knockdown
Cells were seeded into 6-well plates at a density of 1–2 × 105 cells/well. Twenty-four hours later, cells were transfected with 20 nM siRNA against FGFR1 (Dharmacon), DUSP6 (Dharmacon), ZEB1 (Dharmacon and Santa Cruz Biotechnology), SPRY4 (Ambion and Dharmacon), or ERBB3 (Invitrogen) or Stealth RNAi-negative control low GC Duplex #3 (Invitrogen) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions.
Overexpression of SPRY4
The SPRY4 expression construct was obtained from GE Dharmacon. Preparation of lentivirus and infections were performed as previously described (10).
Apoptosis analysis
Cells were seeded at approximately 30%–40% confluence in 6 well plates. After overnight incubation, the media was changed and DMSO or the indicated drugs were added. After 72 hours, floating cells in media and adherent, trypsinized cells were collected in a single tube. Cells were pelleted and washed once with PBS. Apoptotic cells were stained with Annexin V using Annexin V : PE Apoptosis Detection Kit I and assayed on an BD Accuri C6 flowcytometer (BD). All experiments were performed in triplicate.
Xenograft mouse studies
Suspension of 5 × 106 cells was injected subcutaneously into the flanks of 6- to 8-week-old male nude mice (Clea, Tokyo, Japan). The care and treatment of experimental animals were in accordance with institutional guidelines. Mice were randomized (n = 8 in LU99 xenograft and 5 in NCI-H23 and NCI-H358 xenograft) once the mean tumor volume reached approximately 230–300 mm3. Drugs were administered once daily by oral gavage. NVP-BGJ398 was dissolved in acetic acid/acetate buffer pH 4.6/PEG300 1:1; Trametinib and Afatinib were dissolved in 7% DMSO, 13% Tween 80, 4% glucose, and HCl equivalent molar concentration to each drug. Mice were monitored daily for body weight and general condition. Tumors were measured twice weekly using calipers, and volume was calculated using the following formula: length × width2 × 0.52. According to institutional guidelines, mice were sacrificed when their tumor volume reached 1,000 mm3.
Patient-derived xenograft (PDX) experiments
For the PDX study, a xenograft model from a 53 years old female with KRAS G12D mutant lung cancer was purchased from the Jackson Laboratory. Female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice at 6–8 weeks of age were engrafted with tumor fragments at passage P4 at Jackson Laboratory. Then, mice were transferred to the animal facility at VCU and randomized once the mean tumor volume reached approximately 300 mm3.
Agilent microarray analysis
RNA was isolated using RNeasy kit (QIAGEN) following the manufacturer’s recommendations. The microarray analysis was performed using SurePrint G3 Human Gene Expression 8x60K v2 Microarray Kit (Agilent technology). The microarray accession number in GEO is GSE79235.
Gene expression analysis of 39 NSCLC cell lines with KRAS mutation
Gene expression data of lung cancer cell lines was obtained from the CCLE (27). Out of 187 lung cancer cell lines, 39 NSCLC cell lines with KRAS mutation were extracted for further analysis based on the information obtained from CCLE (CCLE_sample_info_file). Unsupervised hierarchical clustering was performed on 39 KRAS mutant NSCLC cell lines using Cluster 3.0 program (54) to perform clustering and JAVA TreeView program (55) to visualize the results. A total of 2635 genes were used for the analysis which demonstrated more than a 5-fold change among cell lines with a median expression value of 6 or more. Three distinct sub-clusters were extracted, which included the marker genes ERBB3, CDH1, FGFR1, and VIM.
Gene expression analysis of patients with KRAS mutant lung adenocarcinoma
Messenger RNA profiling for 230 resected lung adenocarcinomas were obtained from TCGA (34). Out of 230 adenocarcinoma samples, 75 samples with KRAS mutation were extracted for further analysis based on the clinical information. We performed unsupervised hierarchical clustering of 75 adenocarcinoma samples with KRAS mutation based on the 28 genes, which were listed as EMT related genes by Kalluri and Weinberg (35). Two additional gene expression data, FGFR1 and ERBB3, were extracted and ordered based on the order of hierarchical clustering.
Statistics
Linear regression analyses and Student’s t tests were performed where indicated. Linear regression analyses were performed using r software and the slope was considered significantly non-zero when p < 0.05. For Student’s t-tests, populations were considered significantly different at p < 0.05. All statistical tests were two-sided.
Animal study approval
Mouse xenograft experiments were approved by the ethical committee on the Institute for Experimental Animals, Kanazawa University Advanced Science Research Center. The patient derived xenograft experiment was approved by the Virginia Commonwealth University Institutional Animal Care and Use Committee (IACUC protocol #AD10001048).
Supplementary Material
Statement of significance.
Adaptive resistance to MEKi is driven by RTKs specific to the differentiation state of the KRAS mutant NSCLC. In mesenchymal-like KRAS mutant NSCLC, FGFR1 is highly expressed, and MEK inhibition relieves feedback suppression of FGFR1, resulting in re-activation of ERK; suppression of ERK by MEKi/FGFRi combination results in tumor shrinkage.
Acknowledgments
Financial support: This work is supported by Grants-in-Aid for Cancer Research 26830105 (to H.E.) and 21390256 (to S.Y.), Scientific Research on Innovative Areas “Integrative Research on Cancer Microenvironment Network” (22112010A01) (to S.Y.), and Grant-in-Aid for Project for Development of Innovative Research on Cancer Therapeutics (P-Direct) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (to S.Y.), and AstraZeneca R&D Grant Award (to H.E.), and Research grant for developing innovative cancer chemotherapy, Kobayashi Foundation for Cancer Research (to H.E.). ACF is supported by a NCI Transition Career Development Award (K22CA175276), an American Lung Association Lung Cancer Discovery Award and the George and Lavinia Blick Research Fund.
Footnotes
Conflict of interest statement: H. Ebi has received a commercial research grant from AstraZeneca.
References
- 1.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–74. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–81. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–51. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Downward J. RAS Synthetic Lethal Screens Revisited: Still Seeking the Elusive Prize? Clin Cancer Res. 2015;21:1802–9. doi: 10.1158/1078-0432.CCR-14-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Infante JR, Fecher LA, Falchook GS, Nallapareddy S, Gordon MS, Becerra C, et al. Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: a phase 1 dose-escalation trial. Lancet Oncol. 2012;13:773–81. doi: 10.1016/S1470-2045(12)70270-X. [DOI] [PubMed] [Google Scholar]
- 6.Carter CA, Rajan A, Szabo E, Khozin S, Thomas A, Brzezniak CE, et al. Two parallel randomized phase II studies of selumetinib (S) and erlotinib (E) in advanced non-small cell lung cancer selected by KRAS mutations. J Clin Oncol. 2013;31:8026. [Google Scholar]
- 7.Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M, et al. Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells. Sci Signal. 2011;4:ra17. doi: 10.1126/scisignal.2001752. [DOI] [PubMed] [Google Scholar]
- 8.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–21. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer. 2015;15:577–92. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 10.Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest. 2011;121:4311–21. doi: 10.1172/JCI57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molina-Arcas M, Hancock DC, Sheridan C, Kumar MS, Downward J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov. 2013;3:548–63. doi: 10.1158/2159-8290.CD-12-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 13.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–77. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 16.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–51. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–62. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 18.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bremnes RM, Veve R, Gabrielson E, Hirsch FR, Baron A, Bemis L, et al. High-throughput tissue microarray analysis used to evaluate biology and prognostic significance of the E-cadherin pathway in non-small-cell lung cancer. J Clin Oncol. 2002;20:2417–28. doi: 10.1200/JCO.2002.08.159. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs BC, Fujii T, Dorfman JD, Goodwin JM, Zhu AX, Lanuti M, et al. Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res. 2008;68:2391–9. doi: 10.1158/0008-5472.CAN-07-2460. [DOI] [PubMed] [Google Scholar]
- 21.Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatzivassiliou G, Haling JR, Chen H, Song K, Price S, Heald R, et al. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature. 2013;501:232–6. doi: 10.1038/nature12441. [DOI] [PubMed] [Google Scholar]
- 23.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–21. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Salt MB, Bandyopadhyay S, McCormick F. Epithelial-to-mesenchymal transition rewires the molecular path to PI3K-dependent proliferation. Cancer Discov. 2014;4:186–99. doi: 10.1158/2159-8290.CD-13-0520. [DOI] [PubMed] [Google Scholar]
- 27.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guagnano V, Kauffmann A, Wohrle S, Stamm C, Ito M, Barys L, et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2012;2:1118–33. doi: 10.1158/2159-8290.CD-12-0210. [DOI] [PubMed] [Google Scholar]
- 29.Pratilas CA, Solit DB. Targeting the mitogen-activated protein kinase pathway: physiological feedback and drug response. Clin Cancer Res. 2010;16:3329–34. doi: 10.1158/1078-0432.CCR-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki A, Taketomi T, Wakioka T, Kato R, Yoshimura A. Identification of a dominant negative mutant of Sprouty that potentiates fibroblast growth factor- but not epidermal growth factor-induced ERK activation. J Biol Chem. 2001;276:36804–8. doi: 10.1074/jbc.C100386200. [DOI] [PubMed] [Google Scholar]
- 31.Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4:850–8. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- 32.Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet. 2015;47:250–6. doi: 10.1038/ng.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hata AN, Yeo A, Faber AC, Lifshits E, Chen Z, Cheng KA, et al. Failure to induce apoptosis via BCL-2 family proteins underlies lack of efficacy of combined MEK and PI3K inhibitors for KRAS-mutant lung cancers. Cancer Res. 2014;74:3146–56. doi: 10.1158/0008-5472.CAN-13-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 37.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–35. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell. 2012;22:668–82. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tricker EM, Xu C, Uddin S, Capelletti M, Ercan D, Ogino A, et al. Combined EGFR/MEK Inhibition Prevents the Emergence of Resistance in EGFR-Mutant Lung Cancer. Cancer Discov. 2015;5:960–71. doi: 10.1158/2159-8290.CD-15-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hrustanovic G, Olivas V, Pazarentzos E, Tulpule A, Asthana S, Blakely CM, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med. 2015;21:1038–47. doi: 10.1038/nm.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F, Huynh H, Li X, Ruddy DA, Wang Y, Ong R, et al. FGFR-Mediated Reactivation of MAPK Signaling Attenuates Antitumor Effects of Imatinib in Gastrointestinal Stromal Tumors. Cancer Discov. 2015;5:438–51. doi: 10.1158/2159-8290.CD-14-0763. [DOI] [PubMed] [Google Scholar]
- 42.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–72. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mason JM, Morrison DJ, Basson MA, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16:45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Ding W, Bellusci S, Shi W, Warburton D. Genomic structure and promoter characterization of the human Sprouty4 gene, a novel regulator of lung morphogenesis. Am J Physiol Lung Cell Mol Physiol. 2004;287:L52–9. doi: 10.1152/ajplung.00430.2003. [DOI] [PubMed] [Google Scholar]
- 45.Abel EV, Basile KJ, Kugel CH, 3rd, Witkiewicz AK, Le K, Amaravadi RK, et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. J Clin Invest. 2013;123:2155–68. doi: 10.1172/JCI65780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montero-Conde C, Ruiz-Llorente S, Dominguez JM, Knauf JA, Viale A, Sherman EJ, et al. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov. 2013;3:520–33. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483:613–7. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17:547–59. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–39. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–84. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–97. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 53.Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, et al. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell. 2007;12:559–71. doi: 10.1016/j.ccr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 54.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–4. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 55.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.