Abstract
Adult tissues replace lost cells via pools of stem cells. However, the mechanisms of cell self-renewal, commitment, and functional integration into the tissue remain unsolved. Using imaging techniques in live mice, we captured the lifetime of individual cells in the ear and paw epidermis. Our data suggest that epidermal stem cells have equal potential to either divide or directly differentiate. Tracking stem cells over multiple generations reveals that cell behavior is not coordinated between generations. However, sibling cell fate and lifetimes are coupled. We did not observe regulated asymmetric cell divisions. Lastly, we demonstrated that differentiating stem cells integrate into preexisting ordered spatial units of the epidermis. This study elucidates how a tissue is maintained by both temporal and spatial coordination of stem cell behaviors.
Tissue homeostasis requires the ability to replace damaged or lost cells while maintaining tissue structure and function. A model for studying this process is the mouse adult interfollicular epidermis (IFE), where organized layers of progressively differentiated epithelial cells form a barrier from which suprabasal cells are continuously shed and replenished by an underlying proliferative basal layer (1–3). Understanding how basal stem cell proliferation and terminal differentiation remain balanced in homeostasis is a central question in both epithelial and stem cell biology.
Initial models of epidermal maintenance recognized the three-dimensional organization of discrete columns, called epidermal proliferative units (EPUs), which are defined by the perimeter of the most external, terminally differentiated cells (4–6). An important implication of the EPU model is that each unit is autonomously maintained by an asymmetrically dividing, basally located stem cell, with slow-cycling characteristics (7–9). Recent studies support the presence of slow-cycling stem cells in mouse epidermis (10, 11). However, long-term lineage-tracing studies show that basal clones do not strictly adhere to the columnar borders of EPUs and support a model based on a single stem cell population that makes stochastic fate choices, while still relying on mostly (60 to 84%) asymmetric divisions to generate one stem cell and one terminally differentiated cell (12–16). These studies provide critical insights into epidermal homeostasis but remain disconnected and don’t explain how individual stem cells and their progeny are integrated into the existing structure of a tissue.
A major challenge in elucidating cell fate has been the inability to resolve individual cell fate choices within clones. Individual cell behaviors have been indirectly inferred from time series of fixed clonal samples (17). Therefore, we developed an in vivo pulse/chase system for single-cell genetic label retention to continuously track entire lineages across multiple generations and capture the fate of individual basal cells within them (18) (Fig. 1A and fig. S1A). For that, we acquired serial optical sections of the epidermis from the same live adult mice at successive time points and captured the differentiation state of single labeled cells by position and cellular morphology within the entire volume of the IFE (fig. S1B) (19–22). To distinguish between region-specific characteristics and more general epidermal principles, we performed our lineage tracing in both ear and plantar epidermis. Cells that committed to differentiation were scored by their departure from the basal layer and their gradual movement toward the surface of the skin, which was irreversible in all cases (fig. S1C). Cell divisions in the basal layer generated two daughter cells that remained within the basal layer upon division (fig. S1D).
Fig. 1. Subclonal lineage tracing of basal epidermal cells.
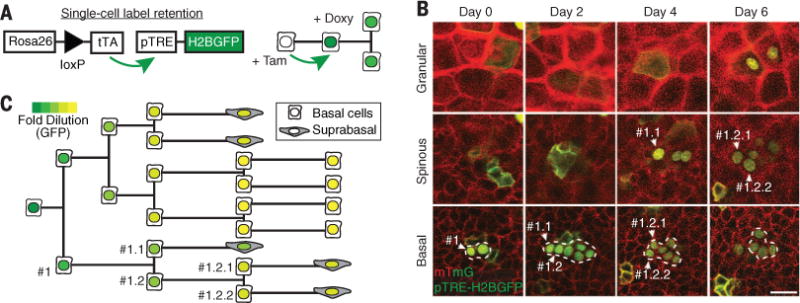
(A) Experimental approach for epidermal fate tracking by single-cell label retention. After clonal induction with tamoxifen, K14CreER;R26flox-stop-tTA/mTmG; pTREH2BGFP mice were treated with doxycycline to evaluate H2BGFP label retention. (B) The fate and subsequent behavior of identified cells were determined by live imaging at 2-day intervals. The epidermis is composed of cellular layers starting from the most external, cornified layer, then moving inward to the granular, spinous, and finally basal layer. The dashed outlines indicate the boundaries of the traced clone. (B) and (C) Representative time sequence of a single-cell label retention experiment, showing the resulting lineage tree (C), with individual cell fate choices identified through label dilution within the same clone. Scale bar, 25 μm.
Analysis of division and differentiation events in clonal lineage trees provided direct access to lifetimes and fate choices of individual basal cells, and revealed fate correlations that could not be addressed from static clonal analysis (Fig. 1, B and C, and fig. S2A). We tested two key hypotheses: First, we asked whether the basal layer is maintained through a proliferative hierarchy by a small population of stem cells (10, 11); if so, mother and daughter cell fates should be correlated, because only stem cells should give rise to daughter stem cells. We performed this multigenerational analysis in the ear epidermis and detected no mother-daughter bias in fate choice [supplementary theory (ST) S5] or in their lifetimes (Pearson correlation R = −0.11, P = 0.2). Second, we tested whether asymmetric fate divisions are the main mode of self-renewal, as widely suggested from static lineage tracing (10, 11, 15, 23). Asymmetric divisions should result in anticorrelated sister cell fates, but we found that sister cell fates were either independent (ear) or positively correlated (paw). In both tissues, we found that sister cells had strongly correlated lifetimes (Fig. 2, A and B, and ST S4). Such sibling correlations could be indicative of coupled activities due to spatial co-localization or co-inheritance.
Fig. 2. Basal stem cells make stochastic fate choices that are temporally coordinated.
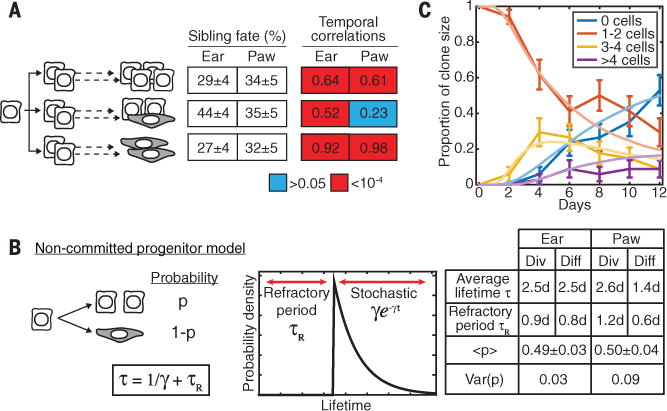
(A) The proportion of divisions leading to symmetric and asymmetric fates, and the magnitude and significance of sister cell lifetime correlations, measured directly from lineage trees (n = 136 divisions across n = 40 trees in the ear, and n =101 divisions across n = 92 trees in the paw). Color shows the statistical significance of correlations: P > 0.05 (blue), P < 10−4 (red). (B) A stochastic model of cell fate, with each cell dividing or directly differentiating after a minimum refractory period, with a fluctuating division probability P balanced at 50% in homeostasis. Spatial or lineage-coupled fluctuations in P between sister cells, measured by the variance in P, lead to correlated sister cell fates. τ, average cell lifetime, γ, stochastic division/differentiation rate after a refractory period. The model is mathematically defined in ST S3. (C) A fit of the model to the distribution of clone sizes (basal cells per clone) over time (n = 40 clones); error bars, SEM.
These results suggest a simple model for stem cell fate without hierarchy or division asymmetry (23) (Fig. 2, B and C; figs. S2, B to E, and S3; and STs S2 and S3). This model excludes fate asymmetry, does not imply fate commitment at birth, and demonstrates the temporal and fate coordination of adult sister stem cell behaviors in vivo.
To date, our work, along with the field in general, has relied on genetic approaches based on Crerecombinase lineage tracing, which could potentially introduce bias (10, 11, 24). We therefore used two additional independent lineage-tracing approaches to track single cells and populations within the epidermis, respectively. First, we engineered a transgenic mouse that expresses a light-activatable fluorescent reporter (26) in the epidermis (fig. S4 and movie S1). Using two-photon illumination to randomly label and track individual basal cells, we found that basal cell behavioral kinetics are comparable to those shown by our Cre lineage-tracing analysis, supporting our proposed stem cell model (Fig. 3, A and B, and fig. S5). Furthermore, we labeled geometrically defined regions within different epidermal layers to track the uniformity of population cell fate over time (movie S2). By these means, we found a global basal layer turnover as well as rapid progression of committed cells through the suprabasal layers within only a few days (Fig. 3C). Second, we used a previously developed tetracycline-inducible H2B-GFP (GFP, green fluorescent protein) label retention system (10, 11, 18) to track cells for up to 2 weeks in the ear epidermis and found global dilution of the H2B-GFP signal already by 1 week. No cells (n = 11,370) retained their GFP label, even after correcting for background signal decay, suggesting that all basal cells cycle at similar rates (fig. S6, A to F). Collectively, our work has demonstrated that the basal layer of the epidermis is composed of a single equipotent stem cell population, through the utilization of three powerful and complementary label retention approaches in live mice.
Fig. 3. Unbiased epidermal fate tracking by single-cell photolabeling.
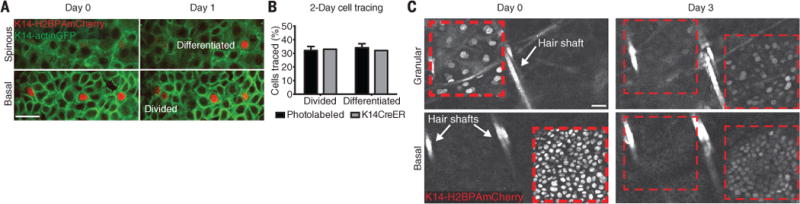
(A) Representative examples of cell division and differentiation fates. (B) Quantification of cell division and differentiation events. (C) Representative time sequence of a region labeled with a photoactivatable reporter. At day 0, two adjacent square areas were scanned to activate the H2BPAmCherry reporter in the granular and basal layer, respectively. Scale bars, 25 μm.
We next sought to understand how these seemingly random cell fate decisions in the basal layer contribute to organized suprabasal differentiated layers. Previous work has demonstrated that cells in the basal layer are not constrained to column boundaries, and it has been proposed that cells could differentiate through self-assembling processes (23, 27, 28). To interrogate the mechanism by which cells transit through suprabasal layers, we used a dual membrane fluorescent reporter to differentially label neighboring cells in the ear epidermis (fig. S1). We find that neighboring cells independently transit through the differentiated epidermal layers before they are shed from the skin (Fig. 4A). This suggests that within layers, the lateral connections between cells are dynamic during differentiation. Analysis of cell borders showed that the global organization in the terminally differentiated granular layer remained relatively unchanged in the short term (fig. S8A).
Fig. 4. Basal cells transit into preexisting epidermal differentiation units.
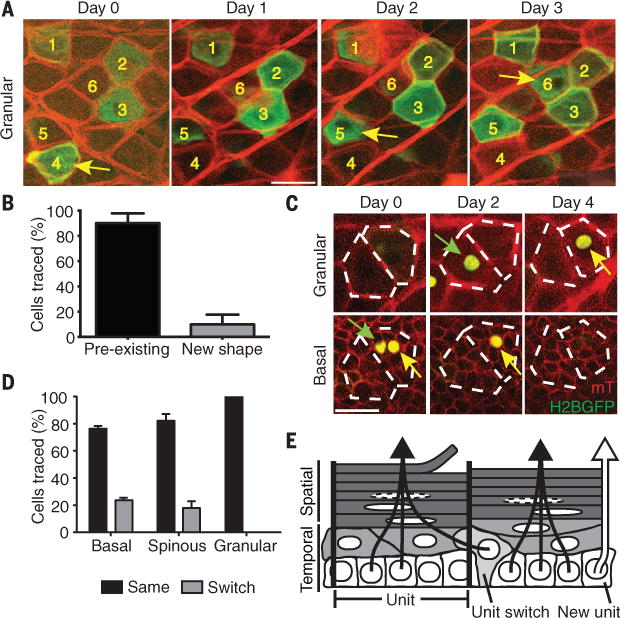
(A) Optical section of a single plane of the top granular layer of the epidermis, taken over 3 days, depicting individually labeled differentiating cells as they transit through at different time points. The majority of differentiating cells arrive at the same space as their predecessors (yellow arrows), as indicated by the unchanging cell boundaries. (B) Quantification of epidermal differentiation behaviors (n = 40 cells); error bars represent SD. (C) Representative examples of differentiating epidermal cells switching or integrating into existing units (green and yellow arrows, respectively). (D) Quantification of the frequency of unit switching for each epidermal layer (n = 40 cells); Error bars represent SD. (E) Schematic for epidermal homeostasis. Basal stem cells stochastically commit to differentiation and transit through the suprabasal layers by predominately using existing columnar units. Scale bars, 25 μm.
To reconcile the highly dynamic reorganization of the basal layer with the seemingly unchanged architecture of the terminally differentiated layers, we examined the trajectory of individual cells from the basal to the cornified layers. We find that the majority of committed cells align into vertical columns, giving rise to the structures that inspired the EPU hypothesis (29) (figs. S7 and S8). Therefore, we propose that such columns are epidermal differentiation units (EDUs), rather than EPUs. These columns persist over time as newly differentiated cells move upward to recycle the same space occupied by preceding cells, but they are not entirely stable because a minority of cells (~10%) do not funnel into preexisting EDUs but instead form new units as they transit through the layers of the epidermis (Fig. 4, C and D). Furthermore, our lineage-tracing data show that although cells inhabiting the basal and spinous layers have the flexibility to switch to neighboring EDUs as cells arrive in the granular layer, their fate is vertically fixed into a supply chain for the cornified layers above (Fig. 4E). Analysis of the distribution of residence times of cells in the suprabasal layers shows that cell maturation in the spinous layer spans the same typical time for almost all cells, and their subsequent exit from the granular layer appears to be stochastic (ST S2). This suggests that after departure from the basal layer, the granular layer spatially coordinates the transition to cornified tissue by acting as a buffer zone. Furthermore, the identification of a small percentage of cells that are capable of creating new differentiation paths suggests a mechanism to confer flexibility for epidermal remodeling over time (Fig. 4E).
These results support a simple model of epidermal maintenance proposed over 50 years ago (18, 24), in which basal cells are born as uncommitted stem cells, with an equal chance to ultimately proliferate or differentiate. Sibling stem cells coordinate fate commitment and temporal execution of their behaviors. Finally, as cells depart from the basal layer, they funnel predominantly into metastable EDUs. Taken together, this study demonstrates how spatiotemporal coordination in both the proliferative and differentiated layers sustains epidermal maintenance and function. This provides a foundation for future work to investigate the largely unknown regulatory mechanisms that coordinate cell-cell interactions on a tissue scale, during homeostasis. Furthermore, such progressive acquisition of cell fate could allow for cells in the epidermis to remain flexible in response to environmental demands, as was recently suggested from work on human keratinocytes in vitro (30), and therefore has potential relevance to pathological states and regeneration.
Acknowledgments
We thank E. Fuchs for K14-actinGFP mice; V. Verkhusha for the PAmCherry construct; and M. Theo, S. Ghazizadeh, and P. Jones for critical manuscript feedback. This work was supported by The New York Stem Cell Foundation; the Edward Mallinckrodt Jr. Foundation; the National Institute of Arthritis and Musculoskeletal and Skin Disease (NIAMS), grants NIH-5R01AR063663-04 and NIH-1R01AR067755-01A1; and the NIH Predoctoral Program in Cellular and Molecular Biology (K.R.M. and S.B., grant T32GM007223). K.R.M is an NSF Graduate Research Fellow. S.P. is supported by CT Stem Cell grant 14-SCA-YALE-05. P.R. is supported by an American Skin Association Research Scholar Award, was a New York Stem Cell Foundation-Druckenmiller Fellow, and was supported by CT Stem Cell grant 13-SCA-YALE-20. V.G. is a New York Stem Cell Foundation Robertson Investigator. A.M.K. is supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and an Edward Mallinckrodt Jr. Foundation grant. The K14-H2BPAmCherry mouse is available from the Greco lab under a materials transfer agreement with Yale.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/352/6292/1471/suppl/DC1
Materials and Methods
Author Contributions
Supplementary Text
Figs. S1 to S8
Movies S1 and S2
REFERENCES AND NOTES
- 1.Levy V, Lindon C, Harfe BD, Morgan BA. Dev Cell. 2005;9:855–861. doi: 10.1016/j.devcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Christophers E, Laurence EB. Curr Probl Dermatol. 1976;6:87–106. doi: 10.1159/000399007. [DOI] [PubMed] [Google Scholar]
- 3.Ito M, et al. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 4.Potten C. Cell Prolif. 1974;7:77–88. [Google Scholar]
- 5.Ghazizadeh S, Taichman LB. EMBO J. 2001;20:1215–1222. doi: 10.1093/emboj/20.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackenzie IC. J Invest Dermatol. 1997;109:377–383. doi: 10.1111/1523-1747.ep12336255. [DOI] [PubMed] [Google Scholar]
- 7.Potten CS, Morris RJ. J Cell Sci Suppl. 1988;1988(suppl 10):45–62. doi: 10.1242/jcs.1988.supplement_10.4. [DOI] [PubMed] [Google Scholar]
- 8.Ghazizadeh S, Taichman LB. J Invest Dermatol. 2005;124:367–372. doi: 10.1111/j.0022-202X.2004.23599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen UB, Lowell S, Watt FM. Development. 1999;126:2409–2418. doi: 10.1242/dev.126.11.2409. [DOI] [PubMed] [Google Scholar]
- 10.Mascré G, et al. Nature. 2012;489:257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 11.Sada A, et al. Nat Cell Biol. 2016;18:619–631. doi: 10.1038/ncb3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clayton E, et al. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 13.Klein AM, Nakagawa T, Ichikawa R, Yoshida S, Simons BD. Cell Stem Cell. 2010;7:214–224. doi: 10.1016/j.stem.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Doupé DP, et al. Science. 2012;337:1091–1093. doi: 10.1126/science.1218835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim X, et al. Science. 2013;342:1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Füllgrabe A, et al. Stem Cell Rep. 2015;5:843–855. doi: 10.1016/j.stemcr.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doupé DP, Klein AM, Simons BD, Jones PH. Dev Cell. 2010;18:317–323. doi: 10.1016/j.devcel.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 18.Klein AM, Simons BD. Development. 2011;138:3103–3111. doi: 10.1242/dev.060103. [DOI] [PubMed] [Google Scholar]
- 19.Tumbar T, et al. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rompolas P, et al. Nature. 2012;487:496–499. doi: 10.1038/nature11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rompolas P, Mesa KR, Greco V. Nature. 2013;502:513–518. doi: 10.1038/nature12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mesa KR, et al. Nature. 2015;522:94–97. doi: 10.1038/nature14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pineda CM, et al. Nat Protoc. 2015;10:1116–1130. doi: 10.1038/nprot.2015.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marques-Pereira JP, Leblond CP. Am J Anat. 1965;117:73–87. doi: 10.1002/aja.1001170106. [DOI] [PubMed] [Google Scholar]
- 25.Kretzschmar K, Watt FM. Cell. 2012;148:33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Subach FV, et al. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda H, Tanemura M, Imayama S. J Invest Dermatol. 1996;106:312–315. doi: 10.1111/1523-1747.ep12342964. [DOI] [PubMed] [Google Scholar]
- 28.Menton DN. J Invest Dermatol. 1976;66:283–291. doi: 10.1111/1523-1747.ep12482234. [DOI] [PubMed] [Google Scholar]
- 29.Potten CS. J Invest Dermatol. 1975;65:488–500. doi: 10.1111/1523-1747.ep12610194. [DOI] [PubMed] [Google Scholar]
- 30.Roshan A, et al. Nat Cell Biol. 2016;18:145–156. doi: 10.1038/ncb3282. [DOI] [PMC free article] [PubMed] [Google Scholar]


