Abstract
A series of novel 2,5-disubstituted-1,3,4-oxadiazole derivatives were synthesized and screened for their antimicrobial and antioxidant activities. The assay indicated that compounds 3c, 3d, and 3i exhibited comparable antibacterial and antioxidant activity with first-line drugs. The structure activity relationship and molecular docking study of the synthesized compounds are also reported.
Keywords: 1,3,4-Oxadiazole; Antimicrobial; Antioxidant; Molecular docking
Introduction
Worldwide, people are being threatened by more and more microbial infectious diseases. Unfortunately, the incidence of systemic microbial infections has increased relentlessly due to increase of immune compromised hosts [1–3]. As per the results, at present, the major global problem is the more pathogenetic bacterial species resistant to conventional antibiotics that has resulted in either high expenses or failure in the treatment of infectious diseases it may attributed due to emergence of multidrug bacterial and fungal strains [4–6]. On the other hand, reactive oxygen species (ROS) and reactive nitrogen species (RNS) are products of normal cellular metabolism [7]. At higher concentration, these radicals extensively cause oxidative damage to the biomolecules and contribute to the pathogenesis of oxidative stress-related diseases like cancer, atherosclerosis, ageing, and diabetes mellitus [8–10]. A proper equilibrium between the ROS generations in humans and components of defense system is needed by supplementing antioxidants via diet or medicine to prevent diseases related to chronic oxidative damages in tissues and cells. The antioxidants are thus believed to protect against oxidative stress-related diseases [11, 12].
Hence, the urgent need of new molecules to combat microbial infections and antioxidants is immense. To meet the drug demands, common strategies adopted by pharmaceutical companies to supply new drugs to the market include changing the molecular structure of the existing medicines in order to make them more efficient or recover the activity lost due to bacterial resistance mechanisms with minimal side effects. In silico molecular docking is an upcoming tool to investigate promising therapeutics for their effective inclination as able leads toward specific pathologies. The docking analysis furnishes the knowledge of the extent of plausible interaction between the target of interest and the drug under investigation. This in turn helps to procure a primary understanding of the viability of the prodrug under scrutiny. The docking study has been advantageous over the in vitro and in vivo analyses in being faster, safer, and having less infrastructural requirements [13].
1,3,4-Oxadiazole and its derivatives have attracted interest over the past few years as they have been widely used in pharmaceuticals, dyes, photographic materials, agrochemicals, and corrosion inhibition [14, 15]. 1,3,4-Oxadiazole cores have a broad spectrum of biological activities including antimicrobial [16], antimalarial [17], anticancer [18], anticonvulsant [19], antiinflammatory [20], antihepatitis B [21], and anti-HIV-1 [22] activities. On the other hand, widespread use of naphthofuran as a scaffold in organic as well as medicinal chemistry has been adopted due to their diverse syntheses and various biological activities including antioxidant, antibacterial, antitubercular, anti-HIV-1, and anticancer properties [23–25].
The extensive literature survey revealed that the presence of heterocyclic rings at the second and fifth positions of the oxadiazole ring increases the biological profile of these compounds to a greater extent [26–29]. These observations stimulated us to synthesize an array of substituted oxadiazoles by fusing naphthofuran at the fifth position and heterocycles at the second position with the intention of synthesizing some novel derivatives with pharmacological relevance. Thus, herein, we report the design, synthesis, bioactive evaluation, and in silico molecular docking studies of a series of novel 2,5-disubstituted-1,3,4-oxadiazoles containing aliphatic, aromatic, and heterocyclic amides at position 2 and naphthofuran ring at position 5.
Materials and methods
Materials
All the reactions were carried out under inert atmosphere of nitrogen. TLC was performed on precoated aluminum sheets of silica (60F254 nm) and visualized by shortwave UV light at λ 254. Melting points were determined on an EZ-Melt automated melting point apparatus. 1H NMR spectra were recorded at 400 MHz using an internal deuterium lock. Chemical shifts were measured in δ (ppm). LC-MS analyses were performed using ESI/APCI, with an ATLANTIS C18 (50 × 4.6 mm–5 μm) column and a flow rate of 1.2 cm3/min.
Experimental procedures
General procedure for synthesis of 4-[5-(naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl]aniline (2)
Naphthofuran-2-hydrazide 1 (1 equiv.) was taken in a round-bottomed flask and PABA (1.2 equiv.) was added followed by the addition of 3 cm3 POCl3 at 0 °C. The reaction mixture was stirred at room temperature for 10 min, and the reaction mixture was heated at 80 °C for 4 h. The reaction mixture was then quenched with crushed ice and basified with NaHCO3. The resulting solution was filtered, and the residue was washed with saturated sodium bicarbonate solution and cold water. The obtained yellow residue was further recrystallized in ethanol to obtain the titled compound as a yellow solid in 85 % yield. MP: 143–145 °C; IR (KBr, v cm−1): 3270 (NH2), 2958, 2688, 1704, 1581, 1442, 1372, 1079;1H NMR (400 MHz, CDCl3): δ 7.84–7.85 (d, J = 7.68 Hz, 2H, Ar-H), 7.52–7.63 (m, 6H, Ar-H) 6.73 (s, 1H, Ar-H), 6.64–6.65 (d, J = 6.24 Hz, 2H, Ar-H), 4.22 (br s, 2H, NH2); LC-MS: m/z 327 [M + 1].
General procedure for the synthesis of compounds 3a-n
4-[5-(Naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl]aniline intermediate, 2 (1 equivalent) was mixed with dichloromethane (DCM) in a round-bottomed flask. To that, HATU (0.3 equiv.), N,N-diisopropyl ethyl amine (2 equiv.) were added subsequently, and the reaction mixture was stirred at room temperature for about 5–10 min. The carboxylic acids (1.2 equiv.) were added, and the stirring was continuous for about 3–5 h. After the completion of reaction, the reaction mixture was diluted with water and extracted with DCM, dried in sodium sulfate, and distilled under reduced pressure. The obtained crude mixture was purified by column chromatography to obtain the final compounds 3a-n.
N-{4-[5-(Naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl]phenyl}benzamide (3a)
C27H17N3O3, yellow solid, yield 90 %; MP 260–262 °C; IR (KBr, v cm−1): 3321 (amide NH), 2998, 1684 (amide I), 1578 (amide II), 1487, 1247(C-O-C); 1H NMR (400 MHz, CDCl3): δ 8.34 (s, 1H, amide-NH), 7.97–7.86 (m, 5H, Ar-H), 7.69–7.54 (m, 6H, Ar-H), 6.8 (s, 1H, furan-H); 13C NMR: 167.9 (CO), 166.4, 160,3, 158.8, 156.4, 140.2, 136.3, 133.6, 130.4, 129.3, 129.0, 128.6, 127.9, 127.3, 124.3, 110.3, 103.8; MS (LCMS): m/z 432 [M + 1].
4-Chloro-N-{4-[5-(naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl]phenyl}benzamide (3b)
C27H16ClN3O3, pale yellow solid, yield 87 %; MP 267–269 °C; IR (KBr, v cm−1): 3254 (amide NH), 3002, 1670 (amide I), 1574 (amide II), 1442, 1242 (C-O-C); 1H NMR (400 MHz, CDCl3): δ 8.37 (m, 1H, Ar-H), 8.06 (m, 1H, Ar-H), 7.97 (s, 1H, NH), 7.58 (m, 1H, Ar-H), 7.49 (d, 1H, Ar-H), 7.27 (m, 2H, Ar-H), 7.23–7.16 (m, 2H, Ar-H), 6.62 (d, 2H, Ar-H), 6.52 (m, 3H, Ar-H); 13C NMR: 170.4, 168.5, 159.3, 158.4, 140.3, 138.4, 134.2, 132.3, 132.0, 130.0, 129.4, 129.1, 126.8, 123.4, 111.6, 102.2; MS (LCMS): m/z 466.9 [M + 1] and 467.9[M + 2].
4-Cyano-N-(4-(5-(naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl)phenyl)benzamide (3c)
C28H16N4O3, pale yellow solid, yield 85 %; MP 264–267 °C; IR (KBr, v cm−1): 3324 (amide NH), 2987, 1704 (amide I), 1542 (amide II), 1451, 1379, 1231 (C-O-C); 1H NMR (400 MHz, CDCl3): δ 8.43–8.48 (m, 2H, ArH), 8.16–8.21 (m, 2H, ArH), 8.10 (s, 1H, NH), 7.66–7.68 (m, 2H, ArH), 7.51–7.54 (d, 2H, ArH), 7.23–7.35(m, 6H, ArH), 6.03 (1H, furan-H); 13C NMR: 103.5, 113.6, 117.9, CN; 123.5, 123.8, 124.2, 125.3, 128.9, 129.1, 129.4, 129.6, 129.9, 130.3, 130.5, 134.1, 136.3, 139.1, 157.2, 157.3, 158.9, 166.1, 167.1, CO; LC-MS: m/z 457 [M + 1].
N-(4-(5-(naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl)phenyl)-2,4-dinitrobenzamide (3d)
C27H15N5O7, yellow solid, yield 86 %; MP 250–252 °C; IR (KBr, v cm−1): 3339 (amide NH), 2992, 1679 (amide I), 1521 (amide II), 1461, 1374, 1249 (C-O-C); 1H NMR (400 MHz, CDCl3): δ 8.62–8.82 (m, 3H, Ar-H), 8.15 (s, 1H, NH), 7.66–7.68 (m, 2H, Ar-H), 7.21–7.44 (d, 2H, Ar-H), 7.13–7.32(m, 6H, Ar-H), 6.41 (1H, furan-H); 13C NMR: 103.8, 112.7, 116.1, 123.4, 123.9, 126.1, 127.8, 129.7, 129.8, 129.9, 130.1, 130.8, 130.9, 132.8, 135.7, 140.1, 152.9, 156.4, 160.2, 166.2, 168.4, CO; LC-MS: m/z 521[M + 1].
4-Methyl-N-(4-(5-(naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl)phenyl)benzamide (3e)
C28H19N3O3, yellow solid, yield 93 %; MP 263–265 °C; IR (KBr, v cm−1): 3308 (amide NH),3048, 2992, 1642 (amide I), 1530 (amide II), 1449, 1375 (C-O-C); 1H NMR (400 MHz, CDCl3): δ 8.82 (s, 1H, NH), 8.16–8.21 (m, 2H, Ar-H), 8.01–8.05 (m, 2H, Ar-H), 7.44–7.48 (m, 2H, Ar-H), 7.14–7.15 (d, J = 7.88 Hz, 2H, Ar-H), 7.03–7.11 (m, 6H, Ar-H), 6.22 (1H, furan-H), 2.55 (s, 3H, CH3); 13C NMR: 25.6, CH3; 101.5, 112.8, 121.7, 121.9, 122.4, 123.4, 127.7, 128.4, 128.4, 128.6, 128.9, 129.2, 129.5, 133.4, 135.8, 138.4, 156.3, 156.6, 157.9, 165.3, 166.1, CO; LC-MS: 445 [M + 1].
5-Methyl-N-(4-(5-(naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl)phenyl)thiophene-2-carboxamide (3f)
C26H17N3O3S, white solid, yield 90 %; MP 266–268 °C; IR (KBr, v cm−1): 3367 (amide NH), 3124, 2974, 1678 (amide I), 1587 (amide II), 1407, 1276 (C-O-C); 1H NMR (400 MHz, CDCl3): δ 7.96 (s, 1H, NH), 7.91–7.98 (m, 2H, Ar-H), 7.62–7.84 (m, 2H, Ar-H), 7.43–7.56 (m, 6H, Ar-H), 7.57–7.55 (d, J = 6.5 Hz, 2H, thiophene-H), 6.92 (1H, furan-H), 2.71 (s, 3H, CH3); 13C NMR: δ 16.4, CH3; 103.7, 113.6, 123.1, 124.3, 128.5, 129.3, 130.9, 131.1, 131.4, 131.6, 131.9, 132.3, 132.5, 134.1, 136.3, 139.1, 157.2, 157.3, 158.9, 166.1, 167.1, CO; LC-MS: m/z 452[M + 1].
5-Methyl-N-(4-(5-(naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl)phenyl)furan-2-carboxamide (3g)
C26H17N3O4, off white solid, yield 95 %; MP 258–260 °C; IR (KBr, v cm−1): 3330 (amide NH), 3057, 2997, 1685 (amide I), 1521 (amide II), 1487, 1246 (C-O-C); 1H NMR (400 MHz, CDCl3): δ 8.04 (s, 1H, NH), 7.86–7.79 (m, 2H, Ar-H), 7.76–7.71 (m, 2H, Ar-H), 7.62–7.57 (m, 6H, Ar-H), 7.53–7.51 (d, J = 7.20 Hz, 2H, furan-H), 6.95 (1H, benzofuran-H), 2.76 (s, 3H, CH3);13C NMR: δ 17.2, CH3; 104.7, 114.8, 123.7, 124.8, 128.8, 130.1, 131.4, 131.6, 131.9, 132.1, 132.4, 132.8, 133.1, 135.4, 136.8, 139.6, 157.7, 157.9, 159.2, 166.5, 167.4, CO;LC-MS: m/z 436 [M + 1].
5-Chloro-N-(4-(5-(naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl)phenyl)thiophene-2-carboxamide (3h)
C25H14ClN3O3S, white solid, yield 94 %; MP 274–276 °C; IR (KBr, v cm−1): 3341 (amide NH), 3124, 2972, 1671 (amide I), 1567 (amide II), 1449, 1267 (C-O-C), 1095; 1H NMR (400 MHz, CDCl3): δ 8.02 (s, 1H, NH), 7.57–7.50 (m, 2H, Ar-H), 7.48–7.42 (m, 2H, Ar-H), 7.33–7.18 (m, 6H, Ar-H), 7.12–7.04 (m, 2H, thiophene-H), 6.84 (1H, Ar-H); 13C NMR: δ 102.1, 112.6, 122.4, 123.3, 127.6, 128.8, 129.9, 130.3, 130.6, 131.3, 131.6, 132.1, 132.4, 133.6, 135.3, 138.5, 156.8, 157.3, 157.9, 165.3, 167.1, CO; HLC-MS: m/z 472 [M + 1] and 473[M + 2].
5-Methyl-N-(4-(5-(naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl)phenyl)pyrazine-2 carboxamide (3i)
C26H17N5O3, yellow solid, yield 80 %; MP 265–267 °C; IR (KBr, v cm−1): 3303 (amide NH), 3047, 2931, 1687 (amide I), 1514 (amide II), 1450, 1239 (C-O-C); 1H NMR (400 MHz, CDCl3): δ 8.50–8.46 (m, 2H, pyrazine-H), 8.16 (s, 1H, NH), 7.95–7.91 (m, 2H, Ar-H), 7.73–7.72 (m,2H, Ar-H), 7.41–7.32 (m, 6H, Ar-H), 6.52 (1H, furan-H), 2.64 (s, 3H, CH3); 13C NMR: δ 26.2, CH3; 104.1, 113.2, 122.6, 122.8, 123.1, 124.7, 128.6, 128.8, 128.9, 129.3, 130.1, 137.1, 142.8, 143.7, 145.1, 156.6, 156.8, 156.9, 158.2, 163.1, 166.1, CO; LC-MS: m/z 448 [M + 1].
N-(4-(5-(Naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl)phenyl)benzofuran-2-carboxamide (3j)
C29H17N3O4, yellow solid, yield 81 %; MP 242–243 °C; IR (KBr, v cm−1): 3241 (amide NH), 3121, 2987, 1680 (amide I), 1578 (amide II), 1468, 1279 (C-O-C); 1H NMR (400 MHz, CDCl3): δ 8.18 (s, 1H, NH), 7.85–7.82 (m,2H, Ar-H), 7.78–7.75 (m, 2H, Ar-H), 7.57–7.42 (m, 5H, benzofuran-H), 7.40–7.33 (m, 6H, naphthofuran-H), 6.61 (s, 1H, furan-H); 13C NMR: δ 103.4, 112.8, 122.4, 122.9, 123.7, 124.5, 127.7, 127.9, 128.7, 129.8, 130.1,132.5, 132.5, 133.5, 133.8, 137.8, 141.9, 142.4, 144.8, 156.8, 156.9, 157.2, 157.8, 161.9, 167.1, CO; LC-MS: m/z 472 [M + 1].
2,2,2-Trichloro-N-{4-[5-(naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl]phenyl}acetamide (3k)
C22H12Cl3N3O3, yellow solid, yield 82 %; MP 205–206 °C; IR (KBr, v cm−1): 3287 (amide NH), 3018, 2988, 1685 (amide I), 1527 (amide II), 1437, 1279 (C-O-C), 806; 1H NMR (400 MHz, CDCl3): δ 7.98 (s, 1H, NH), 7.58–7.51 (m, 2H, Ar-H); 7.49–7.47 (m, 2H, Ar-H), 7.46–7.39 (m, 6H, ArH), 7.1 (s, 1H, furan-H); 13C NMR: δ 93.8 CCl3, 102.8, 112.8, 122.9, 123.1, 123.3, 124.7, 130.1, 133.4, 135.5, 136.7, 156.6, 158.5, 165.6, 166.8, CO; LC-MS: m/z 473 [M + 1] and 474.5 [M + 2].
4,4,4-Trifluoro-N-{4-[5-(naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl]phenyl}butanamide (3l)
C24H16N3F3O3, yellow solid, yield 72 %; MP 198–199 °C; IR (KBr, v cm−1): 3342 (amide NH), 3058, 2971, 1679 (amide I), 1546 (amide II), 1443, 1269 (C-O-C), 1054; 1H NMR (400 MHz, CDCl3): δ 8.27 (s, 1H, NH), 7.49–7.41 (m, 2H, Ar-H); 7.38–7.40 (m, 6H, Ar-H),7.31–7.27 (m, 2H, Ar-H), 6.8 (s, 1H, furan-H), 2.42 (m, 3H), 2.2 (m, 2H); 13C NMR: δ 18.6, 38.9, 103.4, 113.7, 123.4, 123.7, 123.9, 124.2, 128.9 CF3, 131.2, 133.1, 134.3, 135.4, 156.1, 157.3, 166.1, 166.2, CO; LC-MS: m/z 452 [M + 1].
N-{4-[5-(Naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl]phenyl}tetradecanamide (3m)
C34H39N3O3, yellow solid, yield 71 %; MP 172–173 °C; IR (KBr, v cm−1): 3289 (amide NH), 3127, 3087, 2971, 1681 (amide I), 1596 (amide II), 1471, 1245 (C-O-C); 1H NMR (400 MHz, CDCl3): δ 8.20 (s, 1H, NH), 7.47–7.46 (m, 2H, Ar-H), 7.43–7.41 (d, 2H, ArH), 7.42–7.34 (m, 6H, Ar-H), 7.5 (s, 1H, furan-H), 2.43 (t, 2H), 1.75 (m, 2H), 1.40 (m, 18H), 1.38 (m, 2H), 1.2 (t, 3H); 13C NMR: δ 15.8, 23.4, 30.5, 37.9, 38.8, 104.7, 112.9, 122.3, 123.4, 123.8, 124.7, 131.6, 133.8, 133.3, 135.7, 156.2, 166.1, 167.8, CO; LC-MS: m/z 538 [M + 1].
N-{4-[5-(Naphtho[2,1-b]furan-2-yl)-1,3,4-oxadiazol-2-yl]phenyl}stearamide (3n)
C38H47N3O3, yellow solid, yield 70 %; MP 170–171 °C; IR (KBr, ν cm−1): 3365 (amide NH), 3142, 3006, 2994, 1660 (amide I), 1554 (amide II), 1456, 1236 (C-O-C); 1H NMR (400 MHz, CDCl3): δ 8.013 (s, 1H, NH), 7.46–7.45 (m, 2H, Ar-H), 7.42–7.40 (d, 2H, Ar-H), 7.42–7.34 (m, 6H, Ar-H), 7.02 (s, 1H, furan-H), 8.21 (s, 1H, NH), 2.42 (t, 2H), 1.77 (m, 2H), 1.38 (m, 26H), 1.34 (m, 2H), 0.98 (t, 3H); 13C NMR: δ 15.8, 23.4, 30.5, 37.9, 38.8, 104.7, 112.9, 122.3, 123.4, 123.8, 124.7, 131.6, 133.8, 133.3, 135.7, 156.2, 166.1, 167.8, CO; LC-MS: m/z 594 [M + 1].
Biological activity
In vitro antibacterial and antifungal activity
Antimicrobial activity of the synthesized compounds was tested against four bacterial strains and four fungal stains using agar well diffusion method [30]. All bacterial strains were maintained on nutrient agar medium at ±37 °C, and fungi strains were maintained on potato dextrose agar (PDA) at ±25 °C. The test compounds were dissolved in DMSO to get a concentration of 1 and 0.5 mg/cm3. Of this sample, 100 μdm3 was loaded into the wells of agar plates directly. Plates inoculated with the bacteria were incubated at 37 °C for 24 h, and the fungal culture was incubated at 25 °C for 72 h. All determinations were done in triplicates. Ciprofloxacin (0.5 mg/cm3) and fluconazole (0.5 mg/cm3) were used as standard drugs for antibacterial and antifungal activities, respectively. After the incubation period, the inhibition zone at which the microorganism growth was inhibited was measured in millimeters. MIC was performed by serial broth-dilution method at different concentrations of 0.20, 0.40, 0.60, 0.80, and 1 mg/cm3.
In silico molecular docking studies
The structure of synthesized molecules and the standards were drawn in Chem Bio Draw tool (Chem Bio Office Ultra 14.0 suite) assigned with proper 2D orientation, and structure of each was checked for structural drawing error. Energy of each molecule was minimized using ChemBio3D (Chem Bio Office Ultra 14.0 suite). The energy minimized ligand molecules were then used as input for Auto Dock Vina, in order to carry out the docking simulation [31]. The protein data bank (PDB) coordinate file with the name ‘2VF5.pdb’ was used as receptor molecule. All the water molecules were removed from the receptor, and SPDBV Deep view was used to automatically rebuild the missing side chains in receptor. The Graphical User Interface program “MGL Tools” was used to set the grid box for docking simulations. The grid was set so that it surround the region of interest in the macromolecule.
The grid box volume was set to 20, 20, and 20 Å for x, y, and z dimensions, respectively, and the grid center was set to 29.663, 15.499, and −2.754 for x, y, and z center, respectively, which covered all the ten amino acid residues in the considered active pocket. Auto Grid 4.0 Program, supplied with Auto Dock 4.0, was used to produce grid maps [32]. The docking algorithm provided with Auto Dock Vina was used to search for the best docked conformation between ligand and protein. During the docking process, a maximum of ten conformers were considered for each ligand. All the Auto Dock docking runs were performed in Corei7 Intel processor CPU with 8 GB DDR3l RAM. Auto Dock Vina was compiled and run under Windows 8.0 professional operating system. LigPlot+ [33] and PyMol were used to deduce the pictorial representation of interaction between the ligands and the target protein.
Antioxidant activity
1,1-Diphenyl-2-picrylhydrazyl (DPPH) radical scavenging activity
All the synthesized compounds were screened for free radical scavenging activity by DPPH method [34]. In brief, compounds in methanol at different concentrations (20–100 μg/cm3) were added to each test tube and volume was made up to 4 cm3 using methanol. To this, 3 cm3 of 0.004 % DPPH in methanol was added and the mixtures were incubated at room temperature under dark condition for 30 min. Absorbance of the solution was measured at wavelength 517 nm. Radical scavenging activity was calculated using the formula
where Acontrol is the absorbance of the control and Atest is the absorbance of the test sample. The DPPH radical scavenging activity of butylatedhydroxytoluene (BHT) was also assayed for comparison. Test was performed in triplicate, and the results were averaged.
Metal chelating activity
The chelating activity of ferrous ions of the synthesized compounds and standard (EDTA) was estimated by reported procedure [35]. Briefly, 3 cm3 of sample solution at different concentrations (20–100 μg/cm3) was taken, and 0.05 cm3 of 2 mM FeCl2 was added. The reaction was initiated by adding 0.2 cm3 5 mM ferrozine, mixed vigorously, and incubated at room temperature for 10 min. Absorbance of the solution was measured at wavelength 562 nm. The percentage of inhibition of ferrozine-Fe2+ complex formation was calculated by using the formula:
where Acontrol is the absorbance of the control sample and Atest is the absorbance of the test sample. Test was performed in triplicate, and the results were averaged.
Results and discussion
Chemistry
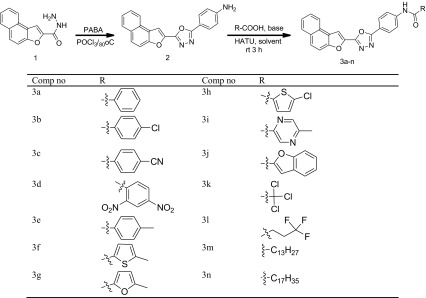
As depicted in Scheme 1, we started our synthetic strategy using the naphthofuran-2-carbohydrazide intermediate 1 which was prepared from its corresponding ester. The intermediate 1 was cyclized to oxadiazole intermediate 2 by using phosphorus oxychloride (POCl3) and para-aminobenzoic acid (PABA). The amino intermediate thus obtained was further coupled with various carboxylic acids to obtain an array of 2,5-disubstituted-1,3,4-oxadiazoles
Scheme 1.
Synthetic route for target compounds (3a–n). Coupling reaction of the intermediate 2 with various acids. Reaction conditions: amine (1 mmol), acid (1.2 mmol), DIPEA (2 mmol), HATU (0.3 mmol), DCM, 3 h, rt.b: isolated yield. Reaction carried out 4–5 h
We started our initial screening by the coupling reaction of intermediate 2 with benzoic acid as it is readily available and less toxic. We carried out the reaction in different bases and solvents by using HATU as the coupling reagent and the reaction optimization was done by monitoring the TLC, and our results are summarized in Table 1. We found that the reaction conditions and nature of the solvent has a determining influence in the formation of the product. HATU in presence of DIPEA was found to be essential for better conversions. DCM was proved to be a better solvent than tetrahydrofuran (THF) and N,N-dimethylformamide (DMF) for effective conversions. Among the various bases screened, DIPEA gave excellent conversions. The usage of inorganic bases did not render any coupled product. The best conversion was obtained when 1.2 equivalent of benzoic acid was used in DCM as solvent and DIPEA as base for 3 h (Table 1).
Table 1.
Effect of various solvents and bases in the coupling reaction of compound 2 with amine
| Entry | Base | Solvent | Yieldb |
|---|---|---|---|
| 1 | DIPEA | DMF | 56 |
| 2 | TEA | DMF | 48 |
| 3 | DABCO | DMF | 45 |
| 4 | NaOH | DMF | Nil |
| 5 | DIPEA | THF | 47 |
| 6 | TEA | THF | 22 |
| 7 | DABCO | THF | Traces |
| 8 | NaOH | THF | Nil |
| 9 | DIPEA | DCM | 90 |
| 10 | TEA | DCM | 64 |
| 11 | DABCO | DCM | 40 |
| 12 | NaOH | DCM | Nil |
With the optimized conditions in hand, we shifted our attention to evaluate the generality of this coupling reaction. Keeping this in mind, we tried the coupling reaction with a variety of aromatic and aliphatic acids (Scheme 1). To our delight, almost all the acids coupled well enough to obtain the required coupled product in satisfactory to excellent yields. Acids possessing electron withdrawing groups at para position reacted a little sluggishly whereas the acids having electron donating groups reacted efficiently to procure the corresponding amides in excellent yield while sterically hindered acids gave slightly lesser yield. In case of aliphatic carboxylic acids, as the carbon chain length increased, the yield of the product was found to be diminished.
The structures of synthesized compounds have been confirmed by IR, NMR, and mass spectroscopic methods. In IR spectrum of compound 2, absence of peak at 1640 cm−1 indicated that the cyclization of compound 1 has occurred to yield interpreted compound 2 and the reappearance of peak at 1680 cm−1 in compound 3 indicated the formation of amide linkage. In 1HNMR spectrum of compound 2, peak at δ 4.22 corresponded to NH2 proton, while in compounds 3a, the downfield signal appeared at δ 8.02 as singlet was assigned to amide NH, this confirms the formation of compound 3a. In 13C NMR spectrum of the compounds 3a-n, appearance of peaks at 167 and 158 ppm indicated the presence of C=O and –N–C–O– groups, this confirmed the presence of amide linkage and oxadiazole ring in the compound 3a–n.
Biological activity
In vitro antibacterial and antifungal activity
Antimicrobial activity and MIC of all synthesized compounds are summarized in Tables 2 and 3, respectively. All the synthesized compounds showed antimicrobial activity in a dose-dependent manner. The compound 3d showed moderate to good activity with MIC ranging between 0.20 to 0.40 mg/cm3 as compared to standard. The compound 3i showed maximum significant activity against Pseudomonas aeruginosa and exhibited comparable activity against Bacillus subtilis, Salmonella typhi, and Escherichia coli, while it exhibited comparable activity against tested fungal organisms with MIC values ranging between 0.40 and 0.60 mg/cm3. The compounds 3c and 3j displayed promising activity with MIC value between 0.20 and 0.40 mg/cm3, while compounds 3f, 3g, 3k, and 3l showed moderate activity with MIC ranging between 0.60 and 0.80 mg/cm3. Compounds 3a, 3b, and 3h showed measurable activity, whereas 3e, 3m, and 3n were found to be inactive against the tested pathogens.
Table 2.
Antimicrobial activity data of synthesized compounds 3a–n
| Comp code | Conc mg/mL | Zone of inhibition in mm (mean ± SD), n = 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Antibacterial | Antifungal | ||||||||
| P.a ± SD | B.s ± SD | S.t ± SD | E.c ± SD | A.f ± SD | C.a ± SD | M.g ± SD | A.t ± SD | ||
| 3a | 1 | 04 ± 0.3 | 05 ± 0.8 | 04 ± 0.1 | 06 ± 0.7 | 08 ± 0.5 | 05 ± 0.9 | 06 ± 0.9 | 04 ± 0.7 |
| 0.5 | 03 ± 0.1 | 03 ± 0.1 | 03 ± 0.2 | 04 ± 0.6 | 03 ± 0.4 | 03 ± 0.5 | 04 ± 0.3 | 03 ± 0.1 | |
| 3b | 1 | 08 ± 0.2 | 09 ± 0.9 | 08 ± 0.9 | 08 ± 0.1 | 11 ± 0.7 | 08 ± 0.1 | 09 ± 0.7 | 06 ± 0.9 |
| 0.5 | 06 ± 0.4 | 05 ± 0.2 | 04 ± 0.4 | 05 ± 0.7 | 06 ± 0.3 | 06 ± 0.3 | 05 ± 0.4 | 02 ± 0.4 | |
| 3c | 1 | 20 ± 0.5 | 17 ± 0.7 | 10 ± 0.8 | 18 ± 0.6 | 09 ± 0.7 | 15 ± 0.7 | 20 ± 0.6 | 14 ± 0.7 |
| 0.5 | 14 ± 0.2 | 12 ± 0.5 | 6 ± 0.2 | 12 ± 0.4 | 04 ± 0.2 | 08 ± 0.3 | 16 ± 0.4 | 09 ± 0.5 | |
| 3d | 1 | 22 ± 0.4 | 21 ± 0.7 | 18 ± 0.2 | 18 ± 0.5 | 20 ± 0.5 | 12 ± 0.4 | 21 ± 0.1 | 10 ± 0.5 |
| 0.5 | 16 ± 0.3 | 14 ± 0.1 | 12 ± 0.3 | 10 ± 0.4 | 11 ± 0.3 | 08 ± 0.9 | 15 ± 0.2 | 06 ± 0.4 | |
| 3e | 1 | 02 ± 0. | 03 ± 0. | 04 ± 0. | 03 ± 0. | – | 02 ± 0. | – | 02 ± 0. |
| 0.5 | – | – | – | – | – | – | – | – | |
| 3f | 1 | 10 ± 0.3 | 08 ± 0.6 | 09 ± 0.1 | 09 ± 0.3 | 11 ± 0.3 | 06 ± 0.2 | 08 ± 0.3 | 09 ± 0.6 |
| 0.5 | 06 ± 0.4 | 06 ± 0.4 | 06 ± 0.3 | 07 ± 0.5 | 09 ± 01. | 04 ± 0.1 | 05 ± 0.5 | 05 ± 0.4 | |
| 3g | 1 | 12 ± 0.7 | 13 ± 0.4 | 12 ± 0.7 | 10 ± 0.8 | 09 ± 0.4 | 08 ± 0.7 | 12 ± 0.7 | 14 ± 0.7 |
| 0.5 | 09 ± 0.6 | 08 ± 0.6 | 13 ± 0.5 | 08 ± 0.5 | 06 ± 0.1 | 06 ± 0.3 | 09 ± 0.5 | 10 ± 0.4 | |
| 3h | 1 | 05 ± 0.5 | 04 ± 0.8 | 07 ± 0.1 | 08 ± 0.3 | 04 ± 0.1 | 09 ± 0.4 | 08 ± 0.3 | 06 ± 0.3 |
| 0.5 | 03 ± 0.4 | 02 ± 0.2 | 06 ± 0.4 | 04 ± 0.1 | 02 ± 0.2 | 05 ± 0.3 | 06 ± 0.4 | 02 ± 0.9 | |
| 3i | 1 | 20 ± 0.6 | 18 ± 0.8 | 11 ± 0.8 | 15 ± 0.7 | 10 ± 0.6 | 12 ± 0.3 | 06 ± 0.7 | 07 ± 0.6 |
| 0.5 | 14 ± 0.4 | 12 ± 0.6 | 06 ± 0.5 | 10 ± 0.5 | 07 ± 0.4 | 08 ± 0.1 | 04 ± 0.5 | 04 ± 0.3 | |
| 3j | 1 | 18 ± 0.6 | 19 ± 0.7 | 15 ± 0.6 | 14 ± 0.5 | 15 ± 0.3 | 14 ± 0.1 | 11 ± 0.7 | 16 ± 0.5 |
| 0.5 | 11 ± 0.4 | 12 ± 0.5 | 10 ± 0.4 | 12 ± 0.3 | 10 ± 0.6 | 09 ± 0.2 | 09 ± 0.2 | 11 ± 0.3 | |
| 3k | 1 | 12 ± 0.3 | 09 ± 0.8 | 11 ± 0.9 | 12 ± 0.4 | 09 ± 0.7 | 10 ± 0.3 | 09 ± 0.3 | 08 ± 0. |
| 0.5 | 09 ± 0.4 | 07 ± 0.4 | 09 ± 0.4 | 10 ± 0.2 | 07 ± 0.4 | 07 ± 0.4 | 07 ± 0.4 | 05 ± 0.3 | |
| 3l | 1 | 10 ± 0.9 | 08 ± 0.7 | 05 ± 0.3 | 06 ± 0.3 | 12 ± 0.6 | 07 ± 0.7 | 09 ± 0.7 | 04 ± 0.1 |
| 0.5 | 07 ± 0.7 | 06 ± 0.4 | 04 ± 0.4 | 04 ± 0.4 | 08 ± 0.4 | 06 ± 0.4 | 07 ± 0.4 | 03 ± 0.4 | |
| 3m | 1 | 02 ± 0.2 | – | – | 03 ± 0.4 | – | 02 ± 0.1 | 04 ± 0.4 | – |
| 0.5 | – | – | – | – | – | – | – | – | |
| 3n | 1 | 03 ± 0.5 | – | 02 ± 0.6 | 02 ± 0.7 | 04 ± 0.1 | 03 ± 0.3 | 02 ± 0.7 | 04 ± 0.6 |
| 0.5 | – | – | – | – | – | – | – | – | |
| Standa | 25 ± 0.40 | 24 ± 0.1 | 21 ± 0.7 | 20 ± 0.1 | – | – | – | ||
| Standb | – | – | – | – | 20 ± 0.40 | 21 ± 0.40 | 22 ± 0.30 | 20 ± 0.1 | |
P.a Pseudomonas aeruginosa, B.s Bacillus subtilis, S.t Salmonella typhi, E.c Escherichia coli, A.f Aspergillus flavus, C.a Candida albicans, M.g Microspora griseous, A.t Aspergillus terreus, SD standard deviation
aStand—ciprofloxacin
bStand—fluconazole
Table 3.
Minimum inhibitory concentration (mg/cm3) of compounds 3a–n
| Comp code | Minimum inhibitory concentration (MIC mg/cm3) | |||||||
|---|---|---|---|---|---|---|---|---|
| P.a | B.s | S.t | E.c | A.f | C.a | M.g | A.t | |
| 3a | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 |
| 3b | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 |
| 3c | 0.40 | 0.40 | 0.40 | 0.40 | 0.40 | 0.40 | 0.40 | 0.40 |
| 3d | 0.20 | 0.20 | 0.40 | 0.40 | 0.20 | 0.60 | 0.40 | 0.40 |
| 3e | NT | NT | NT | NT | NT | NT | NT | NT |
| 3f | 0.60 | 0.80 | 0.80 | 0.60 | 0.60 | 0.80 | 0.80 | 0.80 |
| 3g | 0.60 | 0.60 | 0.60 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 |
| 3 h | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 |
| 3i | 0.40 | 0.40 | 0.60 | 0.40 | 0.60 | 0.60 | 0.60 | 0.40 |
| 3j | 0.20 | 0.20 | 0.40 | 0.40 | 0.40 | 0.40 | 0.60 | 0.40 |
| 3k | 0.60 | 0.80 | 0.80 | 0.60 | 0.80 | 0.80 | 0.80 | 0.60 |
| 3l | 0.60 | 0.60 | 0.80 | 0.60 | 0.80 | 0.80 | 0.60 | 0.60 |
| 3m | NT | NT | NT | NT | NT | NT | NT | NT |
| 3n | NT | NT | NT | NT | NT | NT | NT | NT |
| Ciprofloxacin | 0.20 | 0.20 | 0.20 | 0.20 | NT | NT | NT | NT |
| Fluconazole | NT | NT | NT | NT | 0.20 | 0.20 | 0.20 | 0.20 |
NT not tested
The higher activity profile of compounds 3c and 3d might be plausibly due to the presence of electron withdrawing cyano (CN) and nitro (NO2) groups, and the presence of two ring nitrogens might be the possible reason for the activity profile of compound 3i, whereas in 3j, the activity may be attributed due to the presence of benzofuran ring. The presence of chloro group might be the plausible reason for the enhanced activity of compound 3b. In case of compounds 3k and 3l, the bacterial activity might be due to the presence of chloro and fluro groups on side chain of acids used. The bacterial activity also revealed that the nonpotentiality of compounds 3m and 3n may be due the presence of aliphatic long chain on acids used. Thus, the microbial results revealed that the presence of electron withdrawing groups at the para position of the acids and the presence of ring nitrogens enhanced the antibacterial activity of the newly synthesized molecules to a greater extent with minimal MIC value.
In silico molecular docking studies
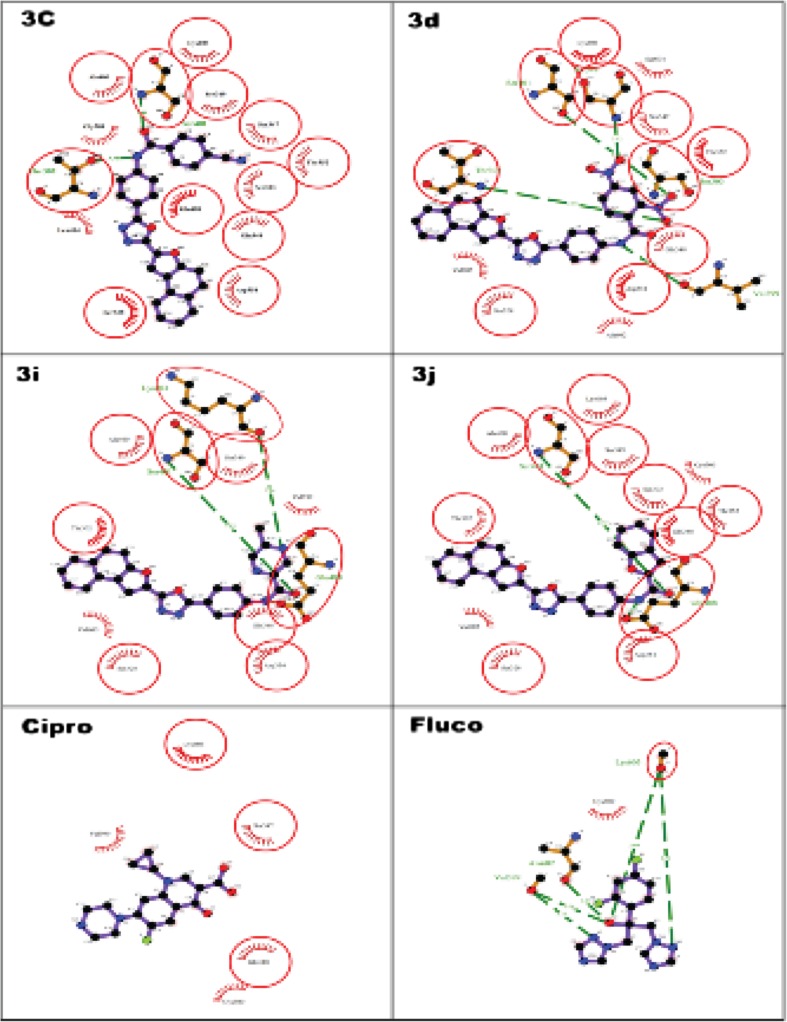
In correlation to in vitro antimicrobial activity, it thought worthwhile to carryout in silico studies to predict the binding affinity and orientation at the active site of the receptor. Automated docking was used to assess the orientation of inhibitors bound in the active pockets of GlcN-6-P synthase. The molecular docking of ligand molecules 3c, 3d, 3i, and 3j with GlcN-6-P synthase revealed that all the tested ligand molecules showed encouraging binding energy and the compounds exhibited hydrogen bonding with one or the other amino acids in the active pockets as shown in Fig. 1 The docked compounds 3c, 3d, 3i, and 3j were found best docked confirmation with least binding affinity (−9.1, −9.3, −8.9, and −9.3 kJ mol−1) than the standard drugs used for docking (Table 4).
Fig. 1.
2D representation of the interaction of the synthesized molecules 3c, 3d, 3i, and 3j and standards ciprofloxacin and fluconazole with glucosamine-6-phosphate synthase
Table 4.
In silico molecular docking results of synthesized compounds 3c, 3d, 3i, and 3j and standard drugs
| Ligand | Affinity (kcal/mol) | H-bonds | H-bond length (Å) | H-bond width | Hydrophobic interactions |
|---|---|---|---|---|---|
| 3c | −9.1 | 2 | 2.81 | 2VF5:Ser404::3c:O2 | Lys603, Ser349, Ser347, Thr352, Ser303, Glu488, Gln348, Asp354, Ser328, Leu484, Gly301, Ala400. |
| 3.35 | 2VF5:Thr302::3c:N2 | ||||
| 3d | −9.3 | 5 | 3.06 | 2VF5:Val399::3d: N3 | Ser328, Ala602, Ser604, Ser347, Thr352, Gln348, Gys603, Val605, Asp354. |
| 3.11 | 2VF5:Ser303::3c:O6 | ||||
| 3.12 | 2VF5:Ser349::3c:O5 | ||||
| 3.16 | 2VF5:Thr302::3c:O7 | ||||
| 3.16 | 2VF5:Ser401::3c:O6 | ||||
| 3i | −8.9 | 3 | 2.85 | 2VF5:Ser401::3i:O2 | Asp345, Val605, Gu348, Ser349, Val399,Ala400, Thr302, Ser328. |
| 3.07 | 2VF5:Glu488::3i:N3 | ||||
| 3.14 | 2VF5:Lys603::3i: N5 | ||||
| 3j | −9.3 | 2 | 2.73 | 2VF5:Ser401::3j:O2 | Asp354, Lys603, Val605, Ser349, Ser303, Gln348, Thr352, Cys300, Thr302. Ala400, Ser328. |
| 3.04 | 2VF5:Glu488::3j:N3 | ||||
| Ciprofloxacin | −8.0 | 4 | 2.71 | 2VF5:Ser303::ciprofloxacin:O2 | Cys300, Leu484, Glu488, Leu601, Val399, Lys603, sSer401. |
| 2.80 | 2VF5:Gln348::ciprofloxacin:O2 | ||||
| 2.97 | 2VF5:Thr352::ciprofloxacin:O3 | ||||
| 3.10 | 2VF5:Ser347::ciprofloxacin:O3 | ||||
| Fluconazole | −7.3 | 6 | 2.70 | 2VF5:Lys603:O | Val605, Cys300, Ser604, Gln348, Ser349, Leu601, Glu488. |
| 2.83 | 2VF5:Lys603::fluconazole:N5 | ||||
| 3.05 | 2VF5:Val399::fluconazole:O | ||||
| 3.09 | 2VF5:Val399::fluconazole:N2 | ||||
| 3.12 | 2VF5:Thr302::fluconazole:F2 | ||||
| 3.35 | 2VF5:Ala602::fluconazole:O |
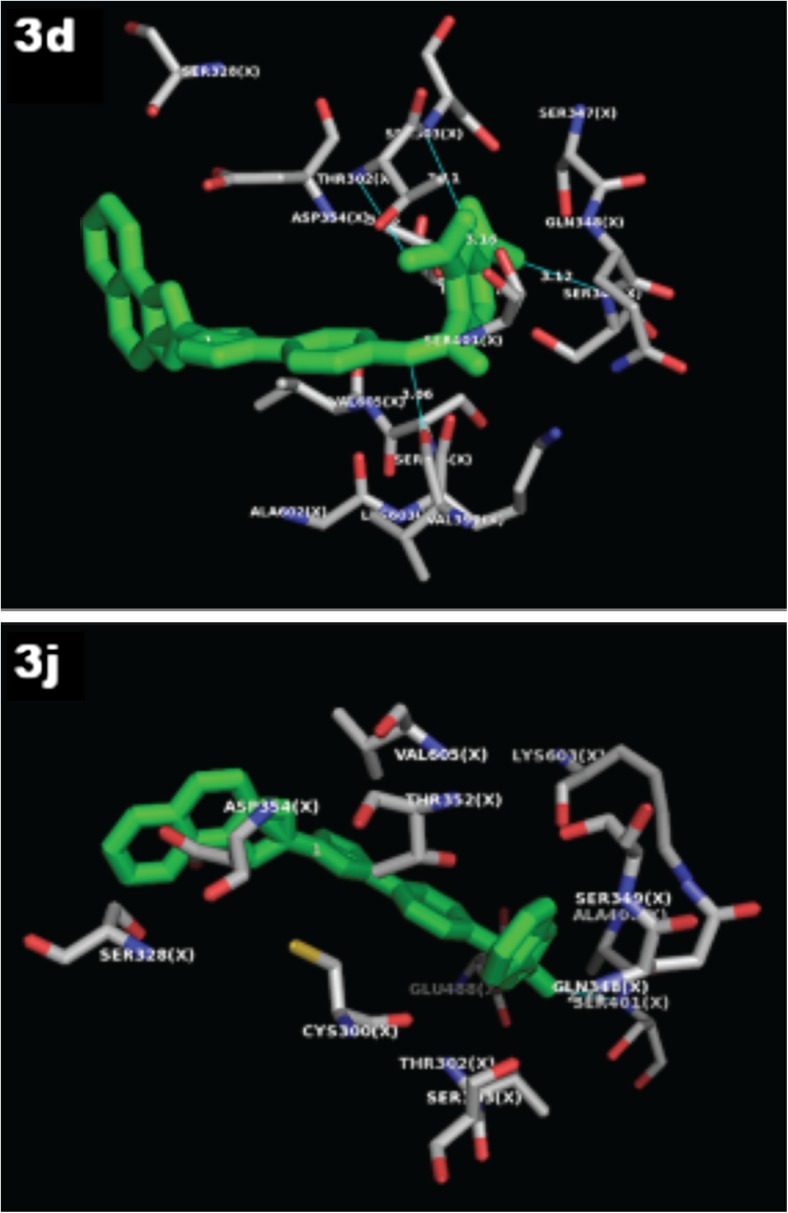
Ligand-protein interactions can be further extrapolated from Fig. 1 representing the 2D and Fig. 2 represents the 3D binding conformation of the best of diclofenac and embelin derived by using LigPlot+. In 2D, ligand is highlighted in purple color and set of conserved residues that are commonly involved in interaction with 3c, 3d, 3i, and 3j are encircled with red color. Figure 2 represents the 3D interaction of 3d and 3j, respectively, with COX2 by using educational version of PyMol. The ligands are represented in green color, H-bonds with their respective distances are represented with pale blue color, and the interacting residues are represented in molecular surface view color model with ball and stick representation.
Fig. 2.
3D representation of the interaction of the synthesized molecules 3d and 3j with glucosamine-6-phosphate synthase
The compound 3c established two hydrogen bonds with Ser404 and Thr302 amino acids in the active site of the target protein with bond length 2.81 and 3.35 A°. The compound 3d established five hydrogen bonds with Val399, Ser303, Ser349, Thr302, and Ser401 amino acids with bond length 3.06, 3.11, 3.12, 3.16, and 3.16 A°, respectively, with the highest affinity and hence is considered as the best dock conformation. Further compounds 3i and 3j exhibited three and two hydrogen bonding with bond length 2.85, 3.07, 3.14, 2.73, and 3.04 with Ser401, Glu488, Lys603, Ser401, and Glu488 amino acids, respectively. In this study, all the docked molecules 3c, 3d, 3i, and 3j showed more hydrophobic interaction than the standards ciprofloxacin and fluconazole. Further, the RMSD has often been used to measure the quality of reproduction of a known binding pose by molecules with ligands. All docked molecules has zero RMSD values; this indicates that the true binding pose of molecules with protein.
Antioxidant activity
The stable DPPH organic radical is widely used for computing the radical scavenging activity of the synthesized compounds. In this assay, the picrylhydrazyl (purple) radical has been reduced to picrylhydrazine (pale yellow) by antioxidant compounds. The decreasing in the absorbance of the solution indicates free radical scavenging capacity of the tested compounds and the absorbance of solution decreased proportionally to the increases of nonradical forms of DPPH. Usually, the compounds with unshared electrons and compounds which are easily ready to share their electron exhibits better DPPH radical scavenging activity. The studied compounds showed significant radical scavenging activity in a concentration-dependent manner, and the results are given in Table 5. As evident from the table, the compound 3c, 3d, and 3i exhibited good and comparable radical scavenging property to the standard with IC50 values 74.23 ± 3.23, 73.42 ± 2.45, and 81.52 ± 1.41 μg/cm3, respectively. On the other hand, 3j and 3g showed moderate activity with IC50 values 92.23 ± 1.61 and 90.42 ± 4.82 μg/cm3, respectively. The compounds 3a and 3 h showed promising activity whereas the compounds3e, 3j, and 3k compounds showed minimal activity. The remaining compounds 3k, 3l, 3m, and 3n did not possess any significant radical scavenging activity when compared to that of the standard.
Table 5.
Antioxidant activity of synthesized compounds
| Comp no | IC50 (μg/cm3) | |
|---|---|---|
| DPPH scavenging | Metal chelating | |
| 3a | 124.05 ± 2.45 | 132.34 ± 0.43 |
| 3b | 168.34 ± 1.43 | 192.52 ± 1.53 |
| 3c | 74.23 ± 3.23 | 84.43 ± 1.08 |
| 3d | 73.42 ± 2.45 | 76.52 ± 2.54 |
| 3e | 167.32 ± 1.85 | 154.42 ± 1.72 |
| 3f | 92.23 ± 1.61 | 105.26 ± 0.72 |
| 3g | 90.42 ± 4.82 | 113.85 ± 3.45 |
| 3h | 141.26 ± 2.33 | 184.03 ± 3.71 |
| 3i | 81.52 ± 1.41 | 93.05 ± 1.51 |
| 3j | 174.72 ± 1.25 | 188.42 ± 1.83 |
| 3k | 192.61 ± 1.41 | 198.22 ± 3.92 |
| 3l | 242.62 ± 2.61 | 165.09 ± 1.18 |
| 3m | 238.25 ± 2.43 | 146.13 ± 3.57 |
| 3n | 217.34 ± 2.83 | 158.31 ± 1.48 |
| Std | 57.05 ± 0.64a | 35.27 ± 3.12b |
aBHA
bEDTA
In nature, iron can be found in either ferrous (Fe2+) or ferric ion (Fe3+) form. In living systems, ferrous iron can facilitate the production of reactive oxygen species. Hence, the ability of substances to chelate iron can be a valuable antioxidant. The chelation of Fe2+ ions was estimated based on the formation of complex between ferrozine and Fe2+ quantitatively. The formation of complex will be distressed if any chelating agents are present in the solution, and subsequently, the color of the solution would be changed. The compounds 3c, 3d, and 3i exhibited good chelating ability with IC50 values 84.43 ± 1.08, 76.52 ± 2.54, and 93.05 ± 1.51 μg/cm3, respectively, while IC50 values 105.26 ± 0.72 and 113.85 ± 3.45 μg/cm3 were exhibited by compounds 3f and 3g, respectively. The reaming compounds showed promising to minimal chelation of Fe2+ ions from the solution.
The antioxidant result of the synthesized compounds gave an idea that the presence of substituents appear to be an important factor in affecting their antioxidant ability. The plausible reason for the enhanced antioxidant activity of the compounds 3c and 3d might be due to the presence of strong electron withdrawing groups. The promising radical scavenging property of the compound 3f and 3g could be due to the presence of heterocyclic rings like thiophene and furan attached to the amide linkage, whereas in 3i, the activity could be attributed to the presence of two nitrogen atoms of pyrazine. The compounds 3k, 3l, 3m, and 3n did not exhibit any activity which could be presumable due to steric hindrance of the molecules.
Conclusion
We have achieved a facile, modified, and efficient access for the synthesis of a series of novel oxadiazole derivatives fused with naphthofuran and evaluated their antimicrobial and antioxidant activities. The values of the MIC against microorganisms showed that these compounds have significant inhibitory effect and the antibacterial data indicated that the synthesized compounds are more effective against bacteria. From the structure activity relationship (SAR) studies, it was evident that the presence of electron withdrawing groups and heterocyclic cores attached to the oxadiazole ring is an indispensible characteristic for their pharmacological action. This work paved the way for the synthesis of a series of oxadiazole derivatives and guided to the various insights of their pharmacological activities. In this context, it is declared that the oxadiazole derivatives are of great importance in the field of drug discovery and research.
Acknowledgments
The authors are thankful to the Department of Industrial Chemistry, Kuvempu University, Shankaraghatta, for providing the laboratory facilities to carry out the research work.
References
- 1.Aksoy DY, Unal S. New antimicrobial agents for the treatment of Gram positive bacterial infections. Clin Micro Biol Infect. 2008;14:411–420. doi: 10.1111/j.1469-0691.2007.01933.x. [DOI] [PubMed] [Google Scholar]
- 2.Chang F, Wan HH, Mark M. Synthesis of anti-inflammatory furan fatty acids from biomass-derived 5-(chloromethyl)furfural. Sustainable Chem Pharm. 2015;1:14–18. doi: 10.1016/j.scp.2015.09.002. [DOI] [Google Scholar]
- 3.Amir M, Javed SA, Hassan MZ. Synthesis and antimicrobial activity of pyrazolinones and pyrazoles having benzothiazole moiety. Med Chem Res. 2012;21:1261–1270. doi: 10.1007/s00044-011-9642-0. [DOI] [Google Scholar]
- 4.Qureshi ST, Skamene E, Malo D. Comparative genomics and host resistance against infectious diseases. Emerg Infect Dis. 1999;5(1):36–47. doi: 10.3201/eid0501.990105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casal M, Vaquero M, Rinder H, Tortoli E, Grosset J, Rusch GS, Gutierrez J, Jarlier V. A case-control study for multidrug-resistant tuberculosis: risk factors in four European countries. Microb Drug Resist. 2013;11(1):62–67. doi: 10.1089/mdr.2005.11.62. [DOI] [PubMed] [Google Scholar]
- 6.Beceiro A, Tomas M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Micro Biol Rev. 2013;26(2):185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David LJ. Nox enzymes, ROS, and chronic disease: an example of antagonistic pleiotropy. Free Radic Biol Med. 2007;43(3):332–347. doi: 10.1016/j.freeradbiomed.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott JA, King GL. Oxidative stress and antioxidant treatment in diabetes. Ann N Y Acad Sci. 2004;1031:204–213. doi: 10.1196/annals.1331.020. [DOI] [PubMed] [Google Scholar]
- 9.Hasan T, Elanur A, Ali A. Xanthoria elegans (Link) (lichen) extract counteracts DNA damage and oxidative stress of mitomycin C in human lymphocytes. Cytotechnology. 2012;64(6):679–686. doi: 10.1007/s10616-012-9447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung-Hee H, Youn-Soo C, Soon-Jae R. Effects of the cellcultured acanthopanax senticosus extract on antioxidative defense system and membrane fluidity in the liver of type 2 diabetes mouse. J Clin Biochem Nutr. 2009;45(1):101–109. doi: 10.3164/jcbn.08-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279(6):1005–1028. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- 12.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 13.Pilger C, Bartolucci C, Lamba D, Tropsha A, Fels G. Accurate prediction of the bound conformation of galanthamine in the active site of torpedo californica acetylcholinesterase using molecular docking. J Mol Graph Model. 2001;19:288–296. doi: 10.1016/S1093-3263(00)00056-5. [DOI] [PubMed] [Google Scholar]
- 14.Sun J, Makawana JA, Zhu HL. 1,3,4-oxadiazole derivatives as potential biological agents. Mini Rev Med Chem. 2013;13(12):1725–1743. doi: 10.2174/13895575113139990071. [DOI] [PubMed] [Google Scholar]
- 15.Khalilullah H, Ahsan MJ, Hedaitullah M, Khan S, Ahmed B. 1,3,4-oxadiazole: a biologically active scaffold. Mini Rev Med Chem. 2012;12(8):789–801. doi: 10.2174/138955712801264800. [DOI] [PubMed] [Google Scholar]
- 16.Sahin G, Palaska E, Ekizoglu M, Ozalp M. Synthesis and antimicrobial activity of some 1,3,4-oxadiazole derivatives. Farmaco. 2002;57(7):539–542. doi: 10.1016/S0014-827X(02)01245-4. [DOI] [PubMed] [Google Scholar]
- 17.Zareef M, Iqbal R, Dominguez NG, Rodrigues J, Zaidi JH, Arfan M, Supuran CT. Synthesis and antimalarial activity of novel chiral and achiral benzenesulfonamides bearing 1, 3, 4-oxadiazole moieties. J Enzyme Inhib Med Chem. 2007;22(3):301–308. doi: 10.1080/14756360601114569. [DOI] [PubMed] [Google Scholar]
- 18.Khalil NA, Kamal AM, Emam SH. Design, synthesis, and antitumor activity of novel 5-pyridyl-1,3,4-oxadiazole derivatives against the breast cancer cell line mcf-7. Biol Pharm Bull. 2015;38(5):763–773. doi: 10.1248/bpb.b14-00867. [DOI] [PubMed] [Google Scholar]
- 19.Zarghi A, Tabatabai SA, Faizi M, Ahadian A, Navabi P, Zanganeh V, Shafiee A. Synthesis and anticonvulsant activity of new 2-substituted-5-(2-benzyloxyphenyl)-1,3,4-oxadiazoles. Bioorg Med Chem Lett. 2005;15(7):1863–1865. doi: 10.1016/j.bmcl.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 20.Arvind KS, Lohani M, Parthsarthya R. Synthesis, characterization and anti-inflammatory activity of some 1, 3,4 -oxadiazole derivatives. Iran J Pharm Res. 2013;12(2):319–323. [PMC free article] [PubMed] [Google Scholar]
- 21.El-Essawy A, El-Sayed WA, El-Kafrawy SA, Morshedy AS, Abdel-Rahman AH. Anti-hepatitis B virus activity of new 1,2,4-triazol-2-yl- and 1,3,4-oxadiazol-2-yl-2-pyridinone derivatives. Z Naturforsch C. 2008;63(10):667–674. doi: 10.1515/znc-2008-9-1010. [DOI] [PubMed] [Google Scholar]
- 22.El-Sayed WA, El-Essawy FA, Ali OM, Nasr BS, Abdalla MM, Abdel-Rahman AA. Anti-HIV activity of new substituted 1,3,4-oxadiazole derivatives and their acyclic nucleoside analogues. Z Naturforsch C. 2009;6412:773–778. doi: 10.1515/znc-2009-11-1203. [DOI] [PubMed] [Google Scholar]
- 23.Santín EP, Khanwalkar H, Voegel J, Collette P, Mauvais P, Gronemeyer H, de Lera AR. Highly potent naphthofuran-based retinoic acid receptor agonists. Chem Med Chem. 2009;4(5):780–791. doi: 10.1002/cmdc.200900015. [DOI] [PubMed] [Google Scholar]
- 24.Kumaraswamy MN, Chandrashekhar C, Shivakumar H, Prathima Mathias DA, Mahadevan KM, Vaidya VP. Synthesis and activity evaluation of 2-(1-naphtho[2,1-b]furan-2-yl-carbonyl)-3,5-disubstituted-2,3-dihydro-1h-pyrazoles. Indian J Pharm Sci. 2008;70(6):715–720. doi: 10.4103/0250-474X.49090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Guevel R, Oger F, Lecorgne A, Dudasova Z, Chevance S, Bondon A, Simonneaux G, Salbert G. Identification of small molecule regulators of the nuclear receptor HNF4alpha based on naphthofuran caffolds. Bioorg Med Chem. 2009;17(19):7021–7030. doi: 10.1016/j.bmc.2009.07.079. [DOI] [PubMed] [Google Scholar]
- 26.Suresh KGV, Rajendra PY, Chandrashekar SM. Synthesis and pharmacological evaluation of novel 4-isopropylthiazole-4-phenyl-1,2,4-triazole derivatives as potential antimicrobial and antitubercular agents. Med Chem Res. 2013;22:938–948. doi: 10.1007/s00044-012-0092-0. [DOI] [Google Scholar]
- 27.Rajesh AR, Pavankumar B, Sheetal DB, Preeti KK. Synthesis and evaluation of novel 4-nitropyrrole-based 1,3,4-oxadiazole derivatives as antimicrobial and anti-tubercular agents. Eur J Med Chem. 2013;70:49–58. doi: 10.1016/j.ejmech.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 28.Wenneng W, Qin C, Anqi T, Guangqi J, Guiping O. Synthesis and antiviral activity of 2-substituted methylthio-5-(4-amino-2-methylpyrimidin-5-yl)-1,3,4-oxadiazole derivatives. Bioorg. Med Chem Lett. 2015;25:2243–2246. doi: 10.1016/j.bmcl.2015.02.069. [DOI] [PubMed] [Google Scholar]
- 29.Islavathu H, Reddymasu S, Surender SJ, Mohamed JA, Rudraraju RR. Synthesis and biological evaluation of 1,3,4-oxadiazole-linked bisindole derivatives as anticancer agents. Monatsh Chem. 2015;146(10):1699–1705. doi: 10.1007/s00706-015-1448-1. [DOI] [Google Scholar]
- 30.Sheelavanth S, Bodke YD, Sundar SM. Synthesis, antioxidant, and antibacterial studies of phenolic esters and amides of 2-(1-benzofuran-2-yl) quinoline-4-carboxylic acid. Med Chem Res. 2013;22:1163–1171. doi: 10.1007/s00044-012-0117-8. [DOI] [Google Scholar]
- 31.Morris GM, Goodsell DS, Halliday RS. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- 32.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 33.Kenchappa R, Yadav DB, Asha B, Sandeep T, Aruna SM. Synthesis, antimicrobial and antioxidant activity of benzofuran barbitone and benzofuran thiobarbitone erivatives. Med Chem Res. 2014;23(6):3065–3081. doi: 10.1007/s00044-013-0892-x. [DOI] [Google Scholar]
- 34.Chandrashekar A, Eswarappa B, Yadav DB, Venkatesh KB, Raghu N, Shivakumar MC, Peethambar SK, Sandeep T. 5-phenyl-1-benzofuran-2-yl derivatives: synthesis, antimicrobial, and antioxidant activity. Med Chem Res. 2013;22:78–87. doi: 10.1007/s00044-012-0017-y. [DOI] [Google Scholar]
- 35.Aruna SM, Yadav DB, Kenchappa R. Isolation, characterization and biological evaluation of N-heptadecanoic acid (margaric acid) isolated from wendlandia thyrsoidea leaves. J Chem Chem Sci. 2015;5(12):663–669. [Google Scholar]