Abstract
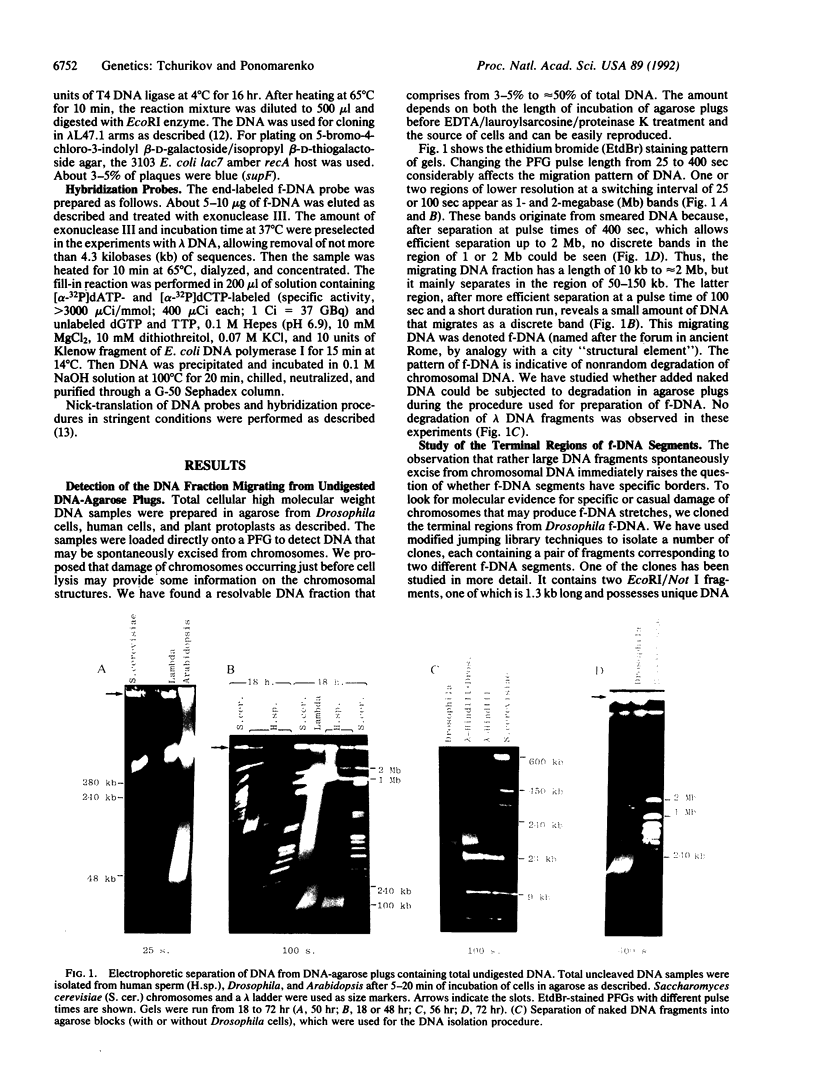
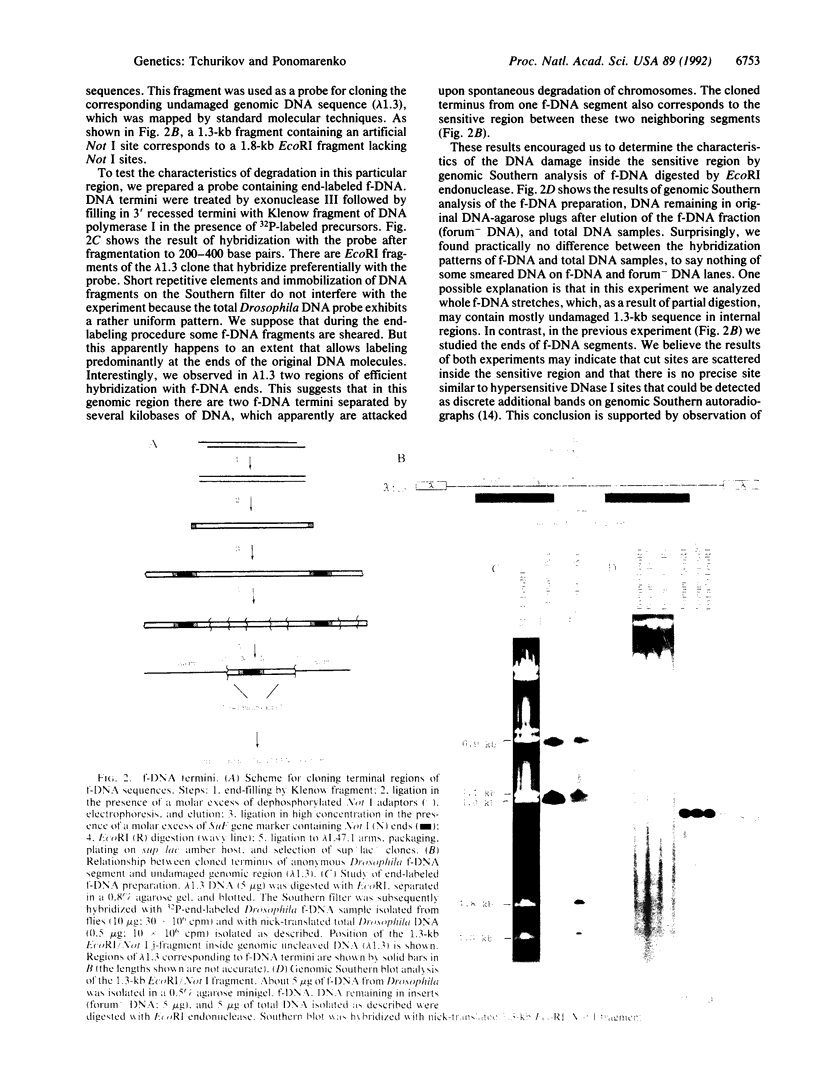
We have used pulsed-field gel electrophoresis of undigested DNA prepared by cell lysis in agarose with proteinase K detergent treatment and found a resolvable DNA fraction, denoted forum DNA (f-DNA). By changing the pulsed-field gel pulse length from 25 to 4500 sec, to obtain optimal separation in different ranges, we have found f-DNA to occupy a rather broad zone from 2 megabases to 10 kilobases (kb), but mainly at a range between 50 and 150 kb. f-DNA seems to appear as a result of nonrandom spontaneous degradation during cell treatment. The terminal regions of f-DNA segments have been cloned by using a jumping library. The molecular analysis of unique DNA sequence from an anonymous Drosophila DNA segment led to the conclusion that f-DNA appears as a result of nonrandom chromosomal DNA cleavage within sensitive regions that occupy a few kilobases. This conclusion was confirmed by detection of rather discrete hybridization bands on pulsed-field gel Southern blots in a region of good separation of undigested f-DNA after hybridization with different unique and repetitive probes. We propose that f-DNA segments may correspond to some regular higher-order structures in the eukaryotic chromosomes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernardi G. The isochore organization of the human genome. Annu Rev Genet. 1989;23:637–661. doi: 10.1146/annurev.ge.23.120189.003225. [DOI] [PubMed] [Google Scholar]
- Blochlinger K., Bodmer R., Jack J., Jan L. Y., Jan Y. N. Primary structure and expression of a product from cut, a locus involved in specifying sensory organ identity in Drosophila. Nature. 1988 Jun 16;333(6174):629–635. doi: 10.1038/333629a0. [DOI] [PubMed] [Google Scholar]
- Butler P. J. The folding of chromatin. CRC Crit Rev Biochem. 1983;15(1):57–91. doi: 10.3109/10409238309102801. [DOI] [PubMed] [Google Scholar]
- Churikov N. A., Ponomarenko N. A., Georgiev G. P. Obnaruzhenie al'ternativnogo splaisinga RNK v lokuse cut D. melanogaster. Dokl Akad Nauk SSSR. 1988;303(4):984–986. [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Conformational constraints in nuclear DNA. J Cell Sci. 1976 Nov;22(2):287–302. doi: 10.1242/jcs.22.2.287. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Shermoen A. W., Beckendorf S. K. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell. 1982 Jun;29(2):601–607. doi: 10.1016/0092-8674(82)90176-3. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Matsumoto T., Niwa O., Klco S., Fan J. B., Yanagida M., Cantor C. R. An electrophoretic karyotype for Schizosaccharomyces pombe by pulsed field gel electrophoresis. Nucleic Acids Res. 1987 Jun 11;15(11):4481–4489. doi: 10.1093/nar/15.11.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. C., Puvion E., Buchholtz L. A., Berezney R. Spatial distribution of DNA loop attachment and replicational sites in the nuclear matrix. J Cell Biol. 1984 Nov;99(5):1794–1802. doi: 10.1083/jcb.99.5.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchurikov N. A., Gerasimova T. I., Johnson T. K., Barbakar N. I., Kenzior A. L., Georgiev G. P. Mobile elements and transposition events in the cut locus of Drosophila melanogaster. Mol Gen Genet. 1989 Oct;219(1-2):241–248. doi: 10.1007/BF00261183. [DOI] [PubMed] [Google Scholar]
- Tchurikov N. A., Naumova A. K., Zelentsova E. S., Georgiev G. P. A cloned unique gene of Drosophila melanogaster contains a repetitive 3' exon whose sequence is present at the 3' ends of many different mRNAs. Cell. 1982 Feb;28(2):365–373. doi: 10.1016/0092-8674(82)90354-3. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Roeder R. G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987 Nov 20;51(4):613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- van der Velden H. M., van Willigen G., Wetzels R. H., Wanka F. Attachment of origins of replication to the nuclear matrix and the chromosomal scaffold. FEBS Lett. 1984 Jun 4;171(1):13–16. doi: 10.1016/0014-5793(84)80451-2. [DOI] [PubMed] [Google Scholar]







