Abstract
Bacterial protein synthesis is the target for numerous natural and synthetic antibacterial agents. We have developed a poly(U) mRNA-directed aminoacylation/translation (A/T) protein synthesis system composed of phenylalanyl-tRNA synthetases (PheRS), ribosomes, and ribosomal factors from Pseudomonas aeruginosa. This system has been used for high-throughput screening of a natural-compound library. Assays were developed for each component of the system to ascertain the specific target of inhibitory compounds. In high-throughput screens, 13 compounds were identified that inhibit protein synthesis with 50% inhibitory concentrations ranging from 0.3 to >80 μM. MICs were determined for the compounds against the growth of a panel of pathogenic organisms, including Enterococcus faecalis, Escherichia coli, Haemophilus influenzae, Moraxella catarrhalis, P. aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Three of the compounds were observed to have broad-spectrum activity and inhibited a hypersensitive strain of P. aeruginosa with MICs of 8 to 16 μg/ml. The molecular target of each of the three compounds was determined to be PheRS. One compound was found to be bacteriostatic, and one compound was bactericidal against both Gram-positive and Gram-negative pathogens. The third compound was observed to be bacteriostatic against Gram-positive and bactericidal against Gram-negative bacteria. All three compounds were competitive with the substrate ATP; however, one compound was competitive, one was uncompetitive, and one noncompetitive with the amino acid substrate. Macromolecular synthesis assays confirm the compounds inhibit protein synthesis. The compounds were shown to be more than 25,000-fold less active than the control staurosporine in cytotoxicity MTT testing in human cell lines.
INTRODUCTION
Bacterial infections are a continuing major worldwide health problem. Infections can be minor, such as skin rashes and common ear infections in infants, or potentially lethal, such as those in many immunocompromised patients. Pseudomonas aeruginosa is an opportunistic Gram-negative bacterial pathogen and the causative agent in a wide range of infections and is responsible for one-seventh of all nosocomial infections (1, 2). Among clinical isolates of P. aeruginosa, antimicrobial resistance is increasing (3) and has become a major problem in the hospital setting (4). However, the most serious medical problem caused by P. aeruginosa is chronic lung colonization associated with cystic fibrosis patients (5), and it is the leading cause of morbidity and mortality in these patients (6).
Antibiotics block cellular processes which are essential for bacterial survival. Many of the current antibiotics, both naturally occurring and synthetic, target protein synthesis as a mechanism of action (for a complete list, see reference 7). Antibiotics that target protein synthesis include the macrolides, clindamycin, chloramphenicol, the aminoglycosides, and the tetracyclines (8–10). Linezolid, one of the newest antibiotics and a protein synthesis inhibitor, is a member of the synthetic oxazolidinone class and is the last line of defense against many bacterial infections (11, 12). The fact that protein synthesis is the most frequent target of naturally occurring antibacterials provides compelling evolutionary evidence for the use of this system in the discovery of new antibacterial compounds (11).
The ribosome has been a central focus for development of inhibitors in drug discovery; however, other components involved in protein synthesis also offer attractive targets. Elongation factor Tu (EF-Tu) (13), elongation factor Ts (EF-Ts) (14), and elongation factor G (EF-G) (15) all play central roles in the elongation phase of protein synthesis. Antibiotics that target the activity of EF-Tu and EF-Ts include kirromycin and pulvomycin and, more recently, a compound series has been identified that includes: indole dipeptides, benzimidazole amidines, 2-arylbenzimidazoles, and N-substituted imidazoles and guanidines (7). EF-G is bifunctional and is also involved in ribosome recycling in a GTP-dependent fashion (16). Fusidic acid is an antibiotic that inhibits protein synthesis by trapping EF-G in the posttranslocation site on the ribosome after hydrolysis of GTP. Indirectly, the amino-acyl tRNA synthetases (aaRS) are essential enzymes involved in protein synthesis and individually provide attractive targets for discovery of antibiotics (17).
Recently, attempts have been made by several groups, with limited success, to screen for inhibitors of bacterial protein synthesis using cell extracts that contain native transcription and translation systems from Escherichia coli, Streptococcus pneumoniae (18), and Staphylococcus aureus (19). The use of cell extracts for screening can be problematic due to numerous problems (20). To avoid these problems we previously developed a polyuridylic acid [poly(U)]-directed aminoacylation/translation (A/T) protein synthesis system composed of PheRS, ribosomes, and ribosomal factors from Escherichia coli. Using this system as a platform for screening, we discovered a compound series (4H-pyridopyrimidine) capable of inhibiting protein synthesis in vitro and in whole-cell assays (21, 22). We describe here the development of an A/T protein synthesis system from P. aeruginosa and its use to screen natural compounds for inhibitors of function. We identified and characterized three compounds that inhibit protein synthesis in vitro, as well as broad-spectrum inhibition of bacteria in cultures.
MATERIALS AND METHODS
Materials.
The MicroSource Natural Products Library (Discovery Systems, Inc., Gaylordsville, CT) is an 800-compound collection of pure natural products and their derivatives. The collection includes simple and complex oxygen heterocycles, alkaloids, sesquiterpenes, diterpenes, pentacyclic triterpenes, sterols, and many other diverse representatives. The compounds have a minimal purity of 95%. Compounds were supplied as 10 mM stocks dissolved in dimethyl sulfoxide (DMSO), stored at −20°C, and thawed immediately before analysis. Radioactive amino acids were from Perkin-Elmer (Waltham, MA). All other chemicals were obtained from either Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburg, PA). E. coli tolC efflux pump mutant, P. aeruginosa PAO200 (efflux pump mutant), and P. aeruginosa hypersensitive strain (ATCC 35151) were kindly provided by Urs Ochsner (Crestone Pharma-Boulder, Boulder, CO). All other bacteria were from American Type Culture Collection (ATCC; Manassas, VA).
Purification of ribosomes and proteins.
Early-phase ribosomes from P. aeruginosa (ATCC 47085) were prepared in the Hill laboratory at the University of Montana (Missoula, MT) as previously described (23). P. aeruginosa EF-Tu and EF-Ts (24), EF-G (25), and PheRS (26) were purified as described previously.
Aminoacylation/translation assays.
A scintillation proximity assay (SPA) was developed for the P. aeruginosa A/T assay as described previously (21). The complete assay (50 μl) contained 50 mM Tris-HCl (pH 7.5), 25 mM KCl, 10 mM MgCl2, 0.03 mM spermine, 1.5 mM ATP, 0.5 mM GTP, 40 μM [3H]phenylalanine (Phe), and 0.3 mg of poly(U) mRNA/ml. To maintain constant levels of ATP and GTP the assay contained a nucleotide regeneration system composed of 4 mM phosphoenolpyruvate and 0.025 U of pyruvate kinase/μl. The concentrations of P. aeruginosa ribosomes and proteins in the assay were as follows: ribosome (0.2 μM), PheRS (0.1 μM), EF-Tu (1 μM), EF-Ts (0.05 μM), and EF-G (0.2 μM). These concentrations were determined through sequential rounds of optimization: each concentration represents the concentration just below the saturation point of the titration.
The screening reactions were carried out in 96-well microtiter plates (Costar). Test compounds were equilibrated by the addition of 33 μl of the protein/substrate mix (without tRNA) to 2 μl of chemical compound (3.3 mM) dissolved in 100% DMSO. This mixture was allowed to incubate at ambient temperature for 15 min, and then reactions were initiated by the addition of 15 μl of E. coli tRNA (20 μM), followed by a 2-h incubation at room temperature (comparable to 1 h at 37°C). Reactions were stopped by the addition of 5 μl of 0.5 M EDTA. A 200-μg portion of SPA beads (RNA binding beads [YSI]; Perkin-Elmer) in 150 μl of 300 mM citrate buffer (pH 6.2) was added. The plates were analyzed using a 1450 Microbeta (Jet) liquid scintillation and luminescence counter (Wallac). Assays to determine the 50% inhibitory concentrations (IC50s) were as described above, with the test compounds serially diluted from 200 to 0.4 μM. The concentration ranges of antibiotic in control plates were as follows: spiramycin (0.02 to 20.0 μM) and fusidic acid (4 to 512 μM).
PheRS assays.
Assays to determine inhibition of PheRS by chemical compounds were as described previously (27), except that the enzyme mix was preincubated with 132 μM compound for 15 min prior to the addition of tRNA (80 μM total tRNA or 2 μM tRNAPhe). The reactions were stopped by the addition of 5 μl of 0.5 M EDTA. A 400-μg portion of yttrium silicate (Ysi) poly-l-lysine-coated SPA beads (Perkin-Elmer) in 150 μl of 300 mM citrate buffer (pH 2.0) was added, and the plates were analyzed as described above. To determine the mechanism of action of the inhibitory compounds with respect to PheRS substrates ATP and Phe, competition assays were carried out as previously described (26).
EF-Tu GDP exchange assay.
Nitrocellulose binding assays were used to determine the inhibition of GDP exchange by EF-Tu, as previously described (28), except that the enzyme (1 μM) was preincubated with 132 μM compound for 15 min prior to the addition of [3H]GDP. EF-Ts stimulates the exchange of GDP bound by EF-Tu. The ability of compounds to inhibit EF-Ts stimulation of GDP exchange by EF-Tu was measured in assays, as described for the EF-Tu/GDP exchange, except that EF-Ts was present at 0.05 μM and the time for the reaction was decreased from 30 min to 30 s (24).
EF-G GTPase assay.
Assays for ribosome-dependent GTP hydrolysis by EF-G were carried out in 50-μl reaction mixtures containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 70 mM NH4Cl, 1 mM dithiothreitol (DTT), 1.8 mM GTP, 0.4 μM P. aeruginosa ribosomes, and 0.2 μM EF-G. Reactions were performed at 37°C for 30 min, and the assays were stopped by the addition of 150 μl of 50 mM EDTA. The amount of GTPase activity was determined by measuring the amount of Pi liberated using a colorimetric GTPase assay kit (Novus Biologicals) according to the manufacturer's directions (25). The final concentrations of compounds in the assays were 132 μM.
Eukaryotic protein synthesis assays.
Reactions to determine the inhibitory effect of compounds on eukaryotic protein synthesis were performed using wheat germ cell extracts as described previously (21). The concentrations of the compounds in these assays ranged from 0.8 to 100 μM, and the concentration of the cycloheximide in the control reactions ranged from 0.3 to 300 μM.
Assays were carried out to test the inhibitory effect of compounds on the activity of human mitochondrial PheRS (hmPheRS). Human mitochondrial PheRS was prepared as described previously (27). The assay mixture (50 μl) contained 50 mM Tris-HCl (pH 7.5), 1 mM spermine, 10 mM magnesium acetate, 2.5 mM ATP, 1 mM DTT, 75 μM [3H]Phe, and 0.5 μM hmPheRS. The mixture was incubated for 15 min with various concentrations of the compound (0.8 to 200 μM), and the reaction was then initiated by the addition of E. coli tRNAPhe (2 μM). The reaction was stopped by dilution into 3 ml of ice-cold 5% trichloroacetic acid and filtration through glass fiber filters as described previously (21).
Microbiological assays.
Broth microdilution MIC testing was performed in 96-well microtiter plates according to Clinical and Laboratory Standards Institute (CLSI) guidelines (29). MIC values were determined for E. coli (ATCC 25922), E. coli tolC efflux pump mutant, Enterococcus faecalis (ATCC 29212), Haemophilus influenzae (ATCC 49766), Moraxella catarrhalis (ATCC 25238), P. aeruginosa (ATCC 47085), P. aeruginosa PAO200 (efflux pump mutant), P. aeruginosa hypersensitive strain (ATCC 35151), S. aureus (ATCC 29213), and Streptococcus pneumoniae (ATCC 49619).
Time-kill experiments were performed according to CLSI guidelines (30). Growth media were brain heart infusion and Trypticase soy broth from Remel (Lenexa, KS). For the experiments, 10 ml of broth medium was inoculated with 0.1 ml of a fresh overnight culture and grown at 35°C with shaking (200 rpm) for 2 to 3 h. Prewarmed flasks containing 10 ml of medium alone or 10 ml of medium containing a test compound at 4× the MIC were then inoculated with 0.1 ml of the exponentially growing cultures. The samples were removed at 0, 2, 4, 6, and 24 h, and serial dilutions were plated on blood agar to allow for colony enumeration and calculation of the live cell density.
Macromolecular synthesis (MMS) assays were performed in cultures of E. faecalis as previously described (21). Levofloxacin, rifampin, tetracycline and vancomycin were used as quality controls in assays to measure inhibition of DNA synthesis, RNA synthesis, protein synthesis and cell wall synthesis, respectively.
In vitro cytotoxicity test.
To determine the effect of hit compounds on the growth of human cell cultures, in vitro cytotoxicity testing was carried out as described previously using human embryonic kidney 293 cells (HEK-293) (31). A Trevigen TACS MTT cell proliferation assay kit (Trevigen, Gaithersburg, MD) was utilized to assess impacts on human cell proliferation and/or viability. The control staurosporine was serial diluted in assays from 1 to 0.001 μg/ml, and the compounds tested were diluted in assays from 800 to 25 μg/ml.
RESULTS
Development and optimization of the P. aeruginosa A/T assay for screening.
An aminoacylation/translation (A/T) system was developed which contained all the components required for translation of poly(U) mRNA from P. aeruginosa: ribosomes, EF-Tu, EF-Ts, EF-G, and PheRS. The coupled A/T reaction was adapted from separate aminoacylation and translation assays as previously described (21). The assay was optimized for the detection of inhibition of any component of the A/T system, and compounds were screened in 96-well microtiter plates. The reactions were monitored using SPA technology. Initially, the ribosomes were titrated into the assay while maintaining saturating amounts of the other components (see Fig. S1A in the supplemental material), and 0.2 μM was chosen as the screening concentration, which yielded sufficient signal over background. At this concentration of ribosomes, all other components of the system were then sequentially titrated into the system to determine the inflection point of saturation on a titration curve. Multiple rounds of titration were carried out to arrive at the final screening concentration of each accessory component. The concentrations were set just below the saturation points to facilitate maximum sensitivity to inhibition of each and every component of the system (see Fig. S1B to E in the supplemental material).
Chemical compounds were dissolved in DMSO in stock concentrations of 3.3 mM, and 2 μl was added to reaction mixtures (50 μl), yielding 132 μM final concentrations. The addition of 2 μl of the DMSO in stock solution to screening reactions resulted in a 4% final DMSO concentration. It was therefore necessary to determine the effect of DMSO on the P. aeruginosa A/T system. To test the effects, DMSO was added so that the final amount of DMSO in the reactions ranged from 1 to 10%. There was no decrease in activity of the A/T system observed up to 5% DMSO (data not shown).
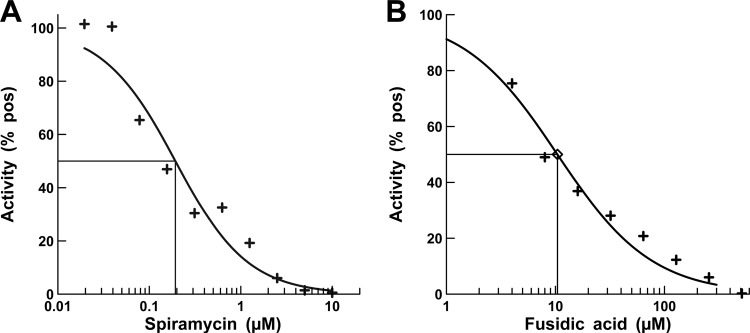
The macrolide spiramycin was selected as a control antibiotic. This antibiotic inhibits protein synthesis by binding the 50S ribosomal subunit near the peptidyl-transferase center, thereby stopping peptide synthesis (32). Fusidic acid inhibits protein synthesis by trapping EF-G in the posttranslocation step during elongation (33, 34) and was also selected as a positive control. Spiramycin and fusidic acid were both effective at inhibiting poly-Phe synthesis in the P. aeruginosa A/T assay with IC50s of 0.20 and 10.4 μM, respectively (Fig. 1).
FIG 1.
IC50 determination for positive controls in the A/T assay. (A and B) Titrations of spiramycin (A) and fusidic acid (B) in an A/T assay. The concentrations of spiramycin ranged from 0.02 to 10.0 μM, and the concentration of fusidic was from 4 to 256 μM. The antibiotics were diluted into DMSO, so that the concentration of DMSO was the same as in the screening assays. Positives were control assays in which there were no antibiotics, and approximately 500 pmol of phenylalanine were synthesized in these assays. The curve fits and IC50s were determined using XLfit 5.3 (IDBS) as part of Microsoft Excel.
Screening of a natural-product library yields confirmed hit compounds specific for individual targets.
A compound library containing over 800 natural products was tested in a high-throughput format. The screening reactions were very robust with an average signal-to-background ratio of 65:1 and a percent coefficient of variation of 0.41. The Z′ and Z factors across all plates averaged 0.69 and 0.4, respectively. The initial screen was conducted as single point assays, and a compound that inhibited >50% of the poly-Phe synthesis was defined as a hit. Hit compounds were retested in triplicate for the confirmation of activity. The confirmed hit compounds were then tested in individual specificity assays against each of the four accessory proteins (EF-Tu, EF-Ts, EF-G, and PheRS) as described in Materials and Methods. Thirteen compounds were found to be specific for a single component of the A/T assay (Table 1). Seven of the hit compounds did not inhibit an accessory protein but did inhibit poly-Phe synthesis in the biochemical assay, leading us to conclude that the likely mechanism of action of these compounds was direct inhibition of the ribosome itself. The activity of EF-Tu or EF-Ts was not observed to be affected in the presence of any of the compounds. The IC50s for these 13 compounds were determined by serially diluting the compounds from 200 to 0.4 μM in the A/T assay (Table 1).
TABLE 1.
Confirmed hit compound, component inhibited, and IC50 in the A/T assay
| Compound | EF-G | PheRS | Ribosome | IC50 (μM) |
|---|---|---|---|---|
| BM02F09 | X | 87 | ||
| BM02H08 | X | 52 | ||
| BM03E08 | X | 34 | ||
| BM03F03 | X | 18 | ||
| BM03F11 | X | 0.3 | ||
| BM04E04 | X | 42 | ||
| BM04E05 | X | 57 | ||
| BM05E06 | X | 28 | ||
| BM06G07 | X | 13 | ||
| BM07F03 | X | 33 | ||
| BM09B09 | X | 13 | ||
| BM10D05 | X | 40 | ||
| BM10G05 | X | 34 |
Microbiological testing of the confirmed hit compounds.
The 13 confirmed hit compounds were tested in broth microdilution assays to determine MICs against 10 bacteria (Table 2). The organisms used in MIC determination were as suggested in CLSI guidelines plus efflux and hypersensitive mutant strains of E. coli and P. aeruginosa. Quality control was maintained by testing against prescribed antibiotics (see Table S1 in the supplemental material). There was no inhibition of wild-type E. coli observed at concentrations below 128 μg/ml by any of the compounds, and only three compounds inhibited the efflux pump mutant strain of E. coli. The two Gram-negative bacteria (H. influenzae and M. catarrhalis) and the three Gram-positive bacteria (E. faecalis, S. aureus, and S. pneumoniae) were observed to be inhibited in culture by several of the compounds. Only one compound, BM09B09, inhibited cultures of wild-type P. aeruginosa and the efflux pump mutant P. aeruginosa strain at 64 μg/ml. However, many of the compounds had good activity against the hypersensitive strain of P. aeruginosa. The compounds showing the highest levels of activity against most of the bacteria were BM03E08, BM04E04, and BM07F03 (Fig. 2). MIC values of 16 μg/ml were observed for BM03E08 against E. faecalis, H. influenzae, M. catarrhalis, and the hypersensitive strain of P. aeruginosa, and an MIC of 64 μg/ml was determined against S. pneumoniae. Compound BM04E04 exhibited MICs of 16, 8, 32, 1, 8, and 64 μg/ml against E. coli tolC, E. faecalis, H. influenzae, M. catarrhalis, the hypersensitive strain of P. aeruginosa, and S. pneumoniae, respectively. MIC values of 2, 32, 16, 16, 16, and 64 μg/ml against E. coli tolC, E. faecalis, H. influenzae, M. catarrhalis, the hypersensitive strain of P. aeruginosa, and S. pneumoniae, respectively, were observed for compound BM07F03. These three compounds demonstrated broad-spectrum activity and were selected for further analysis.
TABLE 2.
MICs of confirmed hit compounds
| Pathogen | MIC (μg/ml)a |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 02F09 | 02H08 | 03E08 | 03F03 | 03F11 | 04E04 | 04E05 | 05E06 | 06G07 | 07F03 | 09B09 | 10D05 | 10G05 | |
| E. coli | >128a | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| E. coli tolC mutant | 64 | >128 | >128 | >128 | >128 | 16 | >128 | >128 | >128 | 2 | >128 | >128 | >128 |
| E. faecalis | >128 | >128 | 16 | >128 | >128 | 8 | >128 | >128 | >128 | 32 | 64 | >128 | >128 |
| H. influenzae | 32 | 32 | 16 | >128 | 4 | 32 | >128 | 128 | 32 | 16 | 32 | >128 | >128 |
| M. catarrhalis | 32 | >128 | 16 | >128 | 128 | 1 | 128 | >128 | 64 | 16 | >128 | 128 | >128 |
| P. aeruginosa | 128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 64 | >128 | >128 |
| P. aeruginosa hypersensitive | 4 | 4 | 16 | 128 | 8 | 8 | 128 | >128 | >128 | 16 | 32 | >128 | >128 |
| P. aeruginosa efflux mutant | 128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 64 | >128 | >128 |
| S. aureus | 128 | >128 | 128 | >128 | 64 | 128 | 128 | >128 | >128 | 128 | >128 | >128 | >128 |
| S. pneumoniae | 64 | 128 | 64 | 128 | 128 | 64 | 128 | >128 | >128 | 64 | 128 | 128 | >128 |
MIC values were determined for each compound in triplicate.
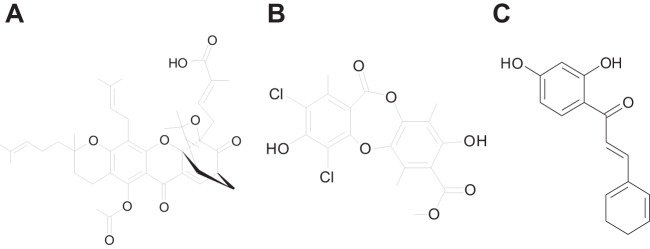
FIG 2.
Chemical structures of BM03E08 (A), BM04E04 (B), and BM07F03 (C). The chemical structure information for these compounds is available at the ChemSpider website through the substance identifier numbers 5257113, 2743198, and 4526121, respectively.
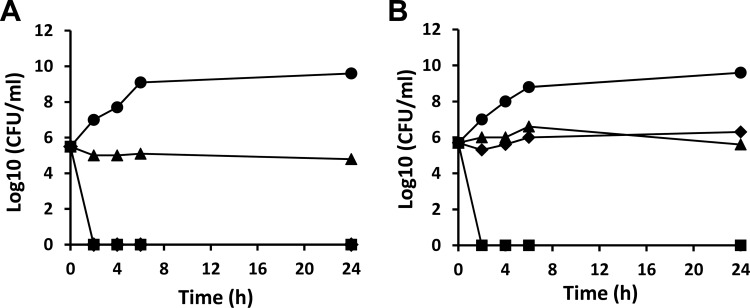
To determine mode of inhibition of bacterial growth, time-kill assays were performed using the compounds at 4× the MIC (Fig. 3). The compounds were tested against cultures of H. influenzae, a Gram-negative respiratory pathogen and E. faecalis, a Gram-positive pathogen causing endocarditis, bacteremia, urinary tract infections, meningitis, and other infections in humans. BM04E04 was observed to be bacteriostatic against both of the bacteria tested. A >1,000-fold decrease in viable cell counts within 24 h is indicative of bactericidal effects. Bactericidal activity was achieved upon exposure of BM07F03 to either of the bacterial growths. BM03E08 exhibited bacteriostatic effects against the Gram-positive organism E. faecalis; however, it was bactericidal against H. influenzae.
FIG 3.
Time-kill kinetics against selected bacteria. The activity of the hit compounds against growths of H. influenzae (A) and E. faecalis (B) was determined. Compounds were added to bacterial cultures at 4× the MIC. Samples were analyzed by plating and determination of the CFU at 0, 2, 4, 6, and 24 h. Filled triangles (▲) represent cultures containing BM04E04, filled diamonds (◆) represent cultures containing BM03E08, and filled squares (■) represent cultures containing BM07F03. Filled circles (●) represent control cultures grown in the absence of compounds.
Activity of the three selected compounds against the specific target.
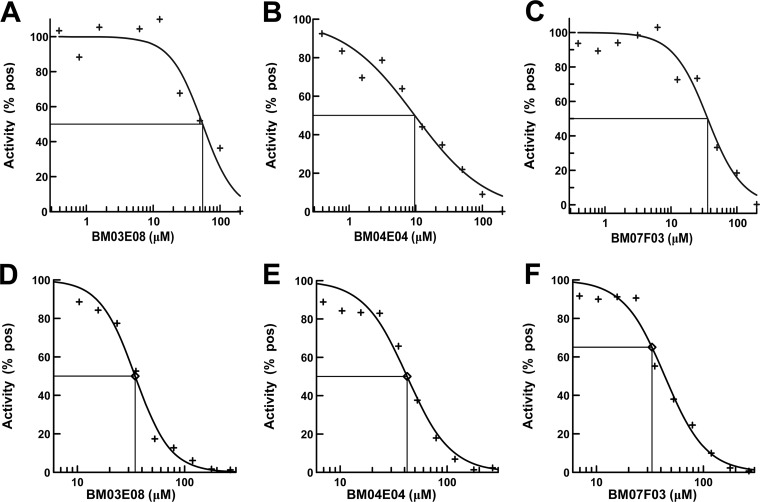
The molecular target of all three of the lead compounds was identified as P. aeruginosa PheRS (Table 1). Assays were carried out using the aminoacylation reaction to determine the IC50s of the compounds against P. aeruginosa PheRS (Fig. 4A to C). The compounds were serially diluted into reactions from 200 to 0.4 μM, and the IC50s for BM03E08, BM04E04, and BM07F03 were determined to be 55, 9.6, and 36 μM, respectively. These values are similar to the IC50s (34, 42, and 33 μM, respectively) determined in the A/T assays (Fig. 4D to F).
FIG 4.
IC50 determination for hit compounds in the aminoacylation assay compared to IC50s determined in the A/T assay. Titrations of BM03M08 (A), BM04E04 (B), and BM07F03 (C) in the aminoacylation assay were performed. The compound concentrations ranged from 0.4 to 200 μM. Titrations of BM03M08 (D), BM04E04 (E), and BM07F03 (F) in the A/T assay were performed. The concentrations ranged from 5 to 275 μM. Positives were control assays in which there were no compound. The curve fits and IC50s were determined using XLfit 5.3 (IDBS) as part of Microsoft Excel.
Compounds specifically inhibit bacterial protein synthesis.
An ideal antibacterial compound would show potent inhibition of its bacterial target but little or no inhibition of the corresponding eukaryotic system. To test for activity of the three compounds against eukaryotic protein synthesis, we first assayed for inhibition using wheat germ cell extract assays. The poly(U) mRNA, yeast tRNAPhe, [3H]phenylalanine, and Mg2+ concentrations were optimized for poly-Phe synthesis in the wheat germ cell extract assays. The lead compounds were tested, along with a known inhibitor of protein synthesis, cycloheximide (35). In these assays cycloheximide inhibited 80% of the protein synthesis at 30 μM (21). In contrast, none of the test compounds inhibited protein synthesis at concentrations up to 100 μM (see Fig. S2A to C in the supplemental material).
Mitochondria, the organelles present in eukaryotic cells, contains their own protein synthesis system, and this system is subject to inhibition, leading to cell death (36). PheRS from human mitochondria (hmPheRS) was previously cloned and characterized (27). To test whether the compounds affected the mitochondrial homolog of PheRS, they were assayed at concentrations from 0.8 to 200 μM in hmPheRS aminoacylation assays (26). In these assays, the activity of hmPheRS was not observed to be affected by any of the compounds (see Fig. S2D to F in the supplemental material).
Mechanism of action for inhibition of PheRS.
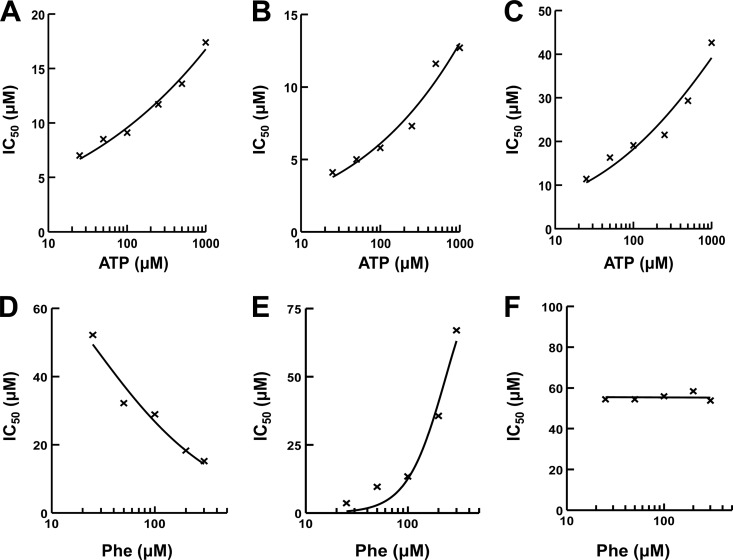
In a two-substrate reaction, the shift in IC50s with respect to various substrate concentrations is the product of the shift due to each individual substrate (37). Therefore, the IC50s for the test compounds were determined using the aminoacylation assay at various concentrations of ATP or Phe to determine the mechanism of action of the inhibitor relative to ATP and the amino acid. The mechanism of the inhibition for P. aeruginosa PheRS with respect to ATP was determined at various ATP concentrations (25, 50, 100, 250, 500, and 1,000 μM) ranging from 8-fold below to 5-fold above the Km (26). The IC50s for each of the three compounds increased with increasing ATP concentration (Fig. 5A to C), which is characteristic of a competitive inhibitor (37). To determine the mechanism of inhibition with respect to the amino acid, the same assay was used, except ATP was held constant at saturating concentrations (2.5 mM), and the IC50 was determined at different concentrations of Phe (25, 50, 100, 200, and 300 μM). The Phe concentrations ranged from ∼2-fold below to 6-fold above the Km (26). The IC50s for interaction of BM03E08 with Phe decreased with increasing Phe concentration (Fig. 5D), which is characteristic of an uncompetitive inhibitor. For the PheRS aminoacylation assay with inhibitors that are phenylalanine uncompetitive and ATP competitive, the relationship is as follows: IC50 = (1 + [ATP]/KmATP)(1 + KmPhe/[Phe])Ki (37). The IC50s for interaction of BM04E04 with Phe increased with increasing Phe concentration (Fig. 5E), which is characteristic of a competitive inhibitor. For the PheRS aminoacylation assay with inhibitors that are competitive with ATP and phenylalanine, the relationship is as follows: IC50 = (1 + [Phe]/KmPhe)(1 + [ATP]/KmATP)Ki (37). There was no apparent increase or decrease in the IC50s for interaction of BM07F03 with Phe, which is characteristic of a noncompetitive inhibitor (Fig. 5F). For the PheRS aminoacylation assay with inhibitors that are competitive with ATP and noncompetitive with phenylalanine, the presence of the noncompetitive inhibitor does not interfere with the competitive inhibition of ATP and the relationship is as follows: IC50 = (1 + [ATP]/KMATP)Ki (37). These equations were used to calculate Ki for the hit compounds with IC50s determined at 100 μM Phe and 2.5 mM ATP. In multiple determinations, BM03E08, BM04E04, and BM07F03 had mean Ki values of 2.75, 0.23, and 2.67 μM when tested against P. aeruginosa PheRS. Characterization of BM03E08 was not considered further for reasons indicated in the Discussion.
FIG 5.
Determination of the mechanism of action of the hit compounds relative to ATP and phenylalanine. IC50s were determined by using aminoacylation assays. The final compound concentrations in the IC50 reactions ranged from 0.4 to 200 μM. In assays to determine competition with ATP, the phenylalanine concentration was fixed at 100 μM. IC50s were determined for BM03E08 (A), BM04E04 (B), and BM07F03 (C) at six different ATP concentrations (25, 50, 100, 250, 500, and 1000 μM). In assays to determine competition with phenylalanine the ATP concentration was fixed at 2.5 mM. IC50s were determined for BM03E08 (D), BM04E04 (E), and BM07F03 (F) at five different Phe concentrations (25, 50, 100, 200, and 300 μM).
MMS assays.
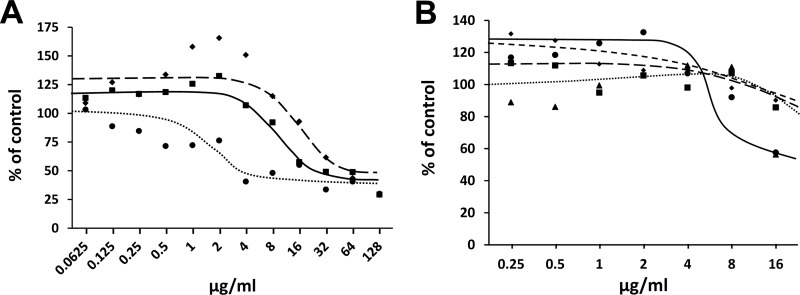
Macromolecular synthesis (MMS) assays can be used to determine the global mechanism of action of an inhibitor on the growth of bacteria. MMS assays were carried out with the test compounds to determine whether the RNA, DNA, cell wall, or protein synthesis was inhibited in bacterial cultures. Assays were carried out in cultures of E. faecalis. Compared to tetracycline, a known inhibitor of protein synthesis, both BM04E04 and BM07F03 were shown to inhibit protein synthesis (Fig. 6A) at concentrations near the previously determined MIC for each compound (see Table 2). In the time-kill assays described above, BM07F03 was shown to be bactericidal and in agreement preferentially inhibited protein synthesis in MMS assays in E. faecalis with an IC50 of ∼8.0 μg/ml (Fig. 6B). Measurements from the inhibition plots of the precursor incorporation assays resulted in IC50s of >32 μg/ml for the synthesis of RNA and DNA and for cell wall synthesis. This indicates that BM07F03 is a specific inhibitor of protein synthesis in the cell. The decreases in the synthesis of RNA, DNA, and cell wall at high compound concentrations seen in Fig. 6B represent indirect effects of the inhibition of protein synthesis. Alternatively, BM04E04 was observed to be bacteriostatic in time-kill experiments, and the expected result is the induction of the stringent response (38). Upon induction of the stringent response, RNA and DNA macromolecular synthesis ceases at the same rate as protein synthesis (39, 40). Also, in agreement, when compound BM04E04 was tested in the same MMS assays multiple times, we could not discriminate the inhibition of protein synthesis from that of RNA, DNA, or cell wall synthesis (data not shown).
FIG 6.
Macromolecular synthesis (MMS) in E. faecalis. (A) Compounds BM04E04 (◆) and BM07F03 (■) are compared to an inhibitor of protein synthesis, tetracycline (●). (B) Compound BM07F03 was tested in MMS assays to determine the effect on protein (●), RNA (▲), DNA (■), and cell wall (◆) synthesis.
Human cell cultures are affected at high concentrations of compounds.
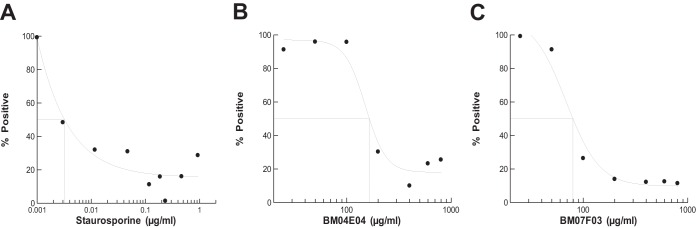
To test for cytotoxicity in human cell lines, HEK-293 cells were dosed with either a control compound, staurosporine, or with the hit compounds BM04E04 and BM07F03 (Fig. 7). Staurosporine is a potent protein kinase inhibitor that is characteristically used as a control inhibitor of cell cultures (41). The control staurosporine was serial diluted in assays from 1 to 0.001 μg/ml, and the IC50 was determined to be 0.003 μg/ml. The test compounds were diluted in assays from 800 to 25 μg/ml. The IC50s for BM04E04 and BM07F03 were determined to be 170 and 80 μg/ml, respectively. This represents 57,000- and 27,000-fold increases in the IC50 compared to the control staurosporine, indicating that substantially greater concentrations of the test compounds can be tolerated by this cell line without causing cytotoxic effects.
FIG 7.
Determination of the toxicity of the hit compounds BM04E04 and BM07F03 relative to the control staurosporine in mammalian cell cultures. MTT assays were performed with the indicated dose of drug for 24 h under standard tissue culture conditions as described in Materials and Methods. (A) The control staurosporine concentration ranged from 0.001 to 1 μg/ml in these assays. The hit compound BM04E04 (B) and BM07F03 (C) concentrations ranged from 25 to 800 μg/ml in these assays. The data points represents an average value of assays carried out in triplicate.
DISCUSSION
The results presented here show that a coupled A/T system constructed using purified components from P. aeruginosa is functional in poly-Phe synthesis and can be used to screen for compounds that inhibit protein synthesis in bacteria in a high-throughput format. We have used this system to screen a natural-compound library containing 800 compounds and identified three compounds that are bacterial protein synthesis inhibitors. Specificity screening identified PheRS as the molecular target in the A/T assay of the three compounds. When assayed in the presence of test compounds the activity of P. aeruginosa PheRS was reduced in a dose-dependent manner. The compounds identified in the A/T screen were subjected to microbiological assays in which they displayed various levels of activity against a broad-spectrum of pathogenic organisms. MIC determination indicated that the compounds had little inhibitory effect against E. coli and P. aeruginosa activity or against the efflux mutant forms of these bacteria. The compounds did exhibit good activity against the hypersensitive strain of P. aeruginosa, suggesting that the lack of activity against the wild-type strains may not be the result of efflux. There were, however, inhibitory effects against other Gram-negative organisms, including H. influenzae and M. catarrhalis. In particular, the three compounds showed good activity against the respiratory pathogen H. influenzae. All three compounds showed mixed levels of activity against the Gram-positive bacteria.
It is worth mentioning that only one compound exhibited inhibitory activity against the wild-type strain of P. aeruginosa. BM09B09 inhibited P. aeruginosa with an MIC of 64 μg/ml and had an IC50 in the P. aeruginosa A/T assay of 13 μM. Unfortunately, we have not been able to locate sources from which to acquire sufficient amounts of this compound to carry out additional testing.
In time-kill assays BM07F03 was shown to have excellent bactericidal activity against H. influenzae and E. faecalis. On the other hand, BM04E04 was bacteriostatic against both Gram-positive and Gram-negative pathogens. BM03E08 appeared to be bactericidal against Gram-negative organisms and bacteriostatic against Gram-positive organisms. Inhibition of the aminoacylation activity of an aaRS likely leads to amino acid starvation in protein biosynthesis, which can elicit the stringent response resulting in static bacterial growth, but not necessarily killing of bacteria (38). However, many aaRS proteins have secondary roles and functions; this is particularly true with eukaryotic aaRS, but it has also been observed in bacteria (42). PheRS has been shown to contain domains not specific for tRNA aminoacylation that may be linked to binding certain regions of DNA. Cell death (bactericidal activity), as seen with BM07F03, may result from inhibition of a secondary function contained within these structural regions of PheRS (43, 44). Interestingly, BM03E08 showed bactericidal activity against Gram-negative bacteria, whereas bacteriostatic activity was observed against Gram-positive bacteria. This phenomenon has also been observed with the use of phenyl-thiazolylurea-sulfonamides, a PheRS inhibitor, in which the stringent response was induced by administration of the compound in certain Gram-positive bacteria; however, in susceptible Gram-negative bacteria a bactericidal effect was observed (45, 46).
Previously, we developed the 16-membered macrolides, spiramycin and tylosin, as controls for the E. coli A/T system in which they exhibited IC50s of 0.022 and 0.038 μM, respectively. In the P. aeruginosa A/T system, IC50s of 0.31 and 0.20 μM were determined for these two macrolides, respectively, which is 1 order of magnitude higher. During the earlier work, we also tested fusidic acid as a control antibiotic against the E. coli A/T system; however, only low levels of inhibition were observed as fusidic acid was titrated at levels up to 1 mM (data not shown). Assuming similar results would be observed in the P. aeruginosa A/T system, we were surprised to find that fusidic acid inhibited protein synthesis in this system with an IC50 near 10 μM. There are two forms of EF-G (the target of fusidic acid) present in P. aeruginosa but only one (EF-G1B) functions in the elongation phase of protein synthesis (25). EF-G1B was the form of EF-G used in the P. aeruginosa A/T assay. In E. coli only one form of EF-G is present and is functional in both elongation and ribosome recycling. The specificity of EF-G in P. aeruginosa may account for the increased inhibitory effect observed for fusidic acid against P. aeruginosa protein synthesis compared to that of E. coli.
Many natural products have been previously identified and studied to different extents and natural-product libraries that are available contain many of these compounds, as well as compounds that have not been investigated. The compound library investigated here is no exception and contains both known and unknown compounds. BM03E08 is a compound known as acetyl isogambogic acid and is a member of a group of compounds known as caged Garcinia xanthones, which is a family of plant metabolites isolated from a variety of Garcinia trees (47). Gambogic acid has been shown to have both anticancer and anti-infective properties. This compound has been shown to affect growth of cancer cells in multiple ways against multiple targets, including gene regulation, as an antiapoptosis agent, by inducing production of reactive oxygen species, and as a cell cycle modifier. This compound has also been shown to inhibit bacterial growth, in particular Gram-positive; however, the mechanism of action is unknown (47). We have shown here that at least one target in bacteria is PheRS. A compound that inhibits a number of biochemical activities may be classified as a “promiscuous” compound (48), and its usefulness as a drug candidate may be limited. This compound will not be pursued further.
BM04E04 has been identified as a compound named leoidin, which is a metabolite obtained from lichens. There is a limited amount of information available for this compound. We searched the PubChem BioAssay database for the biological activity and found one assay, AID 894 from the NIH Molecular Libraries Screening Center Network, which reported a low level of inhibition of human HPGD (15-hydroxyprostaglandin dehydrogenase). Another set of assays, AID 977600-1 and AID 977603-4 from an unknown depositor, reported low to moderate levels of inhibition of sodium fluorescein uptake in OATP1B1- or OATP1B3-transfected CHO cells. Information for this compound is available in the PubChem Substance and Compound database through the unique chemical structure identifier CID 3503223. We will pursue a structure-activity relationship (SAR) analysis of this compound to improve potency.
Finally, BM07F03 has been identified as 2′4′-dihydroxychalcone, a flavonoid isolated from certain leguminous plants (49). Derivatives of this compound have shown inhibitory effects against gastric cancer (50), and in some cases this compound has been shown to have synergistic effects on the activity of antibiotics (nalidixic acid and oxacillin) against both E. coli and S. aureus (51, 52). Information for this compound is available in the PubChem Substance and Compound database through the unique chemical structure identifier CID 5376979. We will also continue to enhance the potency of this compound through SAR development.
To proceed in developing these natural-product compounds as drug candidates will require in depth studies. During follow-up studies, these compounds will be subjected to further medicinal chemistry optimization to maximize antibacterial activity and the development of an SAR series based on each compound. We would like to reduce the IC50s in the A/T protein synthesis system to the nanomolar range and decrease the MICs against our panel of bacteria. The cytotoxicity at high concentration is an issue, and promising derivatives of these compounds will be tested in cell cultures.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the financial support provided by the National Institutes of Health (grant 1SC3GM098173-01A1). The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. A portion of student support was from a departmental grant from the Robert A. Welch Foundation (grant BG-0017).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00800-16.
REFERENCES
- 1.Maschmeyer G, Braveny I. 2000. Review of the incidence and prognosis of Pseudomonas aeruginosa infections in cancer patients in the 1990s. Eur J Clin Microbiol Infect Dis 19:915–925. doi: 10.1007/s100960000410. [DOI] [PubMed] [Google Scholar]
- 2.Giamarellou H, Antoniadou A. 2001. Antipseudomonal antibiotics. Med Clin North Am 85:19–42. doi: 10.1016/S0025-7125(05)70303-5. [DOI] [PubMed] [Google Scholar]
- 3.Sahm DF, Draghi DC, Master RN, Thornsberry C, Jones ME, Karlowsky JA, Critchley IA. 2002. Pseudomonas aeruginosa antimicrobial resistance update: US resistance trends from 1998 to 2001, p 91. Abstr 42nd Intersci Conf Antimicrob Agents Chemother. [Google Scholar]
- 4.Nouer SA, Pinto M, Teixeira L, Nucci M. 2002. Risk factors for multi-drug-resistant Pseudomonas aeruginosa (MDRPa) colonization or infection in hospitalized patients, p 340. Abstr 42nd Intersci Conf on Antimicrob Agents Chemother. [Google Scholar]
- 5.Roussel P, Lamblin G. 2003. The glycosylation of airway mucins in cystic fibrosis and its relationship with lung infection by Pseudomonas aeruginosa. Adv Exp Med Biol 535:17–32. doi: 10.1007/978-1-4615-0065-0_2. [DOI] [PubMed] [Google Scholar]
- 6.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 7.Bryskier A. 2005. Antimicrobial agents: antibacterials and antifungals. ASM Press, Washington, DC. [Google Scholar]
- 8.Auerbach T, Bashan A, Harms J, Schluenzen F, Zarivach R, Bartels H, Agmon I, Kessler M, Pioletti M, Franceschi F, Yonat A. 2002. Antibiotics targeting ribosomes: crystallographic studies. Curr Drug Targets Infect Disord 2:169–186. doi: 10.2174/1568005023342506. [DOI] [PubMed] [Google Scholar]
- 9.Hermann T. 2005. Drugs targeting the ribosome. Curr Opin Struct Biol 15:355–366. doi: 10.1016/j.sbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Yonath A. 2005. Ribosomal crystallography: peptide bond formation, chaperone assistance and antibiotics activity. Mol Cells 20:1–16. doi: 10.1016/j.molcel.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Knowles DJ, Foloppe N, Matassova NB, Murchie AI. 2002. The bacterial ribosome, a promising focus for structure-based drug design. Curr Opin Pharmacol 2:501–506. doi: 10.1016/S1471-4892(02)00205-9. [DOI] [PubMed] [Google Scholar]
- 12.Sutcliffe JA. 2005. Improving on nature: antibiotics that target the ribosome. Curr Opin Microbiol 8:534–542. doi: 10.1016/j.mib.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Ravel JM, Shorey RL, Garner CW, Dawkins RC, Shive W. 1969. The role of an aminoacyl-tRNA-GTP-protein complex in polypeptide synthesis. Cold Spring Harbor Symp Quant Biol 34:321–330. doi: 10.1101/SQB.1969.034.01.039. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Yu NJ, Spremulli LL. 1998. Mutational analysis of the roles of residues in Escherichia coli elongation factor Ts in the interaction with elongation factor Tu. J Biol Chem 273:4556–4562. doi: 10.1074/jbc.273.8.4556. [DOI] [PubMed] [Google Scholar]
- 15.Sander G, Marsh RC, Voigt J, Parmeggiani A. 1975. A comparative study of the 50S ribosomal subunit and several 50S subparticles in EF-T-and EF-G-dependent activities. Biochemistry 14:1805–1814. doi: 10.1021/bi00680a001. [DOI] [PubMed] [Google Scholar]
- 16.Tate WP, Beaudet AL, Caskey CT. 1973. Influence of guanine nucleotides and elongation factors on interaction of release factors with the ribosome. Proc Natl Acad Sci U S A 70:2350–2355. doi: 10.1073/pnas.70.8.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schimmel P, Tao J, Hill J. 1998. Aminoacyl tRNA synthetases as targets for new anti-infectives. FASEB J 12:1599–1609. [PubMed] [Google Scholar]
- 18.Pratt SD, David CA, Black-Schaefer C, Dandliker PJ, Xuei X, Warrior U, Burns DJ, Zhong P, Cao Z, Saiki AY, Lerner CG, Chovan LE, Soni NB, Nilius AM, Wagenaar FL, Merta PJ, Traphagen LM, Beutel BA. 2004. A strategy for discovery of novel broad-spectrum antibacterials using a high-throughput Streptococcus pneumoniae transcription/translation screen. J Biomol Screen 9:3–11. doi: 10.1177/1087057103260876. [DOI] [PubMed] [Google Scholar]
- 19.Murray RW, Melchior EP, Hagadorn JC, Marotti KR. 2001. Staphylococcus aureus cell extract transcription-translation assay: firefly luciferase reporter system for evaluating protein translation inhibitors. Antimicrob Agents Chemother 45:1900–1904. doi: 10.1128/AAC.45.6.1900-1904.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudlicki W, Kramer G, Hardesty B. 1992. High efficiency cell-free synthesis of proteins: refinement of the coupled transcription/translation system. Anal Biochem 206:389–393. doi: 10.1016/0003-2697(92)90383-I. [DOI] [PubMed] [Google Scholar]
- 21.Ribble W, Hill WE, Ochsner UA, Jarvis TC, Guiles JW, Janjic N, Bullard JM. 2010. Discovery and analysis of 4H-pyridopyrimidines, a class of selective bacterial protein synthesis inhibitors. Antimicrob Agents Chemother 54:4648–4657. doi: 10.1128/AAC.00638-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guiles JW, Toro A, Ochsner UA, Bullard JM. 2012. Development of 4H-pyridopyrimidines: a class of selective bacterial protein synthesis inhibitors. Org Med Chem Lett 2:5. doi: 10.1186/2191-2858-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tam MF, Dodd JA, Hill WE. 1981. Physical characteristics of 16 S rRNA under reconstitution conditions. J Biol Chem 256:6430–6434. [PubMed] [Google Scholar]
- 24.Palmer SO, Rangel EY, Montalvo AE, Tran AT, Ferguson KC, Bullard JM. 2013. Cloning and characterization of EF-Tu and EF-Ts from Pseudomonas aeruginosa. Biomed Res Int 2013:585748. doi: 10.1155/2013/585748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer SO, Rangel EY, Hu Y, Tran AT, Bullard JM. 2013. Two homologous EF-G proteins from Pseudomonas aeruginosa exhibit distinct functions. PLoS One 8:e80252. doi: 10.1371/journal.pone.0080252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Palmer SO, Munoz H, Bullard JM. 2015. High-throughput screen identifies natural product inhibitor of phenylalanyl-tRNA synthetase from Pseudomonas aeruginosa and Streptococcus pneumoniae. Curr Drug Discov Technol 11:279–292. doi: 10.2174/1570163812666150120154701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bullard JM, Cai YC, Demeler B, Spremulli LL. 1999. Expression and characterization of a human mitochondrial phenylalanyl-tRNA synthetase. J Mol Biol 288:567–577. doi: 10.1006/jmbi.1999.2708. [DOI] [PubMed] [Google Scholar]
- 28.Bullard JM, Cai YC, Zhang Y, Spremulli LL. 1999. Effects of domain exchanges between Escherichia coli and mammalian mitochondrial EF-Tu on interactions with guanine nucleotides, aminoacyl-tRNA, and ribosomes. Biochim Biophys Acta 1446:102–114. doi: 10.1016/S0167-4781(99)00077-9. [DOI] [PubMed] [Google Scholar]
- 29.Clinical Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. Approved guideline M7-A7. CLSI, Wayne, PA. [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2002. Methods for determining bactericidal activity of antimicrobial agents. Approved guideline M26-A. CLSI, Wayne, PA. [Google Scholar]
- 31.Hu Y, Guerrero E, Keniry M, Manrrique J, Bullard JM. 2015. Identification of chemical compounds that inhibit the function of glutamyl-tRNA synthetase from Pseudomonas aeruginosa. J Biomed Screen 20:1160–1170. doi: 10.1177/1087057115591120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell 10:117–128. doi: 10.1016/S1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 33.Bodley JW, Zieve FJ, Lin L, Zieve ST. 1969. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem Biophys Res Commun 37:437–443. doi: 10.1016/0006-291X(69)90934-6. [DOI] [PubMed] [Google Scholar]
- 34.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. 2009. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satav JG, Katyare SS, Fatterparker P, Sreenivasan A. 1977. Study of protein synthesis in rat liver mitochondria use of cycloheximide. Eur J Biochem 73:287–296. doi: 10.1111/j.1432-1033.1977.tb11318.x. [DOI] [PubMed] [Google Scholar]
- 36.Roodyn DB, Reis PJ, Work TS. 1961. Protein synthesis in mitochondria: requirements for the incorporation of radioactive amino acids into mitochondrial protein. Biochem J 80:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng Y, Prusoff WH. 1973. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 percent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 38.Cozzone AJ. 1980. Stringent control and protein synthesis in bacteria. Biochimie 62:647–664. doi: 10.1016/S0300-9084(80)80022-8. [DOI] [PubMed] [Google Scholar]
- 39.Greenwood RC, Gentry DR. 2002. Confirmation of the antibacterial mode of action of SB-219383, a novel tyrosyl tRNA synthetase inhibitor from a Micromonospora sp. J Antibiot 55:423–426. doi: 10.7164/antibiotics.55.423. [DOI] [PubMed] [Google Scholar]
- 40.Ochsner UA, Young CL, Stone KC, Dean FB, Janjic N, Critchley IA. 2005. Mode of action and biochemical characterization of REP8839, a novel inhibitor of methionyl-tRNA synthetase. Antimicrob Agents Chemother 49:4253–4262. doi: 10.1128/AAC.49.10.4253-4262.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnier JB, Gadbois DM, Nishi K, Bradbury EM. 1994. The kinase inhibitor staurosporine induces G1 arrest at two points: effect on retinoblastoma protein phosphorylation and cyclin-dependent kinase 2 in normal and transformed cells. Cancer Res 54:5959–5963. [PubMed] [Google Scholar]
- 42.Martinis SA, Plateau P, Cavarelli J, Florentz C. 1999. Aminoacyl-tRNA synthetases: a family of expanding functions. EMBO J 18:4591–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lechler A, Kreutzer R. 1998. The phenylalanyl-tRNA synthetase specifically binds DNA. J Mol Biol 278:897–901. doi: 10.1006/jmbi.1998.1744. [DOI] [PubMed] [Google Scholar]
- 44.Dou X, Limmer S, Kreutzer R. 2001. DNA-binding of phenylalanyl-tRNA synthetase is accompanied by loop formation of the double-stranded DNA. J Mol Biol 305:451–458. doi: 10.1006/jmbi.2000.4312. [DOI] [PubMed] [Google Scholar]
- 45.Beyer D, Kroll HP, Endermann R, Schiffer G, Siegel S, Bauser M, Pohlmann J, Brands M, Ziegelbauer K, Haebich D, Eymann C, Brotz-Oesterhelt H. 2004. New class of bacterial phenylalanyl-tRNA synthetase inhibitors with high potency and broad-spectrum activity. Antimicrob Agents Chemother 48:525–532. doi: 10.1128/AAC.48.2.525-532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abibi A, Ferguson AD, Fleming PR, Gao N, Hajec LI, Hu J, Laganas VA, McKinney DC, McLeod SM, Prince DB, Shapiro AB, Buurman ET. 2014. The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability. J Biol Chem 289:21651–21662. doi: 10.1074/jbc.M114.574061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chantarasriwong O, Batova A, Chavasiri W, Theodorakis EA. 2010. Chemistry and biology of the caged Garcinia xanthones. Chemistry 16:9944–9962. doi: 10.1002/chem.201000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baell JB. 2010. Observations on screening-based research and some concerning trends in the literature. Future Med Chem 2:1529–1546. doi: 10.4155/fmc.10.237. [DOI] [PubMed] [Google Scholar]
- 49.Cardillo B, Gennaro A, Merlini L, Nasini G, Servi S. 1970. New natural chalcones from Flemingia chappar Ham. Tetrahedron Lett 11:4367–4368. [Google Scholar]
- 50.Lou C, Yang G, Cai H, Zou M, Xu Z, Li Y, Zhao F, Li W, Tong L, Wang M, Cai B. 2010. 2′,4′-Dihydroxychalcone-induced apoptosis of human gastric cancer MGC-803 cells via downregulation of surviving mRNA. Toxicol In Vitro 24:1333–1337. doi: 10.1016/j.tiv.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Talia JM, Tonn CE, Debattista NB, Pappano NB. 2012. Antibacterial efficacy of dihydroxylated chalcones in binary and ternary combinations with nalidixic acid and nalidixic acid-rutin against Escherichia coli ATCC 25922. Indian J Microbiol 52:638–641. doi: 10.1007/s12088-012-0302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talia JM, Alvarez MA, Debattista NB, Pappano NB. 2009. Susceptibility of Staphylococcus aureus strains toward combinations of oxacillin-2,4-dihydroxychalcone. Folia Microbiol 54:516–520. doi: 10.1007/s12223-009-0074-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.