Abstract
We analyzed the staphylococcal cassette chromosome mec (SCCmec) types of 143 fusidic acid- and methicillin-resistant Staphylococcus epidermidis isolates. The most frequent SCCmec type was SCCmec III/SCCHg (53%), followed by SCCmec IV (29%). Clonal spreading of SCCmec III/SCCHg strains contributed to the increased prevalence of SCCmec III. A novel non-mec SCC structure, SCC7684, adjacent to SCCmec III, which carries a new ccrC allotype (ccrC3 allele 1) and contains heavy metal resistance genes, was identified in 14 isolates.
TEXT
Methicillin resistance in staphylococci results from the production of an alternative penicillin-binding protein 2a (PBP 2a) with low affinity for β-lactam antibiotics, encoded by the mecA gene. The mecA gene is located on a mobile genetic element known as the staphylococcal cassette chromosome mec (SCCmec) (1). It has been suggested that methicillin-susceptible Staphylococcus aureus acquires SCCmec from methicillin-resistant coagulase-negative staphylococci (CoNS) and becomes methicillin-resistant S. aureus (MRSA) (2, 3). The structures and types of SCCmec in CoNS are usually more complex than in S. aureus. In addition to SCCmec, non-mec SCCs composed of ccr genes and resistance genes other than mecA have been reported, such as SCCHg, SCCfusC (4), and SCCpbp4 (5).
We previously found that the majority of fusidic acid-resistant Staphylococcus epidermidis isolates were also resistant to methicillin (6). To understand the distribution of SCCmec types, a total of 155 fusidic acid-resistant S. epidermidis isolates, including 141 clinical isolates that were collected between 2008 and 2010 in the Bacteriology Laboratory of the National Taiwan University Hospital (6) and 14 commensal isolates (7), were tested. The mecA gene was detected in 143 isolates, including 137 clinical isolates and 6 commensal isolates. Among 137 clinical isolates, 132 carried fusB, 4 carried fusC, and 1 had an fusA point mutation, all of which have been studied and published (6). All of the 6 commensal methicillin-resistant isolates carried fusB.
The SCCmec types of 143 mecA-positive isolates were determined by standard methods (1, 8–12), and the results are listed in Table 1. The most frequent SCCmec type was SCCmec III/SCCHg (n = 76, 53%), followed by SCCmec IV (n = 41, 29%). This result is different from those of other reports in which SCCmec IV was usually dominant in methicillin-resistant S. epidermidis (MRSE) (13, 14). However, the isolates tested in the present study were all fusidic acid-resistant rather than general MRSE. There were 17 SCCmec III isolates that lacked SCCHg. Of them, 3 carried only SCCmec III, and 14 contained an additional novel structure, SCC7684 (described later). All of the 6 mecA-positive commensal isolates were SCCmec type IV. An additional ccr gene was found in 2 SCCmec type IV/ccrA1B1, 1 SCCmec type IV/ccrC, and 2 SCCmec type IV/SCCfusC isolates. One SCCmec type VT/SCCfusC was identified. It is not uncommon for mecA-positive CoNS to carry multiple ccr copies or no ccr genes (3, 15, 16). SCCfusC adjacent to SCCmec III was previously found in MRSA (4). In the present study, SCCfusC was linked to SCCmec IV or SCCmec VT in S. epidermidis, which has not been reported before, suggesting that insertions of SCCmec and SCCfusC were independent. In 4 SCCmec nontypeable isolates, no ccr gene was detected. For SCCmec nontypeable isolates, there are two possibilities: the isolates may have a novel type or the target sites for the primers may have been altered. In a study in Norway, ccr-nontypeable S. epidermidis isolates were reported 52% of the time (3).
TABLE 1.
SCCmec types and pulsotypes among 143 fusidic acid-resistant S. epidermidis isolates
| SCCmec type | Other SCC, additional ccr gene, or mec complex | No. of isolates | PFGE pattern(s) (no. of isolates) |
|---|---|---|---|
| III | SCCHg | 76 | A (21), B (6), D (36), E (1), F (8), G (2), AC (1), AM (1) |
| III | 3 | A (2), D (1) | |
| III | SCC7684 | 14 | I (11), J (1), N (1), P (1) |
| IV | 36 | A (1), H (1), K (1), Q (3), T (2),b V (2), W (1), X (1),Y (2), Z (2), AA (4), AB (1),b AD (2), AE (2), AF (3), AG (3), AH (1), AI (1), AQ (3)b | |
| ccrA1B1 | 2 | Q (1), R (1) | |
| ccrC | 1 | L (1) | |
| SCCfusC | 2 | X (1), Y (1) | |
| IV varianta | 3 | L (1), M (1), AA (1) | |
| VT | SCCfusC | 1 | X (1) |
| VIII | 1 | A (1) | |
| NT1 | mec complex A | 3 | I (1), N (1), U (1) |
| NT2 | mec complex B | 1 | O (1) |
| Total | 143 |
This type is the large size of mec complex B.
These are commensal isolates collected from healthy volunteers.
Pulsed-field gel electrophoresis (PFGE) analysis divided 155 S. epidermidis isolates (143 MRSE and 12 methicillin-susceptible S. epidermidis [MSSE] isolates) into 43 clusters (Table 1). Pulsotype D was the most frequent (37/155, 24%), followed by pulsotype A (25/155, 16%). Most of the isolates that carried SCCmec III/SCCHg belonged to pulsotypes A (21/76, 28%) and D (36/76, 47%). Clonal spreading of SCCmec III/SCCHg strains may contribute to the increased prevalence of SCCmec III. Eleven of 14 isolates that contained SCCmec III/SCC7684 were pulsotype I. Isolates of SCCmec IV were distributed among different pulsotypes. Six commensal isolates that carried SCCmec IV were distributed in the following three pulsotypes: T, AB, and AQ. Some isolates with different SCCmec types clustered in the same pulsotype, such as pulsotype A (SCCmec III/SCCHg, 21 isolates; SCCmec III, 2 isolates; SCCmec IV, 1 isolate; SCCmec VIII, 1 isolate), indicating the possibility of the intraspecies transfer of SCCmec.
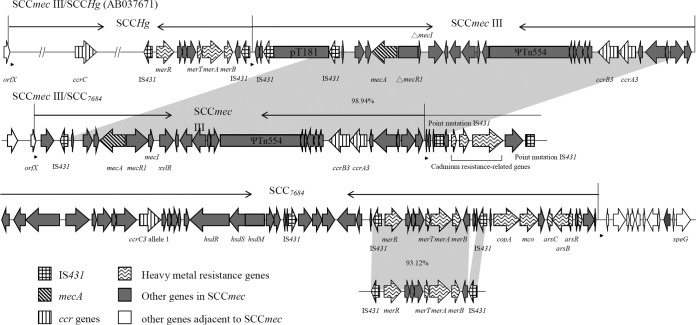
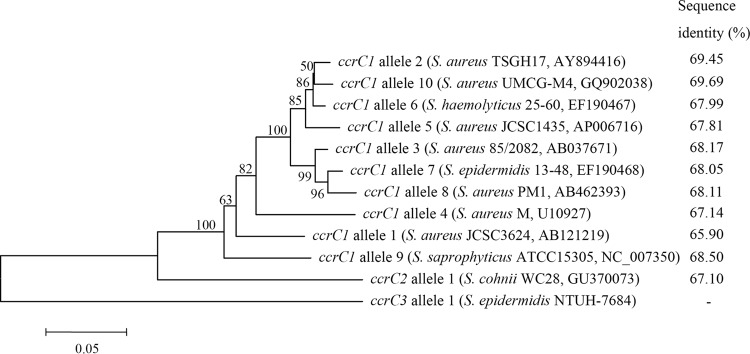
A total of 17 isolates were SCCmec type III but lacked SCCHg, which was the third most frequent type after SCCmec III/SCCHg and SCCmec IV. One isolate, NTUH-7684, was chosen for whole-genome sequencing to determine the possible novel composite SCCmec III-related element. A 56,238-bp element was found abutting the SCCmec III in NTUH-7684 (Fig. 1). The sequence of SCCmec III in NTUH-7684 showed 98.94% identity with that in S. aureus 85/2082, except for the truncated J3 region and the intact mecR1 and mecI, which were truncated in S. aureus 85/2082. A novel allotype of ccrC was identified. The 1,686-bp ccrC sequence was closest to that of the ccrC1 allele 10 in S. aureus TW20 (GenBank accession no. GQ902038; 69.69% identity) (Fig. 2) and showed 69.45% identity to ccrC1 allele 2 in S. aureus TSGH17 (GenBank accession no. AY894416). The ccrC gene in NTUH-7684 is designated ccrC3 allele 1 according to the 85% cutoff value. The phylogenetic tree based on concatenated sequences of ccrC genes indicated that ccrC3 allele 1 was phylogenetically separated from ccrC1 and ccrC2 (Fig. 2). The ccrC3 allele 1 was detected in 14 of 17 isolates with SCCmec type III lacking SCCHg by PCR using a pair of primers (SE-ccrC3-359F, 5′-GCGAAATGATGATAGGAGCA-3′, and SE-ccrC3-1368R, 5′-ATTCATAGCCTCAGCCTGC-3′). Isolates (11/14, 79%) that carried SCCmec III/SCC7684 mostly belonged to pulsotype I, indicating clonal spreading. The presence of the ccrC3 allele 1 indicated that the element was a non-mec SCC, which was referred to as SCC7684.
FIG 1.
Genetic organization of SCCmec III/SCC7684 in S. epidermidis NTUH-7684 (GenBank accession no. LC085180) compared with SCCmec III/SCCHg (GenBank accession no. AB037671) in S. aureus. Genes are shown according to their sequences. The predicted integration site sequences are indicated by arrows. Homologous regions between SCCmec are shown in shaded areas, and the numbers in the shaded areas show percent identities between the corresponding sequences.
FIG 2.
Phylogenetic relationships for the ccrC genes and sequence identity compared with ccrC3 allele 1 in S. epidermidis NTUH-7684. The phylogenetic tree was generated by using the neighbor-joining method with the MEGA6 package. Numbers at nodes are confidence levels expressed as percentages of occurrence in 500 bootstrapped resamplings. Scale bars indicate the evolutionary distance between sequences, determined by measuring the lengths of the horizontal lines connecting two organisms.
SCC7684 contained many heavy metal (cadmium, mercury, copper, and arsenic) resistance genes, genes associated with type I restriction modification systems (hsdR, hsdS, hsdM, IS431), and genes encoding hypothetical proteins. The region containing mercury resistance genes flanked by IS431 showed high similarity (93.12% identity) to the SCCHg region in S. aureus 85/2082. The 18-bp integration site sequences (ISSs), GAAGC(A/T/G)TA(T/C)CA(T/C)AA(A/G)T(A/G)A, were found at the end of SCCmec III and SCC7684 (Fig. 1). The SCCmec III and SCC7684 elements were flanked by 15-bp imperfectly matched direct repeats (DRs), (A/C)GAAGC(A/T/G)TA(T/C)CA(T/C)AA, which have also been found in other SCC elements (17, 18), suggesting that the two SCC elements integrated into the chromosome independently.
In conclusion, this is the first study to investigate the distribution of SCCmec types among fusidic acid-resistant S. epidermidis isolates. PFGE analyses of isolates carrying SCCmec III/SCCHg or SCCmec III/SCC7684 indicate that each showed clonal spreading. The SCC7684 element possessed many genes associated with heavy metal resistance, which may provide an advantage for bacterial survival. Our findings highlight the importance of characterizing the SCC-related elements in S. epidermidis.
Nucleotide sequence accession number.
The SCC7684 sequence from the S. epidermidis clinical isolate NTUH-7684 was deposited in GenBank under accession no. LC085180.
ACKNOWLEDGMENT
The Ministry of Science and Technology of Taiwan provided funding under grant MOST 103-2320-B-002-056-MY3.
REFERENCES
- 1.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berglund C, Soderquist B. 2008. The origin of a methicillin-resistant Staphylococcus aureus isolate at a neonatal ward in Sweden—possible horizontal transfer of a staphylococcal cassette chromosome mec between methicillin-resistant Staphylococcus haemolyticus and Staphylococcus aureus. Clin Microbiol Infect 14:1048–1056. doi: 10.1111/j.1469-0691.2008.02090.x. [DOI] [PubMed] [Google Scholar]
- 3.Hanssen AM, Sollid JU. 2007. Multiple staphylococcal cassette chromosomes and allelic variants of cassette chromosome recombinases in Staphylococcus aureus and coagulase-negative staphylococci from Norway. Antimicrob Agents Chemother 51:1671–1677. doi: 10.1128/AAC.00978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin YT, Tsai JC, Chen HJ, Hung WC, Hsueh PR, Teng LJ. 2014. A novel staphylococcal cassette chromosomal element, SCCfusC, carrying fusC and speG in fusidic acid-resistant methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 58:1224–1227. doi: 10.1128/AAC.01772-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen HJ, Chang YC, Tsai JC, Hung WC, Lin YT, You SJ, Tseng SP, Teng LJ. 2013. New structure of phage-related islands carrying fusB and a virulence gene in fusidic acid-resistant Staphylococcus epidermidis. Antimicrob Agents Chemother 57:5737–5739. doi: 10.1128/AAC.01433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung WC, Chen HJ, Lin YT, Tsai JC, Chen CW, Lu HH, Tseng SP, Jheng YY, Leong KH, Teng LJ. 2015. Skin commensal staphylococci may act as reservoir for fusidic acid resistance genes. PLoS One 10:e0143106. doi: 10.1371/journal.pone.0143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanssen AM, Kjeldsen G, Sollid JU. 2004. Local variants of staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob Agents Chemother 48:285–296. doi: 10.1128/AAC.48.1.285-296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother 48:2637–2651. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi N, Urasawa S, Uehara N, Watanabe N. 1999. Distribution of insertion sequence-like element IS1272 and its position relative to methicillin resistance genes in clinically important staphylococci. Antimicrob Agents Chemother 43:2780–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takano T, Higuchi W, Otsuka T, Baranovich T, Enany S, Saito K, Isobe H, Dohmae S, Ozaki K, Takano M, Iwao Y, Shibuya M, Okubo T, Yabe S, Shi D, Reva I, Teng LJ, Yamamoto T. 2008. Novel characteristics of community-acquired methicillin-resistant Staphylococcus aureus strains belonging to multilocus sequence type 59 in Taiwan. Antimicrob Agents Chemother 52:837–845. doi: 10.1128/AAC.01001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miragaia M, de Lencastre H, Perdreau-Remington F, Chambers HF, Higashi J, Sullam PM, Lin J, Wong KI, King KA, Otto M, Sensabaugh GF, Diep BA. 2009. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS One 4:e7722. doi: 10.1371/journal.pone.0007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wisplinghoff H, Rosato AE, Enright MC, Noto M, Craig W, Archer GL. 2003. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob Agents Chemother 47:3574–3579. doi: 10.1128/AAC.47.11.3574-3579.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahem S, Salmenlinna S, Lyytikainen O, Vaara M, Vuopio-Varkila J. 2008. Molecular characterization of methicillin-resistant Staphylococcus epidermidis strains from bacteraemic patients. Clin Microbiol Infect 14:1020–1027. doi: 10.1111/j.1469-0691.2008.02080.x. [DOI] [PubMed] [Google Scholar]
- 16.Ruppe E, Barbier F, Mesli Y, Maiga A, Cojocaru R, Benkhalfat M, Benchouk S, Hassaine H, Maiga I, Diallo A, Koumare AK, Ouattara K, Soumare S, Dufourcq JB, Nareth C, Sarthou JL, Andremont A, Ruimy R. 2009. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob Agents Chemother 53:442–449. doi: 10.1128/AAC.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zong Z, Lu X. 2010. Characterization of a new SCCmec element in Staphylococcus cohnii. PLoS One 5:e14016. doi: 10.1371/journal.pone.0014016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama Y, Ito T, Hiramatsu K. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 44:1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]