Abstract
Health care-associated infections present a major threat to modern medical care. Six worrisome nosocomial pathogens—Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.—are collectively referred to as the “ESKAPE bugs.” They are notorious for extensive multidrug resistance, yet persistence, or the phenotypic tolerance displayed by a variant subpopulation, remains underappreciated in these pathogens. Importantly, persistence can prevent eradication of antibiotic-sensitive bacterial populations and is thought to act as a catalyst for the development of genetic resistance. Concentration- and time-dependent aminoglycoside killing experiments were used to investigate persistence in the ESKAPE pathogens. Additionally, a recently developed method for the experimental evolution of persistence was employed to investigate adaptation to high-dose, extended-interval aminoglycoside therapy in vitro. We show that ESKAPE pathogens exhibit biphasic killing kinetics, indicative of persister formation. In vitro cycling between aminoglycoside killing and persister cell regrowth, evocative of clinical high-dose extended-interval therapy, caused a 37- to 213-fold increase in persistence without the emergence of resistance. Increased persistence also manifested in biofilms and provided cross-tolerance to different clinically important antibiotics. Together, our results highlight a possible drawback of intermittent, high-dose antibiotic therapy and suggest that clinical diagnostics might benefit from taking into account persistence.
INTRODUCTION
The discovery of antibiotics is commonly regarded as one of the greatest medical breakthroughs of the previous century. Indeed, modern medical care strongly relies on antibiotics for efficient prophylaxis and treatment of bacterial infections. However, due to widespread antimicrobial resistance, an imminent postantibiotic era threatens to nullify this achievement (1). Particularly in hospitals, clinicians are running out of treatment options for infections with antimicrobial-resistant pathogens from the ESKAPE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) (2–4). ESKAPE pathogens rank among the most prevalent causative agents in common health care-associated infections, such as pneumonia, surgical site infection, urinary tract infection, and bloodstream infection (5). Despite concerted efforts targeted at these organisms, few novel antimicrobials have been reported, and therapeutic options remain scarce (6, 7).
In addition to the well-documented burden of antibiotic resistance on health care (8), phenotypic tolerance, mainly caused by specialized survivor cells called persisters (9), is emerging as an important factor complicating treatment of numerous infections (10–12). Free-living persisters are normally killed by the host immune system, but when they are shielded from immune components, as is the case in biofilms, or when the host lacks a competent immune response, persisters can reinitiate infection when antibiotic therapy is ceased (13). Indeed, in many chronic and recalcitrant infections, the causative agents are clearly drug susceptible yet impossible to eradicate (10). For example, P. aeruginosa isolates from recalcitrant lung infections in cystic fibrosis patients treated with long-term intermittent inhaled tobramycin show hardly any increase in MICs (14). In contrast, selection for increased persister fractions under such conditions has been reported (15). Likewise, S. aureus is often involved in chronic cystic fibrosis lung infections, as well as persistent device-associated infections (16). E. faecium, P. aeruginosa, and K. pneumoniae are also commonly isolated from infected medical devices (17), and biofilm-associated chronic wound infections are often colonized with S. aureus, P. aeruginosa, Enterococcus spp., and Enterobacter cloacae (18). Apart from directly complicating antibiotic therapy, it has also been argued that persisters constitute an evolutionary reservoir from which drug-resistant variants can emerge (19, 20).
Evidently, a better understanding of persistence is required to increase the effectiveness of existing and future antibiotic compounds. In recent years, awareness of the clinical importance of persistence has grown, research efforts have increased, and consequently, important mechanistic and other insights have been generated (21–23). For example, a central role for the stringent response and toxin-antitoxin modules in Escherichia coli persistence has emerged (22, 24), and similar mechanisms seem to be implicated in the persistence of P. aeruginosa (25) and Salmonella enterica serovar Typhimurium (26). Additionally, a mechanism for persister formation based on the depletion of ATP levels was recently described in S. aureus (27). However, it remains unclear to what extent these findings can be extrapolated to other Gram-negative or Gram-positive pathogens. Overall, many questions on the molecular mechanisms and the adaptive nature of persistence remain (28).
The objective of the present study was to demonstrate persister formation in the ESKAPE pathogens. We focused on aminoglycosides, which were only last-resort antibiotics in the past 30 years due to toxicity concerns and low oral availability, meaning that resistance to these antibiotics has not yet reached the high levels seen with antibiotics from other classes (29). Consequently, and with the help of new dosing regimens to limit toxicity, aminoglycosides now play an important role in the treatment of infections with multidrug-resistant ESKAPE pathogens (29). Following a recent report of fast selection for extremely high persister levels in E. coli upon repeated rounds of lethal antibiotic dosing (30), we additionally investigated the evolutionary response of ESKAPE pathogens to conditions of periodic, high-dose antibiotic stress. In summary, our results show that all ESKAPE pathogens form persisters and that increased persistence but not resistance is highly selected for under extended-interval antibiotic administration. Furthermore, the high-persistence phenotypes of evolved ESKAPE pathogens are also valid in a biofilm model and extendable to different clinically important antibiotics.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and antibiotics.
The bacterial strains used in this study were P. aeruginosa PA14, S. aureus ATCC 33591, A. baumannii NCTC 13423, K. pneumoniae ATCC 13883, Enterobacter aerogenes ATCC 13048, and E. faecium LMG 8148. All species were routinely grown in Mueller-Hinton broth (MHB) or on lysogeny broth (LB) agar at 37°C. Liquid cultures were incubated with orbital shaking at 200 rpm. Biofilms were grown statically for 24 or 48 h from 1:100-diluted overnight cultures on hanging pegs for P. aeruginosa, A. baumannii, and E. aerogenes (31) or on the bottom of flat-well non-tissue-culture-treated microplates for S. aureus, K. pneumoniae, and E. faecium (32), reflecting the motility or nonmotility and the corresponding in vitro biofilm formation characteristics of these species. Motile bacteria readily form biofilms at the air-liquid interface of immersed pegs, while nonmotile organisms typically form biofilms at the bottom of microplate wells (32).
Amikacin was purchased as a powder from Thermo Scientific. Gentamicin, tobramycin, ciprofloxacin, colistin, and rifampin were purchased as powders from Sigma-Aldrich. Antibiotic stock solutions were prepared in ultrapure water, sterilized by filtration (pore size, 0.2 μm), and stored at 4°C (gentamicin), −80°C (meropenem), or −20°C (all others) for a maximum of 1 month before use. A rifampin stock solution was made in dimethyl sulfoxide (DMSO). Vancomycin was purchased as a ready-made, sterile 100-μg/ml DMSO solution from Sigma-Aldrich and stored at −20°C.
MIC determination.
MICs were determined using the microdilution method (33). Briefly, an overnight culture was diluted to an inoculum of approximately 5 × 105 CFU/ml in MHB and incubated with a 2-fold antibiotic concentration range for 16 to 20 h. The optical density was measured at 595 nm, and the lowest concentration inhibiting growth was reported to be the MIC.
Time- and concentration-dependent killing experiments.
Single colonies were inoculated in 5 ml of MHB and grown overnight. This culture was subsequently diluted 1:100 in 100 ml of fresh medium in an Erlenmeyer flask. After 16 h, the initial cell number was determined by plating using an automated spiral plater (EddyJet spiral plater; IUL Instruments) and colony counter (Flash&Go automated colony counter; IUL Instruments). Aliquots of 1 ml were incubated (37°C, shaking) in glass tubes in the presence of the desired concentration of amikacin or gentamicin. After incubation, the antibiotic was removed by washing with MgSO4 solution (10 mM), and the number of surviving cells was determined by plating. For concentration-dependent kill curves, the following concentrations of antibiotics were applied for 5 h: 10, 20, 50, 100, 200, 400, and 800 μg/ml. For time-dependent kill curves, cultures were incubated with 400 μg/ml of antibiotic for 0.5, 1, 2, 3, 5, 8, 12, and 24 h.
Evolution experiments.
All evolution experiments were founded from an independent clone. A preculture was grown overnight in 5 ml MHB and subsequently diluted 1:100 in 100 ml fresh MHB. This culture was incubated overnight (16 h) to start the first cycle of the evolution experiment. Each cycle consisted of three phases: (i) antibiotic killing for 5 h, (ii) removal of the antibiotic and transfer to fresh medium, and (iii) batch growth to stationary phase (34). Antibiotics were removed by three washing steps with MgSO4 solution (10 mM). Cultures were maintained in 100 ml MHB in Erlenmeyer flasks throughout, except during antibiotic treatment, which was carried out in 1-ml volumes in glass tubes. For S. aureus, E. aerogenes, and E. faecium, each cycle took 24 h. For P. aeruginosa, A. baumannii, and K. pneumoniae, we noted difficulties in regrowth to stationary phase within 24-h cycles. Therefore, we opted to use 48-h cycles for these species instead, extending only the duration of batch growth after treatment, and under these conditions, growth until stationary phase was consistently observed within each cycle. Gram-negative species were evolved using amikacin (400 μg/ml). Because amikacin was ineffective at killing Gram-positive pathogens, these were evolved using gentamicin (400 μg/ml) instead. After nine cycles, evolution experiments were terminated and glycerol stocks from endpoint populations were prepared and kept at −80°C.
Planktonic and biofilm persistence assays.
Persistence assays were carried out as described above for the time-kill experiments using a fixed treatment duration of 5 h and antibiotics at the following concentrations: amikacin and gentamicin, 400 μg/ml; ciprofloxacin and levofloxacin, 30 μg/ml; meropenem, rifampin, and colistin, 100 μg/ml; and vancomycin, 200 μg/ml. These concentrations always correspond to at least six times the MIC for the least susceptible species, except for A. baumannii, which is levofloxacin resistant and which was treated with 3.75 times the levofloxacin MIC. For cell wall-acting agents (meropenem and vancomycin), stationary-phase cultures were diluted 1:10 in fresh medium before adding antibiotics. For the quantification of persisters in biofilms, initial cell numbers were determined by washing the biofilms with phosphate-buffered saline (PBS), dislodging adherent cells in MHB–0.5% Tween 20 by gentle sonication (10 min), diluting in phosphate-buffered saline, and plating. The number of persisters in the biofilm was determined by transferring intact, washed biofilms to spent medium containing antibiotics. Spent medium was prepared separately for each strain by filter sterilization (pore size, 0.2 μm) of stationary-phase cultures. After treatment, the biofilms were washed in PBS and adherent cells were dislodged, diluted, and plated out as described above. To more closely mimic the conditions experienced during the evolution experiment, the incubation time before initiating antibiotic treatment was adjusted to 40 h instead of 16 h for planktonic tests and 48 h instead of 24 h for biofilm assays for species evolved in 48-h cycles.
Statistics.
All statistical calculations were done on log10-transformed data using GraphPad Prism (version 6.0) software. When the number of surviving cells was near the detection limit, some replicates yielded zero colonies from the lowest dilution. When this was the case, a value of half the detection limit was assigned and used for statistical calculations. Wild-type and evolved strain persister fractions for different antibiotics were analyzed using a repeated-measure two-way analysis of variance (ANOVA) and Šidák post hoc testing (significance level, α = 0.05). Biofilm persister levels for wild-type and evolved strains were compared using unpaired, two-tailed t tests (α = 0.05).
RESULTS
Persistence in the ESKAPE pathogens.
First, we assessed the susceptibilities of the ESKAPE pathogens to three commonly used aminoglycoside antibiotics: amikacin, gentamicin, and tobramycin (Table 1). According to the most recent clinical breakpoint values defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST; clinical breakpoints, version 5.0, available at http://www.eucast.org), S. aureus ATCC 33591 is clinically resistant to amikacin at a breakpoint of an MIC of >16 μg/ml and tobramycin at a breakpoint of >1 μg/ml, and A. baumannii NCTC 13423 shows clinical resistance to gentamicin at a breakpoint of >4 μg/ml.
TABLE 1.
Overview of MICs for wild-type strains and evolved high-persistence clones of the ESKAPE pathogens
| ESKAPE pathogen | MICa (μg/ml) |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK |
GEN |
TOB |
MEM |
CIP |
LEV |
RIF |
CST |
VAN |
||||||||||
| wt | hip | wt | hip | wt | hip | wt | hip | wt | hip | wt | hip | wt | hip | wt | hip | wt | hip | |
| P. aeruginosa | 1 | 1 | 1 | ND | 0.5 | 0.5 | 1.024 | 0.512 | 0.032 | 0.032 | ND | ND | ND | ND | 0.5 | 0.5 | ND | ND |
| S. aureus | 32 | ND | 2 | 2 | 512 | 512 | 16b | 16b | 0.5 | 2 | 0.25 | 0.5 | 0.008 | 0.008 | ND | ND | 0.5 | 0.5 |
| A. baumannii | 4 | 2 | >256 | ND | 2 | 1 | 4.096 | 2.048 | >32 | >32 | 8 | 8 | ND | ND | 0.5 | 0.5 | ND | ND |
| K. pneumoniae | 1 | 2 | 0.125 | ND | 0.5 | 0.5 | 0.256 | 0.256 | 0.128 | 0.128 | ND | ND | ND | ND | 2 | 2 | ND | ND |
| E. aerogenes | 4 | 4 | 0.5 | ND | 2 | 1 | 1.024 | 1.024 | 0.064 | 0.032 | ND | ND | ND | ND | 1 | 1 | ND | ND |
| E. faecium | 8b | ND | 8c | 8c | 64c | 64c | 2.048b | 2.048b | 8 | 8 | 2 | 2 | 0.008b | 0.008b | ND | ND | 0.5 | 0.5 |
Bold values indicate clinical resistance according to EUCAST guidelines (clinical breakpoints, version 5.0, available at http://www.eucast.org). Abbreviations: AMK, amikacin; GEN, gentamicin; TOB, tobramycin; MEM, meropenem; CIP, ciprofloxacin; LEV, levofloxacin; RIF, rifampin; CST, colistin; VAN, vancomycin; wt, wild type; hip, high-persistence clone; ND, not determined.
No breakpoints have been defined.
EUCAST defines no aminoglycoside breakpoints for enterococci but instead classifies strains as having either low- or high-level intrinsic resistance to aminoglycosides on the basis of their gentamicin MICs. The aminoglycoside MICs of the E. faecium strain used in this study fell below the upper limit for low-level resistance (MIC ≤ 128 μg/ml).
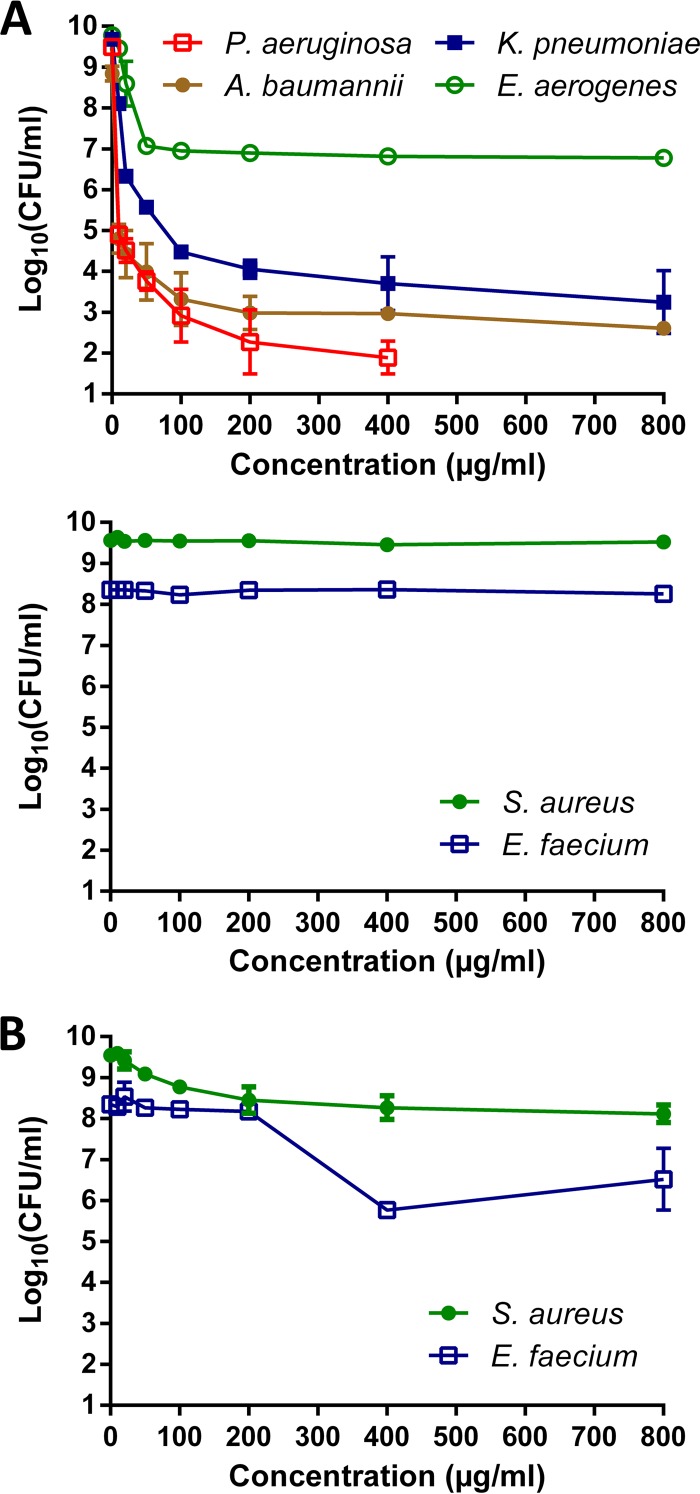
On the basis of the MICs, amikacin seemed to be the most effective agent overall. To assess its bactericidal activity, we generated concentration-kill curves from stationary-phase cultures. Cultures at the nongrowing, stationary phase were chosen over exponentially growing cultures because the latter are likely of little clinical relevance, especially in the context of chronic infections (10). During stationary phase, bacteria are usually more tolerant toward antibiotics, owing to the absence of active growth and division, high cell density, and nutritional starvation. Notably, they share these physiological aspects with biofilms, which are strongly linked with persistent infections (35). Amikacin showed strong bactericidal activity against all Gram-negative pathogens, while no killing was observed in S. aureus or E. faecium cultures (Fig. 1A). This was not unexpected, and the high tolerance of stationary-phase cultures of S. aureus has been described before (36, 37). Because gentamicin MICs for S. aureus and E. faecium indicated high susceptibility, we additionally tested concentration-dependent killing with this antibiotic. After 24 h of treatment, the number of viable cells decreased approximately 1 log for E. faecium and 4 logs for S. aureus (Fig. 1B). On the basis of the susceptibility and concentration-kill data, amikacin and gentamicin were used for the evolution experiments with Gram-negative and Gram-positive ESKAPE pathogens, respectively.
FIG 1.
Concentration-kill curves of the ESKAPE pathogens show various degrees of bactericidal activity. (A) Gram-negative ESKAPE pathogens but not Gram-positive ESKAPE pathogens are readily killed by amikacin (400 μg/ml). (B) Gentamicin possesses a modest bactericidal effect on the Gram-positive ESKAPE pathogens. Missing data points at high concentrations indicate that the number of surviving cells fell below the detection limit (10 CFU/ml). Error bars represent standard deviations (n ≥ 2).
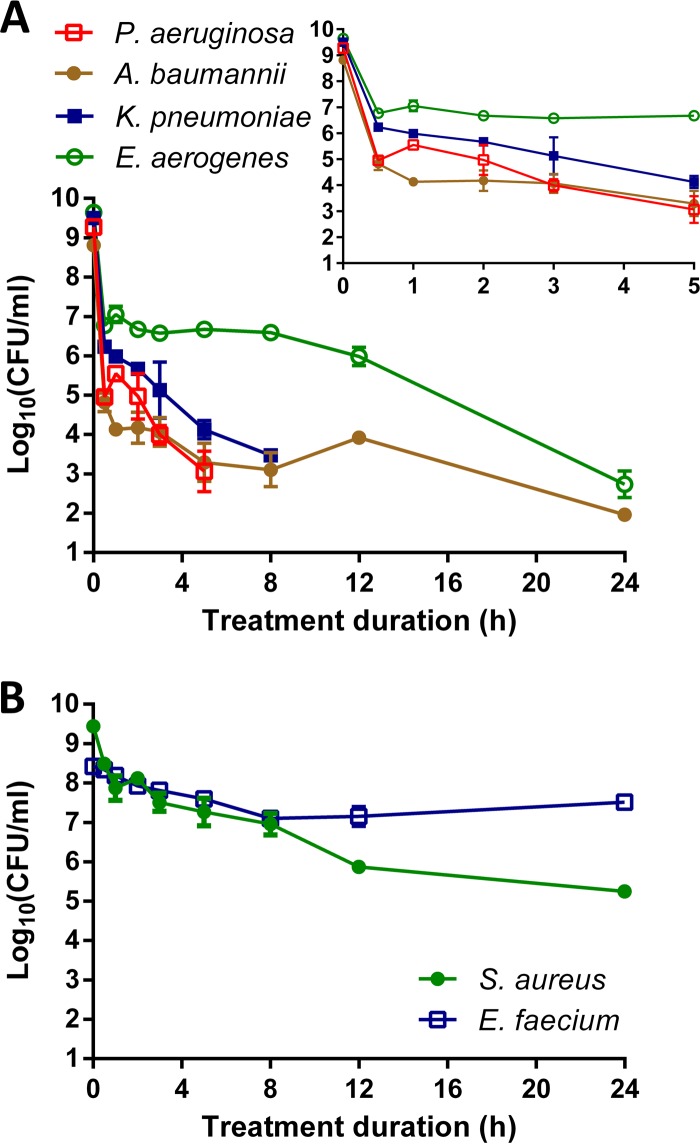
Next, we assessed the presence of persister cells. Biphasic killing kinetics are an indication of the presence of persisters, as the bulk of the population is killed rapidly, followed by a phase of slow killing due to the presence of tolerant persisters. We performed detailed time-based killing experiments using an antibiotic concentration of 400 μg/ml, which corresponds to at least 50 times the MIC and falls within the plateau phase of the concentration-kill curves in Fig. 1. Persister plateaus were clearly identifiable for all pathogens (Fig. 2). To exclude the possibility that surviving cells were resistant mutants, we confirmed that the MICs for the surviving cells were unchanged and, hence, that they were genuine persisters.
FIG 2.
Time-kill curves of the ESKAPE pathogens are biphasic and indicate the presence of persister cells. (A) Amikacin (400 μg/ml) was used for the Gram-negative pathogens K. pneumoniae, E. aerogenes, P. aeruginosa, and A. baumannii. (Inset) Magnification of the killing during the first 5 h. (B) Gentamicin (400 μg/ml) was used for the Gram-positive species S. aureus and E. faecium. Missing data points at long treatment durations indicate that the number of surviving cells fell below the detection limit (10 CFU/ml). Error bars represent standard deviations (n ≥ 2).
Periodic lethal challenge with aminoglycosides selects for increased antibiotic persistence but not antibiotic resistance.
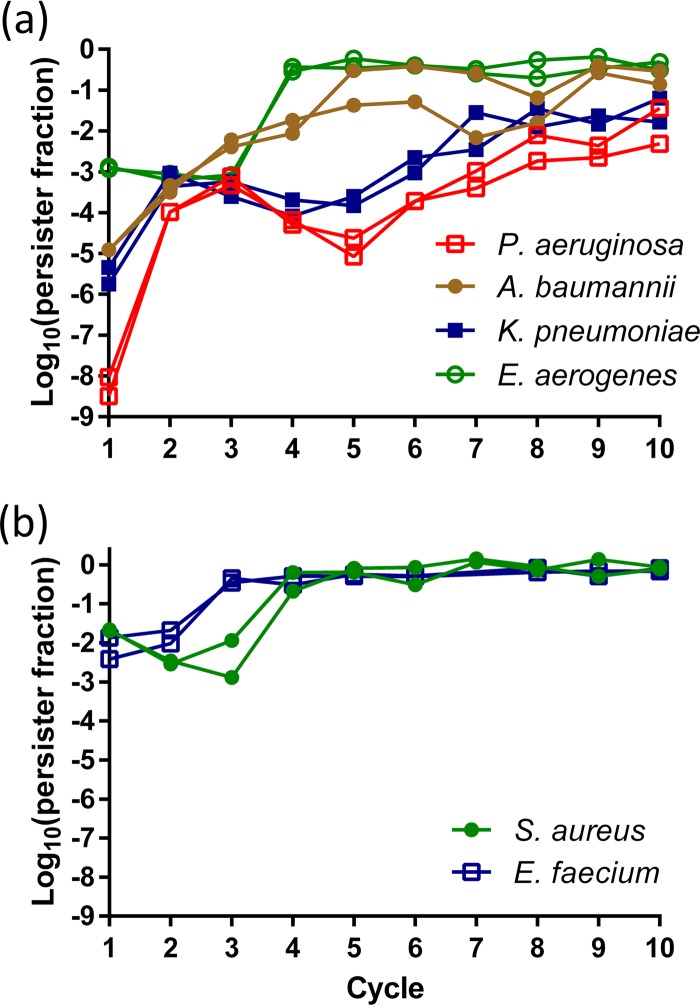
Current clinical guidelines suggest high-dose extended-interval administration of aminoglycosides, reconciling toxicity concerns with efficacy (29, 38). Theoretically, such fluctuating stress conditions are predicted to promote the evolution of antibiotic tolerance through increased persister cell formation, rather than to select for resistant mutants (39). In agreement with our earlier observations in nonpathogenic E. coli (30), all ESKAPE pathogens showed a rapid evolution toward high persistence under cyclic aminoglycoside treatment (Fig. 3). To confirm that our experimental design successfully avoided the selection of resistant mutants, endpoint populations were regrown from glycerol stocks to test for increased MICs. None of the populations' MICs deviated more than 2-fold from the ancestor's MIC.
FIG 3.
ESKAPE pathogens evolve rapidly toward high persister levels under in vitro intermittent aminoglycoside therapy. The antibiotics used were amikacin (Gram-negative pathogens) (a) or gentamicin (Gram-positive pathogens) (b) at 400 μg/ml for 5 h. Each cycle consisted of (i) antibiotic killing for 5 h, (ii) removal of the antibiotic and transfer to fresh medium, and (iii) batch growth to stationary phase. Cycles were repeated every 24 h (S. aureus, E. aerogenes, and E. faecium) or 48 h (P. aeruginosa, A. baumannii, and K. pneumoniae). Two independent replicates per species are shown.
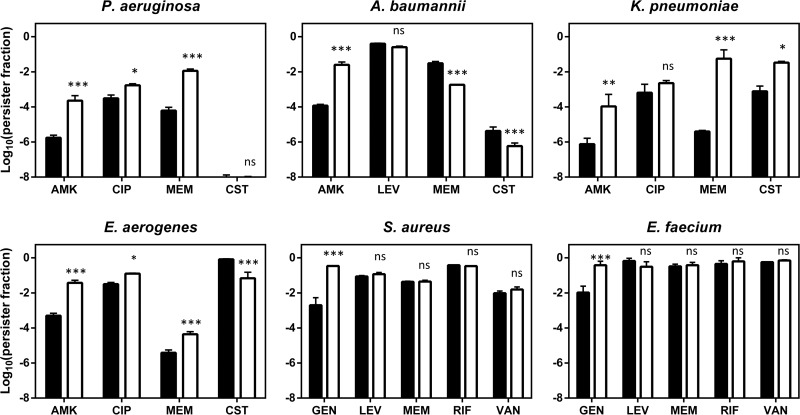
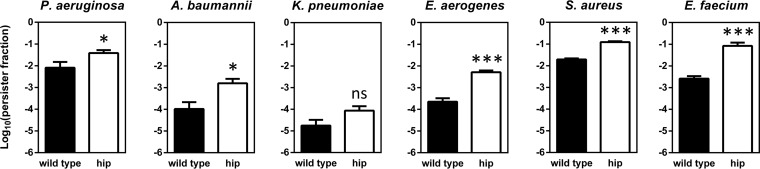
Since the evolved populations are likely heterogeneous, a random clone from one of the evolved populations was isolated for further analysis. None of these clones showed an increased MIC for the antibiotic used during evolution, nor did we observe cross-resistance or hypersensitivity to any other tested antibiotic, including aminoglycosides (amikacin, gentamicin, tobramycin), fluoroquinolones (ciprofloxacin, levofloxacin), a carbapenem β-lactam (meropenem), colistin, vancomycin, and rifampin, except for a single case where the S. aureus evolved clone showed a modest 4-fold increase in the ciprofloxacin MIC (Table 1). Confirming the evolutionary trajectories in Fig. 3, the levels of persisters among the evolved clones were significantly increased upon treatment with the antibiotic used during evolution, ranging from a 37-fold increase for E. faecium to a 213-fold increase for A. baumannii (Fig. 4).
FIG 4.
Persister levels for clinically important antibiotics in the wild type (solid bars) and clones isolated from evolved populations (open bars). Stationary-phase cultures were incubated with a high dose of antibiotic for 5 h to determine persister levels. Antibiotics were used at the following concentrations: amikacin and gentamicin, 400 μg/ml; ciprofloxacin and levofloxacin, 30 μg/ml; meropenem, rifampin, and colistin, 100 μg/ml; and vancomycin, 200 μg/ml. Error bars represent the standard errors of the means (n = 3). Statistical significance was determined using a repeated-measure two-way ANOVA with Šidák post hoc testing (α = 0.05). Symbols and abbreviations: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; LEV, levofloxacin; MEM, meropenem; CST, colistin; RIF, rifampin; VAN, vancomycin.
Evolved high-persistence clones show cross-tolerance to clinically important antibiotics.
Persistence is often denominated multidrug tolerance, as persister cells can withstand the activities of multiple antibiotics (40) and even host-related stresses, such as complement-mediated lysis (41). Therefore, we compared persister levels among the wild-type and evolved high-persistence clones with other important bactericidal antibiotics. For each pathogen we selected a representative fluoroquinolone (ciprofloxacin for all Gram-negative pathogens except A. baumannii and levofloxacin for Gram-positive pathogens and A. baumannii) and β-lactam (meropenem). For Gram-negative pathogens, we also included the last-resort antibiotic colistin, and Gram-positive species were additionally tested with rifampin and vancomycin. Except for A. baumannii, all Gram-negative pathogens showed significantly increased persister fractions with most tested antibiotics (Fig. 4). For S. aureus and E. faecium, the rate of survival of the wild type to stationary phase was already very high with all antibiotics, and no significant cross-tolerance could therefore be observed.
Increased persistence is also observed in a biofilm environment.
In vivo, persisters surviving antibiotic treatment are normally efficiently eliminated by the immune system. Therefore, persisters are clinically problematic only in those situations where they can escape the action of immune components, e.g., by residing in the protective environment of a self-produced biofilm matrix (13). This fact prompted us to investigate if the high-persistence clones also showed increased persister levels in biofilms. All evolved pathogens except K. pneumoniae showed significantly increased persister levels in a biofilm environment (Fig. 5), strengthening the clinical relevance of the high-persistence phenotype.
FIG 5.
Increased persister levels of evolved ESKAPE clones in biofilms. The antibiotics used were amikacin for P. aeruginosa, A. baumannii, K. pneumoniae, and E. aerogenes and gentamicin for S. aureus and E. faecium. Biofilms were challenged with antibiotic in spent MHB medium. Error bars represent the standard errors of the means (n ≥ 6). Statistical significance was determined using unpaired t tests (α = 0.05). Symbols and abbreviations: *, P < 0.05; ***, P < 0.001; ns, nonsignificant; hip, high-persistence clone.
DISCUSSION
Despite increased efforts in recent years, the problem of antibiotic resistance continues to grow (1, 8, 42). It has been postulated that persistence is an important contributor to the emergence of resistance because it produces a continuous reservoir of viable cells in the presence of antibiotics (19). Additionally, many of the processes involved in persistence, e.g., multiple stress responses, have also been found to accelerate adaptive evolution by increasing mutation rates and promoting horizontal gene transfer (20). In this light, a better understanding of the persistence phenomenon not only is required to fight chronic and recurrent infections but also has the added benefit to yield strategies that limit the spread of resistant pathogens.
Persistence has previously been described in P. aeruginosa (37, 43), S. aureus (37, 44), A. baumannii (45, 46), and very recently, K. pneumoniae (47), but to our knowledge, no reports on persistence in E. aerogenes or E. faecium have been published. Our results thus expand the growing list of pathogens for which persistence has been observed, and it is indeed expected that persistence is a universal trait of bacteria as insurance against unforeseeable harsh environments (39). We note that the observed levels of persisters of the different ESKAPE species varied substantially. Indeed, it is known that persister levels differ strongly between species or even between strains of the same species, for example, in A. baumannii (45).
In previous work, we showed strong selection for nonpathogenic E. coli mutants with extremely high levels of persistence under periodic aminoglycoside treatment (30). Here, we show that this is not only of fundamental clinical interest but also of important clinical interest by extending this observation to six of the most challenging human pathogens. Antibiotic dosing in our in vitro evolution experiments shares certain aspects with the current clinical practice of once-daily aminoglycoside dosing, underscoring the relevance of these findings. Although we acknowledge that our experiments do not truly simulate in vivo pharmacokinetics, they do recapitulate two important characteristics: very high, lethal peak doses followed by a considerable time window with no or only low concentrations of antibiotic. We used amikacin and gentamicin at 400 μg/ml, which is a concentration higher than what is usually achieved clinically. However, recent guidelines suggest that gentamicin and amikacin peak serum concentrations in critically ill patients should reach up to 40 and 80 μg/ml, respectively (48), and pathogens might encounter even higher concentrations upon administration through inhalation (49).
Two other recent studies have reported the evolution of antibiotic tolerance without resistance using ampicillin in E. coli (50) and daptomycin in S. aureus (51), indicating that these observations are not restricted to aminoglycosides. Studies of longitudinal isolates from long-term chronic infections have also delivered indirect evidence that the evolution of persistence occurs in vivo. In long-term P. aeruginosa lung infections, selection for increased persister levels without an increase in resistance was reported (15), and similar observations were made in uropathogenic E. coli (52) and the fungal pathogen Candida albicans (53). However, no selection for increased persistence in Burkholderia species was seen during prolonged treatment of infected lungs (54). Remarkably, increased E. coli persister levels decreased again during in vitro evolution in the absence of antibiotic stress (30), but whether this also applies to other pathogens, antibiotics, or clinical situations remains unknown.
A noteworthy finding was that, similar to what we described previously in E. coli (30), the tolerance of high-persistence clones isolated from endpoint populations was not restricted to the antibiotic used during evolution. With the exception of A. baumannii, all Gram-negative pathogens showed cross-tolerance toward at least one other antibiotic. This is remarkable, because these antibiotics were never present during evolution and have very different mechanisms of action. These results indicate that the high-persistence clones evolved a generic strategy conferring multidrug tolerance rather than an antibiotic-specific adaptation. This could have important clinical consequences, as unsuccessful antibiotic treatments might select for cross-tolerance and limit the efficacy of subsequent treatments with other antibiotics. It is also interesting to note how our observations differ from the results of recent work on the experimental evolution of aminoglycoside resistance (55). In this work, researchers describe how increased aminoglycoside resistance correlates with enhanced susceptibility to a whole range of other antibiotics, including ciprofloxacin, vancomycin, and β-lactams. The contrasting effects on cross-protection against unrelated antibiotics during the controlled evolution of aminoglycoside persistence described in our work and aminoglycoside resistance described elsewhere (55) could, hypothetically, favor the evolution of persistence over resistance in real-life situations when aminoglycosides are alternated or combined with other antibiotics.
The high-persistence phenotype, which was evolved under strictly planktonic conditions, was retained when the bacteria adopted a sessile biofilm lifestyle, suggesting that similar mechanisms underlie increased antibiotic persistence under planktonic and biofilm conditions. Biofilm persisters are clinically highly important, as they have been shown to be a major contributor to the antibiotic recalcitrance of biofilms (36, 56). A recent, large-scale survey found that 25.6% of all hospital-acquired infections were due to device-associated infections (i.e., ventilator-associated pneumonia, catheter-associated urinary tract infection, and central catheter-associated bloodstream infection) (5). These kinds of infections are usually accompanied by the formation of biofilms (57), showing that there is an urgent need for a better understanding of persister cell formation by nosocomial pathogens.
Recent studies have investigated the role of global regulators and the genetic basis of aminoglycoside persistence in E. coli (58, 59). Analysis of the genetic basis for the high-persistence phenotype of the isolated clones is outside the scope of this report, but based on our and others' previous work (30, 50, 51), we expect that the high-persistence phenotype results from only one or a few discrete mutations. In ongoing work, we are trying to identify these causal mutations and explain how they lead to increased persistence.
Taken together, we showed that ESKAPE pathogens form persisters and that this trait is under strong selective pressure during periodic, high-dose antibiotic administration, hence revealing a new adaptive strategy employed by pathogens to withstand antibiotic eradication. Moreover, the evolved high-persistence phenotype was retained in biofilms and with different classes of antibiotics. Importantly, these high-persistence mutants would go undetected in routine clinical tests and thus might form the bacterial pathogens' actual, but invisible, first line of defense against antibiotic therapy. Thus, our work adds to the emerging view that persistence is an overlooked and underestimated adaptive strategy allowing pathogens to survive antibiotic treatment.
ACKNOWLEDGMENTS
We thank Rob Lavigne and Pierre Cornelis for providing us with A. baumannii NCTC 13423 and P. aeruginosa PA14 wild-type strains.
J.E.M. and B.V.D.B. are recipients of a fellowship from the Agency for Innovation by Science and Technology (IWT) and the Research Foundation Flanders (FWO), respectively.
REFERENCES
- 1.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Rice LB. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 3.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.Pendleton JN, Gorman SP, Gilmore BF. 2013. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 5.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassetti M, Merelli M, Temperoni C, Astilean A. 2013. New antibiotics for bad bugs: where are we? Ann Clin Microbiol Antimicrob 12:12–22. doi: 10.1186/1476-0711-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. 2015. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov 14:529–542. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 8.O'Neill J. 2014. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. In Review on antimicrobial resistance. Government of the United Kingdom, London, United Kingdom. [Google Scholar]
- 9.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 10.Fauvart M, De Groote VN, Michiels J. 2011. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol 60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 11.Claudi B, Spröte P, Chirkova A, Personnic N, Zankl J, Schürmann N, Schmidt A, Bumann D. 2014. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 158:722–733. doi: 10.1016/j.cell.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser P, Regoes RR, Dolowschiak T, Wotzka SY, Lengefeld J, Slack E, Grant AJ, Ackermann M, Hardt W-D. 2014. Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol 12:e1001793. doi: 10.1371/journal.pbio.1001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 14.Burns JL, Van Dalfsen JM, Shawar RM, Otto KL, Garber RL, Quan JM, Montgomery AB, Albers GM, Ramsey BW, Smith AL. 1999. Effect of chronic intermittent administration of inhaled tobramycin on respiratory microbial flora in patients with cystic fibrosis. J Infect Dis 179:1190–1196. doi: 10.1086/314727. [DOI] [PubMed] [Google Scholar]
- 15.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant SS, Hung DT. 2013. Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence 4:273–283. doi: 10.4161/viru.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donlan RM. 2001. Biofilms and device-associated infections. Emerg Infect Dis 7:277–281. doi: 10.3201/eid0702.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siddiqui AR, Bernstein JM. 2010. Chronic wound infection: facts and controversies. Clin Dermatol 28:519–526. doi: 10.1016/j.clindermatol.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Levin BR, Rozen DE. 2006. Non-inherited antibiotic resistance. Nat Rev Microbiol 4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 20.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maisonneuve E, Gerdes K. 2014. Molecular mechanisms underlying bacterial persisters. Cell 157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 22.Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro CA, Marchal K, Beirlant J, Versées W, Hofkens J, Jansen M, Fauvart M, Michiels J. 2015. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell 59:9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Helaine S, Kugelberg E. 2014. Bacterial persisters: formation, eradication, and experimental systems. Trends Microbiol 22:417–424. doi: 10.1016/j.tim.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. 2014. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K. 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 28.Balaban NQ, Gerdes K, Lewis K, McKinney JD. 2013. A problem of persistence: still more questions than answers? Nat Rev Microbiol 11:587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- 29.Poulikakos P, Falagas ME. 2013. Aminoglycoside therapy in infectious diseases. Expert Opin Pharmacother 14:1585–1597. doi: 10.1517/14656566.2013.806486. [DOI] [PubMed] [Google Scholar]
- 30.Van den Bergh B, Michiels JE, Wenseleers T, Windels EM, Vanden Boer P, Kestemont D, De Meester L, Verstrepen KJ, Verstraeten N, Fauvart M, Michiels J. 2016. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat Microbiol 1:16020. doi: 10.1038/nmicrobiol.2016.20. [DOI] [PubMed] [Google Scholar]
- 31.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Toole GA. 2011. Microtiter dish biofilm formation assay. J Vis Exp 47:e2437. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 34.Van den Bergh B, Michiels JE, Michiels J. 2016. Experimental evolution of Escherichia coli persister levels using cyclic antibiotic treatments, p 131–143. In Methods in molecular biology. Springer Science, New York, NY. [DOI] [PubMed] [Google Scholar]
- 35.Fux CA, Costerton JW, Stewart PS, Stoodley P. 2005. Survival strategies of infectious biofilms. Trends Microbiol 13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Conlon BP, Nakayasu ES, Fleck LE, Lafleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keren I, Kaldalu N, Spoering AL, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 38.Stankowicz MS, Ibrahim J, Brown DL. 2015. Once-daily aminoglycoside dosing: an update on current literature. Am J Health Syst Pharm 72:1357–1364. doi: 10.2146/ajhp140564. [DOI] [PubMed] [Google Scholar]
- 39.Kussell E, Kishony R, Balaban NQ, Leibler S. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169:1807–1814. doi: 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiuff C, Zappala R, Regoes RR, Garner K, Baquero F, Levin BR. 2005. Phenotypic tolerance: antibiotic enrichment of noninherited resistance in bacterial populations. Antimicrob Agents Chemother 49:1438–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Putrinš M, Kogermann K, Lukk E, Lippus M, Varik V, Tenson T. 2015. Phenotypic heterogeneity enables uropathogenic Escherichia coli to evade killing by antibiotics and serum complement. Infect Immun 83:1056–1067. doi: 10.1128/IAI.02725-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 43.De Groote VN, Verstraeten N, Fauvart M, Kint CI, Verbeeck AM, Beullens S, Cornelis P, Michiels J. 2009. Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol Lett 297:73–79. doi: 10.1111/j.1574-6968.2009.01657.x. [DOI] [PubMed] [Google Scholar]
- 44.Johnson PJT, Levin BR. 2013. Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS Genet 9:e1003123. doi: 10.1371/journal.pgen.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barth VC, Rodrigues BÁ Bonatto GD, Gallo SW, Pagnussatti VE, Ferreira CAS, de Oliveira SD. 2013. Heterogeneous persister cells formation in Acinetobacter baumannii. PLoS One 8:e84361. doi: 10.1371/journal.pone.0084361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhargava N, Sharma P, Capalash N. 2014. Pyocyanin stimulates quorum sensing-mediated tolerance to oxidative stress and increases persister cell populations in Acinetobacter baumannii. Infect Immun 8:3417–3425. doi: 10.1128/IAI.01600-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren H, He X, Zou X, Wang G, Li S, Wu Y. 2015. Gradual increase in antibiotic concentration affects persistence of Klebsiella pneumoniae. J Antimicrob Chemother 70:3267–3272. doi: 10.1093/jac/dkv251. [DOI] [PubMed] [Google Scholar]
- 48.Roger C, Nucci B, Molinari N, Bastide S, Saissi G, Pradel G, Barbar S, Aubert C, Lloret S, Elotmani L, Polge A, Lefrant J-Y, Roberts JA, Muller L. 2015. Standard dosing of amikacin and gentamicin in critically ill patients results in variable and subtherapeutic concentrations. Int J Antimicrob Agents 46:21–27. doi: 10.1016/j.ijantimicag.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 49.Quon BS, Goss CH, Ramsey BW. 2014. Inhaled antibiotics for lower airway infections. Ann Am Thorac Soc 11:425–434. doi: 10.1513/AnnalsATS.201311-395FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. 2014. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 99:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 51.Mechler L, Herbig A, Paprotka K, Fraunholz M, Nieselt K, Bertram R. 2015. A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob Agents Chemother 59:5366–5376. doi: 10.1128/AAC.00643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goneau LW, Yeoh NS, Macdonald KW, Cadieux PA, Burton JP, Razvi H, Reid G. 2014. Selective target inactivation rather than global metabolic dormancy causes antibiotic tolerance in uropathogens. Antimicrob Agents Chemother 58:2089–2097. doi: 10.1128/AAC.02552-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LaFleur MD, Qi Q, Lewis K. 2010. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob Agents Chemother 54:39–44. doi: 10.1128/AAC.00860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Acker H, Sass A, Bazzini S, De Roy K, Udine C, Messiaen T, Riccardi G, Boon N, Nelis HJ, Mahenthiralingam E, Coenye T. 2013. Biofilm-grown Burkholderia cepacia complex cells survive antibiotic treatment by avoiding production of reactive oxygen species. PLoS One 8:e58943. doi: 10.1371/journal.pone.0058943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lázár V, Pal Singh G, Spohn R, Nagy I, Horváth B, Hrtyan M, Busa-Fekete R, Bogos B, Méhi O, Csörgő B, Pósfai G, Fekete G, Szappanos B, Kégl B, Papp B, Pál C. 2013. Bacterial evolution of antibiotic hypersensitivity. Mol Syst Biol 9:700. doi: 10.1038/msb.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Percival SL, Suleman L, Vuotto C, Donelli G. 2015. Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J Med Microbiol 64(Pt 4):323–334. doi: 10.1099/jmm.0.000032. [DOI] [PubMed] [Google Scholar]
- 58.Mok WWK, Orman MA, Brynildsen MP. 2015. Impacts of global transcriptional regulators on persister metabolism. Antimicrob Agents Chemother 59:2713–2719. doi: 10.1128/AAC.04908-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shan Y, Lazinski D, Rowe S, Camilli A, Lewis K. 2015. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio 6:e000078–15. doi: 10.1128/mBio.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]