Abstract
For the past decades, an acidic pH has been used to render Mycobacterium tuberculosis susceptible to pyrazinamide for in vitro testing. Here, we show that at the standard breakpoint concentration and reduced culture temperatures, pyrazinamide (PZA) is active against tuberculosis (TB) at neutral pH. This finding should help unravel the mechanism of action of PZA and allow drug susceptibility testing (DST) methods to be optimized.
INTRODUCTION
Pyrazinamide (PZA) is an important drug for TB treatment. PZA is used in standard first- and second-line therapies and is also included in many new regimens due to its unique ability to shorten therapy (1, 2).
The mechanism of action of PZA is unresolved (3), but it is commonly assumed that a low pH is required for PZA activity against Mycobacterium tuberculosis. In a widely accepted model proposed by Zhang and Mitchison (4), low pH causes the protonation of extracellular pyrazinoic acid (POA; the enzymatically activated form of PZA) required for POA to reenter mycobacteria and exert its antimicrobial effect. In addition, the reduced membrane potential at low pH was proposed to facilitate energy depletion by PZA (5). However, the activity of PZA in vivo and in vitro is directed against nonmetabolizing, or slowly metabolizing, mycobacteria (1, 6), and the role of low pH on the transcriptional remodeling of M. tuberculosis known to occur under those conditions (7–9) might also be related to the antimicrobial effects of PZA at low pH. We believe the relative contribution of the protonation and metabolic effects deserves investigation and might help elucidate PZA's mechanism of action in vivo.
Due to the incompletely resolved mechanism, developments in drug susceptibility testing (DST) have been limited to testing at reduced pH. Partly due to the suboptimal growth of the bacteria at low pH, the conditions are difficult to control, and PZA DST results in more failures and a lower test accuracy and reproducibility than those of other first-line drugs (10–12).
It was previously demonstrated that under acidic conditions, PZA activity is enhanced by lowering the temperature (13), but the effect of low temperature alone was not assessed. To investigate how dependent the action of PZA is on low pH, we determined the susceptibility of TB to PZA at reduced temperature at neutral pH.
MATERIALS AND METHODS
Strains.
The tested strains are presented in Table 1. M. tuberculosis strains 12-17995 and 12-17889 are clinical isolates from Georgia (14) from the Beijing lineage. Strain 12-17889 is closely related to the previously described clade A strains sharing a pncA I6L mutation (15). Apart from the pncA I6L mutation, no additional mutation in pncA is present in this strain.
TABLE 1.
Descriptions of the tested strains
| M. tuberculosis strain | PZA DST resulta | Test siteb | pncA mutation | pncA amino acid substitution |
|---|---|---|---|---|
| 12-17995 | Rc | KIT | A212G | H71R |
| 12-17889 | Sc | KIT | A16C | I6L |
| 72 (ATCC 35801) | S | KIT | WTd | |
| H37Ra (ATCC 25177) | S | KIT | WT | |
| ATCC 27294 (H37Rv) | S | BD | ||
| ATCC 25177 (H37Ra) | S | BD | ||
| ATCC 35801 | S | BD | ||
| BD-201 | Moderate R | BD | ||
| ATCC 35828 | R | BD | ||
| DIS529 | R | BD |
R, resistant; S, susceptible.
KIT, KIT Biomedical Research, Amsterdam, The Netherlands; BD, BD Laboratories, Sparks, MD, USA.
Based on DST (standard MGIT protocol using low pH) at Netherlands TB Reference laboratory (RIVM), Bilthoven, The Netherlands.
WT, wild type.
Microcolony-based growth rate determination.
Measurement of the effect of antimicrobials on TB microcolonies on solid medium was performed essentially as previously described (16). In short, aliquots of liquid cultures, sieved through a 5-μm-pore filter, were inoculated on 8 by 8-mm squares of porous supports on nonselective MB7H11 agar (BD, Sparks, MD, USA) supplemented with 0.5% glycerol and 10% oleic acid-albumin-dextrose-catalase (OADC) (BD). After 6 days of incubation at 37°C, the supports containing microcolonies consisting of ∼102 cells were transferred to MB7H11 agar at neutral pH with or without 100 μg/ml PZA or 100 μg/ml POA, and cultivation continued at 28°C. The supports were repetitively imaged using an automatic microscope (LumiByte BV, Nuenen, The Netherlands) before and during PZA/POA/control exposure.
MGIT PZA susceptibility testing.
M. tuberculosis strains ATCC 27294 (H37Rv), ATCC 25177 (H37Ra), ATCC 35801, BD-201, ATCC 35828, and DIS529 were harvested from Coletsos slopes in saline. After 2 sedimentation steps, suspensions were adjusted to a 0.5 McFarland standard. After further 5× dilution, a 0.5-ml suspension was inoculated in duplicate in standard MGIT G&D tubes (neutral medium [pH 6.6]) and MGIT PZA test tubes (medium at pH 5.9) supplemented with MGIT culture supplement (BD) and 100 μg/ml PZA (BD). For the growth controls, tubes were inoculated identically but with a 10-fold lower cell density without PZA. The tubes were incubated at 28 or 37°C and measured 3 times per week with a manual MGIT reader. Time to positivity was defined as the first day on which a signal of 13 or higher was obtained. Interpretation of the results was performed as follows: when the tubes with PZA became positive before the control tubes, strains were classified as resistant. When the time to positivity (TtP) of PZA tubes was delayed compared to the control tubes, strains were classified as susceptible.
RESULTS
Microcolony-based growth rate determination.
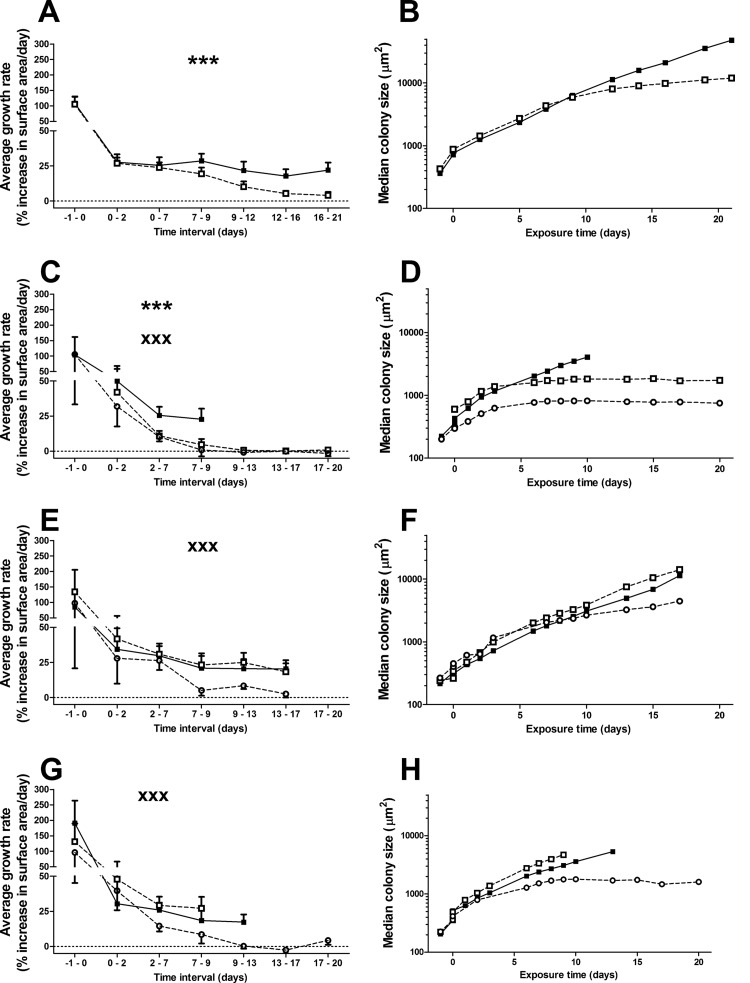
The decrease in temperature from 37°C to 28°C at the start of drug/control exposure (day 0) caused a reduction in growth rate between 70 and 80% for all strains (Fig. 1A, C, E, and G). In the presence of PZA, the growth rates of strains H37Ra and Mtb72 significantly decreased compared to the drug-free control (Fig. 1A and C). This is also visible as a flattening of the growth curves in the presence of PZA (Fig. 1B and D). Both pncA mutant strains were unaffected by PZA and had similar growth rates and growth curves with and without PZA (Fig. 1E to H). POA inhibited the growth of strain 72 and both pncA mutant strains (H37Ra data not available), as expected.
FIG 1.
Growth rates (left) and colony size (right) of PZA-susceptible and -resistant M. tuberculosis strains exposed to PZA or POA at 28°C. Strains were inoculated on solid porous supports on nonselective MB7H11 agar after 6 days of preexposure culturing at 37°C, and established microcolonies of strains H37Ra (A and B), 72 (C and D), M. tuberculosis 12-17889 (E and F), and M. tuberculosis 12-17995 (G and H) were exposed to 100 μg/ml PZA (dashed line, open squares), 100 μg/ml POA (dashed line, open circles), or no drug (continuous line, filled squares) at 28°C. (A, C, E, and G) The average daily growth rates (± the standard deviation [SD]) preexposure (measured at day −1 to 0) and during exposure (day 0 and further) of the four strains are presented. Note that the initial decreases in growth rates between days −1 to 0 and days 0 to 2 are due to the lowering of the temperature. In the presence of PZA, the growth of strains H37Ra and 72 (as indicated by ***, P < 0.0001, 2-sided t test), but not M. tuberculosis 12-17995 and M. tuberculosis 12-17889, was significantly inhibited. Growth of M. tuberculosis 72, M. tuberculosis 12-17995, and M. tuberculosis 12-17889 was significantly decreased in the presence of POA but not PZA compared to the unexposed controls (XXX, P < 0.0001). No data are available for strain H37Ra on POA exposure. (B, D, F, and H) Growth curves based on the median colony size extracted from the same images used to determine growth rates.
MGIT assay.
In the results of the standard MGIT assay (low pH, 37°C), all six strains, four of which tested susceptible, matched the known reference results of these strains. When tested at neutral pH at 28°C, identical results were obtained as those in the standard MGIT assay (Table 2): growth of the borderline-resistant strain and the three susceptible strains was inhibited in the presence of PZA compared to the unexposed controls. Cultures of two of these strains remained negative until the end of the experiment (day 26) in the presence of PZA. The cultures of the resistant strains in PZA-containing medium both became positive before their controls. The time to positivity of the controls was longer at low temperature than in the low-pH assay (10.4 days versus 6.8 days, respectively; P < 0.01) (Table 2). Identical susceptibility results were also obtained when both temperature and pH were reduced (Table 2).
TABLE 2.
Susceptibility of MTBC strains in liquid cultures to PZA at 37°C and 28°C, at neutral and acidic pHa
| Strain | Phenotype | Inoculation density G+C content (bacteria/ml) | PZA DST results by tube type and temp |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 37°C |
28°C |
|||||||||
| MGIT G&D tubes |
MGIT low-pH tubesb |
MGIT G&D tubes |
MGIT low-pH tubes |
|||||||
| Result | TtP4 (control/+PZA) (days) | Result | TtP (control/+PZA) (days) | Result | TtP (control/+PZA) (days) | Result | TtP (control/+PZA) (days) | |||
| ATCC 27294 (H37Rv) | S | 1.0 × 104 | R | 7/5 | S | 8.5/22.5 | S | 14/Nc | S | 12/N |
| ATCC 25177 (H37Ra) | S | 1.1 × 104 | R | 7/3 | S | 7/10 | S | 10/12 | S | 13/N |
| ATCC 35801 | S | 1.0 × 103 | R | 5/3 | S | 5/18 | S | 10/N | S | 10/N |
| BD-201 | Moderate Rd | 3.3 × 102 | R | 6/4 | S | 7/15.5 | S | 10/25 | S | 10/N |
| ATCC 35828 | R | 4.3 × 103 | R | 5/3 | R | 5/3 | R | 8.5/5 | R | 8.5/5 |
| DIS529 | R | 4.7 × 103 | R | 7/5 | R | 8.5/6 | R | 10/7 | R | 11/8.5 |
MTBC, M. tuberculosis complex.
MGIT PZA tubes (with or without PZA) are indicated here as “MGIT low-pH tubes” to avoid confusion.
N, both duplicates remained negative at day 26.
Normally classifies as susceptible in MGIT PZA testing.
As expected, at neutral pH and 37°C, no activity of PZA was detectable on any of the six strains tested.
DISCUSSION
Since its initial introduction, PZA susceptibility testing has routinely been performed at low pH, which is generally assumed to be a prerequisite for PZA activity. However, we demonstrate that M. tuberculosis is also susceptible to PZA at neutral pH at reduced temperature. When cultured at 28°C, pncA wild-type strains were susceptible to PZA, without acidification, at the recommended breakpoint of 100 μg/ml both in Middlebrook broth and on agar. This striking observation is supported by the recent report of Peterson et al. (17), who also demonstrated PZA activity under nonacidic conditions, and in fact by the earliest studies on PZA testing, which reported that while PZA activity was indeed absent at neutral pH, it could be detected at both acidic and basic pH (18).
The mechanism of PZA activity is not fully understood. The in vitro activity of PZA at reduced pH initially reported by McDermott and Tompsett (18) became the standard for DST. Later, Zhang and Mitchison (4) described a model for the antimycobacterial effect of PZA, in which extracellular protonation of POA facilitates its reentry into the mycobacteria, causing membrane damage and acidification of the cytoplasm. This model is not regarded as complete, and alternative mechanisms and targets, e.g., ATP depletion (19), inhibition of ribosomal protein S1 (20), aspartate decarboxylase (21), and quinolinic acid phosphoribosyltransferase (22) have been proposed, but the requirement for low pH has hardly ever been questioned. The data presented here, together with the historical data (18) and those of the recent study of Peterson et al. (17), where antimicrobial activity of PZA at neutral pH was detected in starved cultures in vitro, clearly demonstrate that an extracellular acidic pH is not a prerequisite. As PZA susceptibility can also be obtained under neutral conditions with specific adaptations, we conclude that metabolic and energetic changes that occur at reduced pH (5, 7, 8) but also under starvation (17) and other metabolic stresses slowing bacterial growth, such as low temperature, as shown here, can be sufficient to establish a PZA-susceptible bacterial phenotype. This is supported by the predominant activity of PZA against slowly or nondividing mycobacterial populations (1, 6) in vivo. The relative importance for PZA susceptibility of extracellular pH per se and bacterial phenotype, in vitro and in vivo, deserves increased attention.
PZA is an important drug for which it is notoriously difficult to perform DST and interpret DST results, partly due to the requirement for testing at low pH. Despite a prevalence of PZA resistance in multidrug-resistant (MDR)-TB of >40% in many settings (23), PZA DST is often not performed and rarely used to inform treatment. As a result of the sometimes poor reproducibility of PZA DST between laboratories, the WHO considers it acceptable practice to prescribe PZA even when a resistant DST result is obtained (24). As PZA resistance-associated pncA mutations are highly diverse, and sequencing of pncA and possibly additional genes is required to determine the presence of resistance-associated mutations, PZA DST cannot simply be replaced by practical rapid molecular assays (line probe assays, etc.) (25–28).
Developments in PZA DST have been hindered by the often-assumed need to test at a low pH. Although our testing at reduced temperature was somewhat slower than that of the standard commercial liquid medium (Table 2), we believe the data demonstrate that conditions apart from low pH should be considered for PZA DST.
Furthermore, the single discrepancy between the routine and low-temperature microcolony results was from the pncA I6L mutant strain 12-17889, which previously tested susceptible at low pH but in this study tested resistant at 28°C. This strain differs at <10 single-nucleotide polymorphisms (SNPs) from the large Russian cluster of clade A strains previously reported by Casali et al. (15, and N. Casali and S. Sengstake, personal communication). Although in MGIT under standard acidic conditions, the majority of this cluster shows PZA susceptibility, this mutation is strongly suspected to have a role in clinical resistance (15).
Further studies are urgently needed to understand PZA's mechanism of action and to link in vitro susceptibility (e.g., that at low temperature) to clinical activity. Recognizing that PZA activity is not exclusively dependent on an extracellular acidic pH (this study and reference 17) may help facilitate such studies.
ACKNOWLEDGMENTS
Clinical strains used were collected within a collaborative study with the National TB Reference Laboratory (NRL), National Center for Tuberculosis and Lung Diseases, Tbilisi, Georgia, and the National Institute for Public Health and the Environment, RIVM, Bilthoven, The Netherlands. We acknowledge Nestani Tukvadze, Nino Bablishvili, and Rusudan Aspindzelashvili (NRL) for the strain collection and Dick van Soolingen and Jessica de Beer (RIVM) for providing DST results. We acknowledge Sarah Sengstake and Indra Bergval (KIT) for typing the strains and helping to select strains for our experiments and Nicola Casali and Francis Drobniewski (Queen Mary University of London and Imperial College London, London, United Kingdom) for comparing the sequence data of the I6L mutant strain included in this study with the previously published panel of I6L mutants (Casali et al. [15]). We acknowledge LumiByte for providing access to the microscope system for colony monitoring.
BD produces and sells systems and consumables for growth, detection, and susceptibility testing for the mycobacteriology laboratory.
Salman H. Siddiqi is a consultant for Becton Dickinson Diagnostic Systems.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Hu Y, Coates AR, Mitchison DA. 2006. Sterilising action of pyrazinamide in models of dormant and rifampicin-tolerant Mycobacterium tuberculosis. Int J Tuberc Lung Dis 10:317–322. [PubMed] [Google Scholar]
- 2.Murray JF, Schraufnagel DE, Hopewell PC. 2015. Treatment of tuberculosis. A historical perspective. Ann Am Thorac Soc 12:1749–1759. doi: 10.1513/AnnalsATS.201509-632PS. [DOI] [PubMed] [Google Scholar]
- 3.Stehr M, Elamin AA, Singh M. 2015. Pyrazinamide: the importance of uncovering the mechanisms of action in mycobacteria. Expert Rev Anti Infect Ther 13:593–603. doi: 10.1586/14787210.2015.1021784. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Mitchison D. 2003. The curious characteristics of pyrazinamide: a review. Int J Tuberc Lung Dis 7:6–21. [PubMed] [Google Scholar]
- 5.Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. 2003. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother 52:790–795. doi: 10.1093/jac/dkg446. [DOI] [PubMed] [Google Scholar]
- 6.McCune RM Jr, McDermott W, Tompsett R. 1956. The fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. II. The conversion of tuberculous infection to the latent state by the administration of pyrazinamide and a companion drug. J Exp Med 104:763–802. doi: 10.1084/jem.104.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher MA, Plikaytis BB, Shinnick TM. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol 184:4025–4032. doi: 10.1128/JB.184.14.4025-4032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abramovitch RB, Rohde KH, Hsu F-F, Russell DG. 2011. aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol Microbiol 80:678–694. doi: 10.1111/j.1365-2958.2011.07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker JJ, Johnson BK, Abramovitch RB. 2014. Slow growth of Mycobacterium tuberculosis at acidic pH is regulated by phoPR and host-associated carbon sources. Mol Microbiol 94:56–69. doi: 10.1111/mmi.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Permar S, Sun Z. 2002. Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J Med Microbiol 51:42–49. doi: 10.1099/0022-1317-51-1-42. [DOI] [PubMed] [Google Scholar]
- 11.Chedore P, Bertucci L, Wolfe J, Sharma M, Jamieson F. 2010. Potential for erroneous results indicating resistance when using the Bactec MGIT 960 system for testing susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Clin Microbiol 48:300–301. doi: 10.1128/JCM.01775-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffner S, Angeby K, Sturegård E, Jönsson B, Johansson A, Sellin M, Werngren J. 2013. Proficiency of drug susceptibility testing of Mycobacterium tuberculosis against pyrazinamide: the Swedish experience. Int J Tuberc Lung Dis 17:1486–1490. [DOI] [PubMed] [Google Scholar]
- 13.Coleman D, Waddell SJ, Mitchison DA. 2011. Effects of low incubation temperatures on the bactericidal activity of anti-tuberculosis drugs. J Antimicrob Chemother 66:146–150. doi: 10.1093/jac/dkq414. [DOI] [PubMed] [Google Scholar]
- 14.Tukvadze N, Bergval IL, Bablishvili N, Bzekalava N, Schuitema ARJ, de Beer J, de Zwaan R, Alba S, van Soolingen D, Aspindzelashvili R, Anthony RM, Sengstake S. 2016. Evaluation of SNP-based genotyping to monitor tuberculosis control in a high MDR-TB setting. bioRxiv. doi: 10.1101/044370. [DOI] [Google Scholar]
- 15.Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, Kontsevaya I, Corander J, Bryant J, Parkhill J, Nejentsev S, Horstmann RD, Brown T, Drobniewski F. 2014. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet 46:279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.den Hertog AL, Menting S, Smienk ET, Werngren J, Hoffner S, Anthony RM. 2014. Evaluation of a microcolony growth monitoring method for the rapid determination of ethambutol resistance in Mycobacterium tuberculosis. BMC Infect Dis 14:380. doi: 10.1186/1471-2334-14-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson ND, Rosen BC, Dillon NA, Baughn AD. 2015. Uncoupling environmental pH and intrabacterial acidification from pyrazinamide susceptibility in Mycobacterium tuberculosis. Antimicrob Agents Chemother 59:7320–7326. doi: 10.1128/AAC.00967-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDermott W, Tompsett R. 1954. Activation of pyrazinamide and nicotinamide in acidic environments in vitro. Am Rev Tuberc 70:748–754. [DOI] [PubMed] [Google Scholar]
- 19.Lu P, Haagsma AC, Pham H, Maaskant JJ, Mol S, Lill H, Bald D. 2011. Pyrazinoic acid decreases the proton motive force, respiratory ATP synthesis activity, and cellular ATP levels. Antimicrob Agents Chemother 55:5354–5357. doi: 10.1128/AAC.00507-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi W, Zhang X, Jiang X, Ruan H, Barry CE, Wang H, Zhang W, Zhang Y. 2011. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis: a potential mechanism for shortening the duration of tuberculosis chemotherapy. Science 333:1630–1632. doi: 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi W, Chen J, Feng J, Cui P, Zhang S, Weng X, Zhang W, Zhang Y. 2014. Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosis. Emerg Microbes Infect 3:e58. doi: 10.1038/emi.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H, Shibayama K, Rimbara E, Mori S. 2014. Biochemical characterization of quinolinic acid phosphoribosyltransferase from Mycobacterium tuberculosis H37Rv and inhibition of its activity by pyrazinamide. PLoS One 9:e100062. doi: 10.1371/journal.pone.0100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.den Hertog AL, Sengstake S, Anthony RM. 2015. Pyrazinamide resistance in Mycobacterium tuberculosis fails to bite? Pathog Dis 73. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization. 2014. Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/130918/1/9789241548809_eng.pdf?ua=1&ua=1. [PubMed] [Google Scholar]
- 25.Daum LT, Fourie PB, Bhattacharyya S, Ismail NA, Gradus S, Maningi NE, Omar SV, Fischer GW. 2014. Next-generation sequencing for identifying pyrazinamide resistance in Mycobacterium tuberculosis. Clin Infect Dis 58:903–904. doi: 10.1093/cid/cit811. [DOI] [PubMed] [Google Scholar]
- 26.Miotto P, Cabibbe AM, Feuerriegel S, Casali N, Drobniewski F, Rodionova Y, Bakonyte D, Stakenas P, Pimkina E, Augustynowicz-Kopeć E, Degano M, Ambrosi A, Hoffner S, Mansjö M, Werngren J, Rüsch-Gerdes S, Niemann S, Cirillo DM. 2014. Mycobacterium tuberculosis pyrazinamide resistance determinants: a multicenter study. mBio 5(5):e01819-14. doi: 10.1128/mBio.01819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitfield MG, Warren RM, Streicher EM, Sampson SL, Sirgel FA, van Helden PD, Mercante A, Willby M, Hughes K, Birkness K, Morlock G, van Rie A, Posey JE. 2015. Mycobacterium tuberculosis pncA polymorphisms that do not confer pyrazinamide resistance at a breakpoint concentration of 100 micrograms per milliliter in MGIT. J Clin Microbiol 53:3633–3635. doi: 10.1128/JCM.01001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seiner DR, Hegde SS, Blanchard JS. 2010. Kinetics and inhibition of nicotinamidase from Mycobacterium tuberculosis. Biochemistry 49:9613–9619. doi: 10.1021/bi1011157. [DOI] [PMC free article] [PubMed] [Google Scholar]