Abstract
To improve antibiotic prescribing, we sought to establish the probability of a resistant organism in urine culture given a previous resistant culture in a setting endemic for multidrug-resistant (MDR) organisms. We performed a retrospective analysis of inpatients with paired positive urine cultures. We focused on ciprofloxacin-resistant (cipror) Gram-negative bacteria, extended-spectrum-beta-lactamase (ESBL)-producing Enterobacteriaceae, carbapenem-resistant Enterobacteriaceae (CRE), and carbapenem-resistant nonfermenters (CRNF). Comparisons were made between the frequency of each resistance phenotype following a previous culture with the same phenotype and the overall frequency of that phenotype, and odds ratios (ORs) were calculated. We performed a regression to assess the effects of other variables on the likelihood of a repeat resistant culture. A total of 4,409 patients (52.5% women; median age, 70 years) with 19,546 paired positive urine cultures were analyzed. The frequencies of cipror bacteria, ESBL-producing Enterobacteriaceae, CRE, and CRNF among all cultures were 47.7%, 30.6%, 1.7%, and 2.6%, respectively. ORs for repeated resistance phenotypes were 1.87, 3.19, 48.25, and 19.02 for cipror bacteria, ESBL-producing Enterobacteriaceae, CRE, and CRNF, respectively (P < 0.001 for all). At 1 month, the frequencies of repeated resistance phenotypes were 77.4%, 66.4%, 57.1%, and 33.3% for cipror bacteria, ESBL-producing Enterobacteriaceae, CRE, and CRNF, respectively. Increasing time between cultures and the presence of an intervening nonresistant culture significantly reduced the chances of a repeat resistant culture. Associations were statistically significant over the duration of follow-up (60 months) for CRE and for up to 6 months for all other pathogens. Knowledge of microbiology results in the six preceding months may assist with antibiotic stewardship and improve the appropriateness of empirical treatment for urinary tract infections (UTIs).
INTRODUCTION
Appropriate empirical treatment of urinary tract infections (UTIs) is important for successful treatment and prevention of complications. However, with the increasing prevalence of antibiotic-resistant urinary pathogens, the selection of an appropriate empirical agent is increasingly difficult. This is reflected in 2010 clinical practice guidelines which recommend using nitrofurantoin as a first-line agent in place of cotrimoxazole, owing to a rise in the occurrence of organisms resistant to the latter (1, 2). Resistance to other antimicrobials used in the treatment of UTIs, particularly fluoroquinolones, is also increasing, as is the prevalence of extended-spectrum-beta-lactamase (ESBL)-producing Enterobacteriaceae and multidrug-resistant (MDR) Pseudomonas aeruginosa (3–5).
The problem is particularly prominent in the hospital setting, where UTIs can present as severe infections and MDR organisms are frequent. Although inappropriate initial treatment of cystitis seldom leads to significant morbidity or mortality, failing to treat patients with severe infections may have more serious consequences. A study of hospitalized patients with UTIs due to ESBL-producing pathogens found that inappropriate empirical therapy was associated with longer treatments and lengths of hospital stay as well as higher costs (6). Among bacteremic patients, inappropriate empirical treatment is associated with higher mortality (7). One technique commonly practiced by clinicians is basing therapy on the results of previous urine cultures. Previous cultures are frequently available in the health care setting, and with the computerization of health care records, such information can easily be obtained at the point of care. However, to use this information, data are needed on the predictive value of previous cultures stratified by the time elapsed prior to the current infection.
Given the challenge of determining a priori which patients will ultimately have infection due to a resistant organism, we sought to determine the value of a previous culture with a resistant uropathogen for predicting growth of resistant bacteria in a subsequent culture among hospitalized patients in a setting where MDR organisms are endemic.
MATERIALS AND METHODS
We performed a retrospective study of all patients with repeat positive urine cultures at Rambam Health Care Campus, a 900-bed primary and tertiary hospital in northern Israel, from 2011 to 2015. All the urine cultures were taken for a clinical indication (suspicion of infection). We included patients with polymicrobial cultures and cultures yielding growth of Gram-positive bacteria and yeasts; multiple cultures taken on the same date were included and assessed similarly to polymicrobial cultures. Negative (sterile) cultures were excluded from the analysis. Cultures were analyzed per patient based on the presence or absence of resistance profiles on a given date, regardless of the number of organisms present. Patients could be considered positive for more than one resistance profile simultaneously on a given date.
Data were obtained from the electronic registry of the hospital, which records demographic, administrative, and microbiological data. Patients were identified through the microbiology reports. Antibiotic susceptibility testing was performed using a Vitek 2.0 system and Etest based on CLSI criteria. ESBL-producing Enterobacteriaceae were defined as Enterobacteriaceae resistant to all third-generation cephalosporins, including ceftazidime.
Cultures were assessed for the presence of any ciprofloxacin-resistant (cipror) Gram-negative bacteria, ESBL-producing Enterobacteriaceae, carbapenem-resistant Enterobacteriaceae (CRE), or carbapenem-resistant nonfermenters (CRNF; including Pseudomonas and Acinetobacter species). In the first analysis, we compared the presence of the resistance phenotype in the repeat urine culture and the immediately previous positive culture. In the second analysis, we compared resistances in urine cultures with those in any previous positive culture. The chance of a subsequent culture being positive for the same phenotype of resistance as the previous culture was determined. We compared it to the prevalence of resistance in our population (for the same phenotype) among all repeat urine cultures by using the χ2 test. Odds ratios (ORs) with 95% confidence intervals (CIs) and other test characteristics (sensitivity, specificity, positive predictive value [PPV], and negative predictive value [NPV]) are reported for a repeat resistant organism given a known prior resistant organism compared to the baseline prevalence of resistance. Additionally, we analyzed the likelihood of an antibiotic-sensitive organism (i.e., no cipror bacteria, ESBL-producing Enterobacteriaceae, CRE, or CRNF) given prior cultures with only sensitive organisms. A multivariate analysis was performed to determine risk factors for repeated growth of resistant organisms in patients with a previous resistant organism. A sensitivity analysis was performed by excluding cultures taken within 14 days of one another. All analyses were performed using SPSS 23 and VassarStats.
RESULTS
A total of 4,409 patients with 19,546 paired positive urine cultures were analyzed from a total of 26,692 positive urine cultures during the study period. About half (52.5%) of the patients were female, with a median (interquartile range [IQR]) age of 70 (55 to 81) years. The department of admission at the time when the final culture was taken was the emergency room (ER) for 15.0% of patients, medicine for 36.6%, surgery for 46.2%, and intensive care unit (ICU) for 2.1%. The overall frequencies of cipror bacteria, ESBL-producing Enterobacteriaceae, CRE, and CRNF in the cultures included in the analysis were 49.9%, 26.5%, 1.7%, and 2.8%, respectively.
All four resistance phenotypes were significantly more frequent following a previous culture with the same phenotype, regardless of organism identity. For patients whose last urine culture grew a resistant organism, the ORs for a culture positive for cipror bacteria, ESBL-producing Enterobacteriaceae, CRE, and CRNF were 1.87 (95% CI, 1.74 to 2.01), 3.19 (95% CI, 2.91 to 3.49), 48.25 (95% CI, 34.13 to 68.22), and 19.02 (95% CI, 14.38 to 25.17), respectively (P < 0.001 for all), for the overall follow-up duration, with a median (IQR) of 26 (7 to 104) days between the immediately previous culture and the final urine culture. For patients with any previously positive culture with a resistance phenotype, the ORs were 1.64 (95% CI, 1.54 to 1.75), 2.42 (95% CI, 2.23 to 2.61), 31.74 (95% CI, 23.66 to 42.58), and 9.82 (95% CI, 7.80 to 12.36), respectively (P < 0.001 for all), with a median (IQR) of 34 (9 to 122) days between cultures for cipror bacteria, 45 (11 to 156) days for ESBL-producing Enterobacteriaceae, 55 (11 to 190) days for CRE, and 49 (14 to 189) days for CRNF. Except for ciprofloxacin resistance, specificity and NPVs were high, while all other diagnostic performance values were low (Table 1).
TABLE 1.
Test characteristics for effects of previous resistance profiles
| Culture group and resistance phenotype | Median (95% CI) |
|||
|---|---|---|---|---|
| Sensitivity | Specificity | Positive predictive value | Negative predictive value | |
| Immediately previous culture | ||||
| cipror | 0.38 (0.37–0.39) | 0.76 (0.75–0.77) | 0.68 (0.66–0.69) | 0.47 (0.46–0.48) |
| ESBL producer | 0.33 (0.31–0.34) | 0.87 (0.86–0.88) | 0.56 (0.54–0.58) | 0.72 (0.71–0.72) |
| CRE | 0.29 (0.24–0.35) | 0.99 (0.99–0.99) | 0.48 (0.40–0.56) | 0.98 (0.98–0.98) |
| CRNF | 0.24 (0.20–0.29) | 0.98 (0.98–0.99) | 0.37 (0.31–0.43) | 0.97 (0.97–0.97) |
| Sensitive to all antibiotics tested | 0.66 (0.64–0.67) | 0.64 (0.62–0.65) | 0.58 (0.57–0.60) | 0.71 (0.69–0.72) |
| Any previous culture | ||||
| cipror | 0.42 (0.41–0.43) | 0.69 (0.68–0.70) | 0.65 (0.63–0.66) | 0.47 (0.46–0.48) |
| ESBL producer | 0.38 (0.37–0.40) | 0.80 (0.79–0.80) | 0.49 (0.47–0.51) | 0.72 (0.71–0.72) |
| CRE | 0.34 (0.29–0.40) | 0.98 (0.98–0.99) | 0.38 (0.32–0.44) | 0.98 (0.98–0.98) |
| CRNF | 0.30 (0.26–0.35) | 0.96 (0.95–0.96) | 0.23 (0.20–0.27) | 0.97 (0.97–0.97) |
| Sensitive to all antibiotics tested | 0.54 (0.53–0.56) | 0.75 (0.74–0.76) | 0.62 (0.61–0.64) | 0.68 (0.67–0.69) |
Excluding cultures taken within 14 days of one another yielded similar results. For patients whose last urine culture grew a resistant organism, the ORs for a culture positive for cipror bacteria, ESBL-producing Enterobacteriaceae, CRE, and CRNF were 1.86 (95% CI, 1.70 to 2.05), 3.30 (95% CI, 2.94 to 3.71), 41.41 (95% CI, 26.17 to 65.53), and 12.64 (95% CI, 8.32 to 19.22), respectively (P < 0.001 for all). For patients with any previously positive culture with a resistance phenotype, the ORs were 1.94 (95% CI, 1.79 to 2.10), 2.78 (95% CI, 2.54 to 3.05), 31.67 (95% CI, 22.24 to 45.11), and 7.30 (95% CI, 5.41 to 9.86), respectively (P < 0.001 for all).
For patients whose previous urine culture grew a sensitive organism, i.e., lacking all resistance phenotypes assessed, the OR for a sensitive organism in the next culture was 3.37 (95% CI, 3.10 to 3.67) (P < 0.001). For patients in whom all previous cultures grew sensitive organisms, the value was 3.55 (95% CI, 3.26 to 3.86) (P < 0.001). The sensitivity, specificity, PPV, and NPV were low (Table 1).
As shown in Fig. 1, the frequency of a repeat culture positive for cipror bacteria or ESBL-producing Enterobacteriaceae increased in the first 2 weeks after the previous culture and thereafter decreased, whereas the frequencies of repeat CRE and CRNF cultures decreased continuously with time from the initial culture. The initially lower frequencies of cipror bacteria and ESBL-producing Enterobacteriaceae seen in cultures taken within 7 days of the previous culture reflected the predominance of cultures taken during the same admission; among repeat cultures taken on new admissions, the rates of cipror bacteria and ESBL-producing Enterobacteriaceae were 72.1% and 63.0% (versus 54.6% and 40.5% in cultures from the same admission), respectively. The frequencies of all four resistance phenotypes decreased steadily starting 2 weeks following the initial culture. Among cultures taken approximately 1 and 3 months following previous cultures with a resistance phenotype, the frequencies of cultures with the same phenotype were 77.4%, 66.4%, 57.1%, and 33.3% at 1 month and 73.8%, 47.1%, 33.3%, and 0% at 3 months for cipror bacteria, ESBL-producing Enterobacteriaceae, CRE, and CRNF, respectively.
FIG 1.
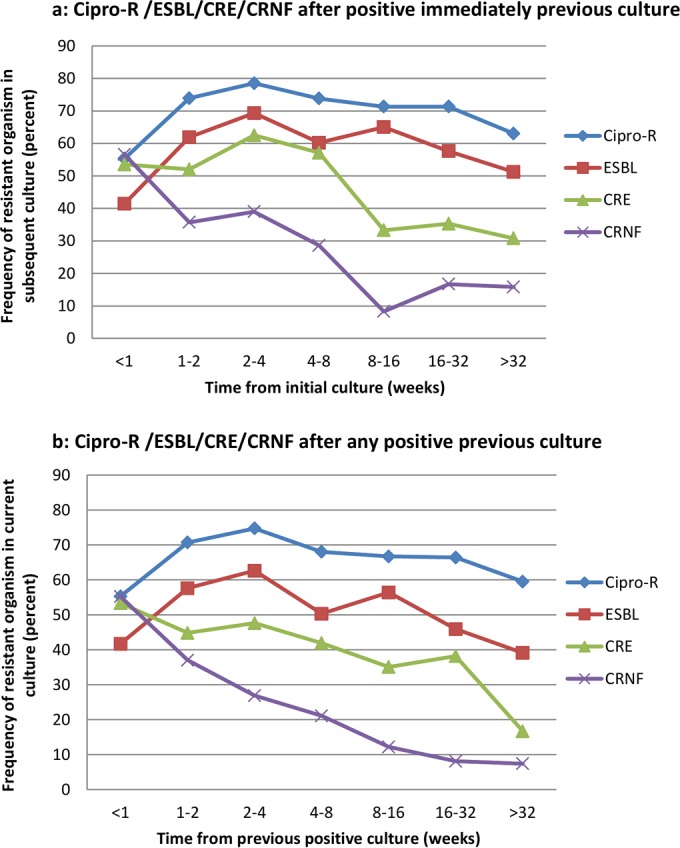
(a) Frequencies of cipror bacteria, ESBL-producing Enterobacteriaceae, CRE, and CRNF after positive immediately previous culture. (b) Frequencies of cipror bacteria, ESBL-producing Enterobacteriaceae, CRE, and CRNF after any positive previous culture. Cipro-R, ciprofloxacin-resistant bacteria; CRE, carbapenem-resistant Enterobacteriaceae; CRNF, carbapenem-resistant nonfermenters; ESBL, extended-spectrum-beta-lactamase-producing Enterobacteriaceae.
Multivariate analyses were performed for each of the resistance phenotypes (Table 2). Increasing time between cultures and the presence of at least one intervening culture negative for the resistance phenotype decreased the risk of a subsequent culture being positive for a resistant organism for all four resistance phenotypes. For cipror bacteria, increasing age and male gender were associated with increased risk; for ESBL-producing Enterobacteriaceae, male gender and emergency room admission increased risk; for CRE, no other factors significantly affected risk; and for CRNF, increasing age decreased risk. Results were similar for exclusion of repeat cultures performed within 14 days of each other, although increasing time was not significantly associated with a drop in the risk of CRE, age was not associated with the risk of CRNF, and hospital department was positively associated with the risk of cipror bacteria (data not shown).
TABLE 2.
Factors associated with a repeat resistant pathogen, by resistance phenotype
| Resistance phenotype | Risk factora | All isolates |
Isolates excluding cultures taken within 14 days of each other |
||||
|---|---|---|---|---|---|---|---|
| Univariate P value | Multivariate P value | Multivariate OR (95% CI) | Univariate P value | Multivariate P value | Multivariate OR (95% CI) | ||
| cipror | Age | 0.033 | 0.019 | 1.004 (1.001–1.007) | 0.009 | 0.002 | 1.006 (1.002–1.010) |
| Male gender | <0.001 | <0.001 | 1.288 (1.151–1.440) | <0.001 | 0.001 | 1.274 (1.108–1.464) | |
| Time between cultures | 0.014 | 0.016 | 1.000 (0.999–1.000) | <0.001 | <0.001 | 0.999 (0.999–1.000) | |
| Intervening cultures without cipror | <0.001 | <0.001 | 0.565 (0.496–0.645) | <0.001 | <0.001 | 0.455 (0.392–0.528) | |
| ER | 0.024 | 0.508 | 0.610 | ||||
| Surgery | 0.009 | 0.406 | <0.001 | 0.292 | |||
| ICU | 0.017 | 0.115 | 0.019 | 0.008 | 0.397 (0.201–0.783) | ||
| Medicine | <0.001 | 0.283 | <0.001 | 0.022 | 0.796 (0.655–0.968) | ||
| ESBL producer | Age | 0.011 | 0.069 | 0.008 | 0.062 | ||
| Male gender | <0.001 | <0.001 | 1.386 (1.206–1.593) | 0.003 | 0.005 | 1.271 (1.075–1.501) | |
| Time between cultures | 0.013 | <0.001 | 0.999 (0.999–1.000) | <0.001 | <0.001 | 0.999 (0.999–0.999) | |
| Intervening cultures without ESBL producer | <0.001 | <0.001 | 0.449 (0.385–0.524) | <0.001 | <0.001 | 0.354 (0.298–0.420) | |
| ER | <0.001 | <0.001 | 1.552 (1.264–1.905) | 0.001 | 0.018 | 1.314 (1.048–1.648) | |
| Surgery | 0.017 | 0.663 | 0.019 | 0.957 | |||
| ICU | 0.215 | 0.606 | |||||
| Medicine | 0.487 | 0.900 | |||||
| CRE | Age | 0.222 | 0.904 | ||||
| Male gender | 0.662 | 0.072 | |||||
| Time between cultures | <0.001 | 0.044 | 0.999 (0.997–1.000) | 0.001 | 0.059 | ||
| Intervening cultures without CRE | <0.001 | 0.002 | 0.374 (0.200–0.699) | 0.001 | 0.012 | 0.417 (0.210–0.827) | |
| ER | 0.805 | 0.949 | |||||
| Surgery | 0.740 | 0.679 | |||||
| ICU | 1.000 | 1.000 | |||||
| Medicine | 0.841 | 0.566 | |||||
| CRNF | Age | 0.001 | <0.001 | 0.978 (0.967–0.990) | 0.359 | ||
| Male gender | 0.548 | 0.718 | |||||
| Time between cultures | <0.001 | <0.001 | 0.996 (0.994–0.998) | <0.001 | 0.032 | 0.998 (0.996–1.000) | |
| Intervening cultures without CRNF | <0.001 | <0.001 | 0.287 (0.175–0.471) | <0.001 | <0.001 | 0.333 (0.182–0.609) | |
| ER | 0.359 | 0.935 | |||||
| Surgery | 0.025 | 0.297 | 0.027 | 0.243 | |||
| ICU | 0.231 | 1.000 | |||||
| Medicine | 0.019 | 0.333 | 0.020 | 0.235 | |||
Odds ratios for the time between cultures are values per day. “ER,” “surgery,” “ICU,” and “medicine” indicate patient locations at the times of second culture.
DISCUSSION
We analyzed whether the presence of a resistant organism in a previous urine culture predicts the growth of an organism with the same resistance phenotype in a subsequent urine culture. In a population with high frequencies of resistance to ciprofloxacin and extended-spectrum beta-lactams, we found that a previous culture with a resistant organism was predictive of a repeat resistant organism, with the highest risk if the previous organism was carbapenem resistant. Conversely, previous cultures with sensitive organisms were predictive of repeat culture of an antibiotic-sensitive organism. The existence of at least one intervening culture negative for organisms with a resistance phenotype as well as increasing time from the original culture significantly reduced the chances of a repeat resistant organism. The association remained significant at 6 months for cipror bacteria, ESBL-producing Enterobacteriaceae, and CRNF, whereas the association between CRE and time between cultures was weak, indicating that CRE in urine persist for prolonged durations; in the sensitivity analysis excluding 14-day repeat cultures, the time between cultures was not a significant modifier of the risk for recurrent CRE infection. The effects of other factors on risk, including age, gender, and department of hospitalization, varied between resistance phenotypes.
In general, the association between cultures decreased with time. However, we observed increasing associations for frequencies of cipror bacteria and ESBL-producing Enterobacteriaceae in cultures taken within 1 week of previous cultures. This reflected the rates of these phenotypes in cultures which were taken within the same hospitalization and which, unsurprisingly, made up the majority of repeated cultures in this short period. This may be due to the fact that in the hospital, the natural history of colonization/persistence is disturbed by antibiotic treatment, which masks the association between previous and recurrent resistances.
A recent study examining the added value of previous cultures found that they predicted both bacterial species and antibiogram, and a subanalysis of susceptibility to ciprofloxacin found that a previous culture with a ciprofloxacin-susceptible pathogen was predictive of sensitivity to the antibiotic in a subsequent culture (8). That study consisted primarily of cultures taken from outpatients with relatively low rates of antibiotic resistance; the number of cultures analyzed for antibiotic susceptibility was similar to that in the present work. In comparison, our analysis was performed on a mostly hospital-based population with widespread resistance and provides information on several resistance phenotypes. We chose to analyze bacterial groups rather than specific bacterial species (although a separation was maintained between the Enterobacteriaceae and P. aeruginosa/Acinetobacter spp.), focusing on susceptibility patterns of resistant rather than sensitive organisms, since this information is more helpful to antibiotic prescribers. While the previous study benefited from having information on antibiotic treatment in the interim since the previous culture, which is information we lacked, there were no details provided on said treatment, including whether it covered the organism in the prior culture.
A second retrospective study assessing methods to improve the accuracy of empirical antibiotics among patients who had UTIs with multidrug-resistant organisms found that employing an antimicrobial concordant with prior culture results, where applicable, increased the likelihood of appropriate therapy 7-fold (9). That study had access to information on prior antibiotic treatment and its appropriateness as well as the virtue of simplicity and easy introduction of its findings into clinical practice. However, the analysis involved a small number of cultures (126), which may explain why there was no significant association between time from initial culture and likelihood of accurate therapy; additionally, 90% of the patients were male, which fails to represent the typical cohort for UTIs. MDR was defined as resistance to three or more classes of antibiotics, which would fail to include ESBL-producing Enterobacteriaceae.
Prescription of appropriate empirical antibiotic treatment necessitates consideration of the probability of bacterial infection, its source, and the bacterial epidemiology of that infection source (types of bacteria and resistance patterns) given the circumstances of infection acquisition (degree of exposure to the health care setting). The knowledge gained from previous cultures can also contribute to these baseline probabilities. We quantified this contribution for UTIs. It is likely that other cultures can contribute similarly to the process of selecting empirical antibiotics (e.g., using results of previous sputum cultures to prescribe antibiotics for pneumonia). It is also likely that the association differs by type of previous culture and the current infection, since the natural histories of colonization in the urinary, gastrointestinal, and respiratory tracts differ. It is difficult to compute this matrix of probabilities in clinical practice and to incorporate ORs for previous cultures. Sensitivity, specificity, PPV, and NPV were mostly unhelpful, with none of the PPVs exceeding 70%. Only a lack of carbapenem resistance was predictive of no such resistance in subsequent cultures with a high degree of certainty. Computerized decision support systems might make better use of baseline probabilities of infection and susceptibilities by accounting for quantitative factors, such as the time since previous culture (10).
Our study has several advantages. By including all urine cultures, including polymicrobial cultures, we reflected a “real-world” setting in which physicians have to deal with multiple culture results and it is not always possible to ascertain if previous cultures were taken properly. By analyzing different resistance phenotypes independently, we showed differences in the timing and risk factors for repeat resistant organisms, presumably reflecting differing baseline prevalences of these resistance patterns. Our main limitation is that we had no means of determining if patients had received antibiotics, particularly covering antibiotics, in the interval since their previous culture. However, it is not always possible to obtain such information from patients in clinical settings. A previous study showed that use of any antibiotic (regardless of coverage of the initial isolate) in the interval between urine cultures decreased the rates of concordance between cultures, but the effect was still significant after antibiotic treatment, and the trends with time were conserved (8). Our sensitivity analysis excluding repeat cultures taken within 14 days of the initial culture showed results similar to those of the main analysis. Further studies should establish the association with covering and noncovering intervening antibiotics. We also lacked information on clinical symptoms, the presence of urinary catheters and other devices (which might explain the continued persistence of CRE), and cultures taken outside our hospital (11). While this is a serious limitation to answering questions on the mechanism of resistance formation, the present study was an effort to provide a pragmatic solution to the bedside clinician, who may also lack information regarding the circumstances under which previous cultures were taken, prior antibiotics used, and cultures taken outside his or her hospital system. A third potential disadvantage was that our analysis did not include negative culture results. Since in our institution urine cultures are frequently taken without suspicion of UTI, inclusion of the culture-negative samples would have biased the ORs toward the null. Thus, our results apply to patients with clinically suspected UTI for whom empirical antibiotics are considered necessary. A final limitation was the possibility that analyzing paired cultures biased the study toward a patient population with an increased frequency of antibiotic resistance. However, such a bias is presumably true for any cohort with repeated cultures and does not affect the predictive value of the earlier culture for resistant organisms in subsequent cultures. Furthermore, the use of the same methodology as that used in a previous study allows for better comparison of our findings (8).
In conclusion, in a location with high rates of antibiotic resistance and without information on antibiotic therapy and other potential confounders, a urine culture positive for a resistant Gram-negative organism was predictive of a subsequent culture with the same resistance phenotype. Physicians should prescribe empirical antibiotics for patients with severe urinary tract infections that cover resistant organisms if these were identified in a previous urine culture, particularly when there are no intervening negative cultures and the time since the preceding culture is less than 6 months. The protective nature of intervening negative cultures is an interesting finding inviting further study.
ACKNOWLEDGMENTS
We acknowledge the support of The Dalia and Eli Hurvitz Foundation in the implementation of Treat Steward at the Beilinson and Rambam hospitals. This work was performed as part of the system development.
Steen Andreassen is employed part time at Treat Systems, a company developing a software solution for antimicrobial stewardship called Treat Steward. Treat Systems did not financially support the work.
REFERENCES
- 1.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, Moran GJ, Nicolle LE, Raz R, Schaeffer AJ, Soper DE, Infectious Diseases Society of America, European Society for Microbiology and Infectious Diseases. 2011. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis 52:e103–e120. doi: 10.1093/cid/ciq257. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez GV, Master RN, Bordon J. 2011. Trimethoprim-sulfamethoxazole may no longer be acceptable for the treatment of acute uncomplicated cystitis in the United States. Clin Infect Dis 53:316–317. doi: 10.1093/cid/cir345. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez GV, Master RN, Karlowsky JA, Bordon JM. 2012. In vitro antimicrobial resistance of urinary Escherichia coli isolates among U.S. outpatients from 2000 to 2010. Antimicrob Agents Chemother 56:2181–2183. doi: 10.1128/AAC.06060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sader HS, Flamm RK, Jones RN. 2014. Frequency of occurrence and antimicrobial susceptibility of Gram-negative bacteremia isolates in patients with urinary tract infection: results from United States and European hospitals (2009–2011). J Chemother 26:133–138. doi: 10.1179/1973947813Y.0000000121. [DOI] [PubMed] [Google Scholar]
- 5.Zilberberg MD, Shorr AF. 2013. Secular trends in gram-negative resistance among urinary tract infection hospitalizations in the United States, 2000–2009. Infect Control Hosp Epidemiol 34:940–946. doi: 10.1086/671740. [DOI] [PubMed] [Google Scholar]
- 6.MacVane SH, Tuttle LO, Nicolau DP. 2014. Impact of extended-spectrum β-lactamase-producing organisms on clinical and economic outcomes in patients with urinary tract infection. J Hosp Med 9:232–238. doi: 10.1002/jhm.2157. [DOI] [PubMed] [Google Scholar]
- 7.Paul M, Shani V, Muchtar E, Kariv G, Robenshtok E, Leibovici L. 2010. Systematic review and meta-analysis of the efficacy of appropriate empiric antibiotic therapy for sepsis. Antimicrob Agents Chemother 54:4851–4863. doi: 10.1128/AAC.00627-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacFadden DR, Ridgway JP, Robicsek A, Elligsen M, Daneman N. 2014. Predictive utility of prior positive urine cultures. Clin Infect Dis 59:1265–1271. doi: 10.1093/cid/ciu588. [DOI] [PubMed] [Google Scholar]
- 9.Linsenmeyer K, Strymish J, Gupta K. 2015. Two simple rules for improving the accuracy of empiric treatment of multidrug-resistant urinary tract infections. Antimicrob Agents Chemother 59:7593–7596. doi: 10.1128/AAC.01638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul M, Andreassen S, Tacconelli E, Nielsen AD, Almanasreh N, Frank U, Cauda R, Leibovici L, TREAT Study Group. 2006. Improving empirical antibiotic treatment using TREAT, a computerized decision support system: cluster randomized trial. J Antimicrob Chemother 58:1238–1245. doi: 10.1093/jac/dkl372. [DOI] [PubMed] [Google Scholar]
- 11.Bart Y, Paul M, Eluk O, Geffen Y, Rabino G, Hussein K. 2015. Risk factors for recurrence of carbapenem-resistant Enterobacteriaceae carriage: case-control study. Infect Control Hosp Epidemiol 36:936–941. doi: 10.1017/ice.2015.82. [DOI] [PubMed] [Google Scholar]


