LETTER
The BKC-1 (Brazilian Klebsiella carbapenemase-1) class A β-lactamase, a weak carbapenemase, was recently reported in Klebsiella pneumoniae ST1781 isolates from São Paulo, Brazil (1). blaBKC-1 was found between the novel insertion sequence ISKpn23 (IS1380 family) and an aphA6 [aph(3)-VI] gene on an IncQ plasmid, p60136 (GenBank accession no. KP689347) (1). As in some other IncQ plasmids (2, 3), the resistance segment is located between two oriV regions (Fig. 1A). Nicoletti et al. (1) proposed transposition of a genetic element carrying ISKpn23, blaBKC-1, and aphA6.
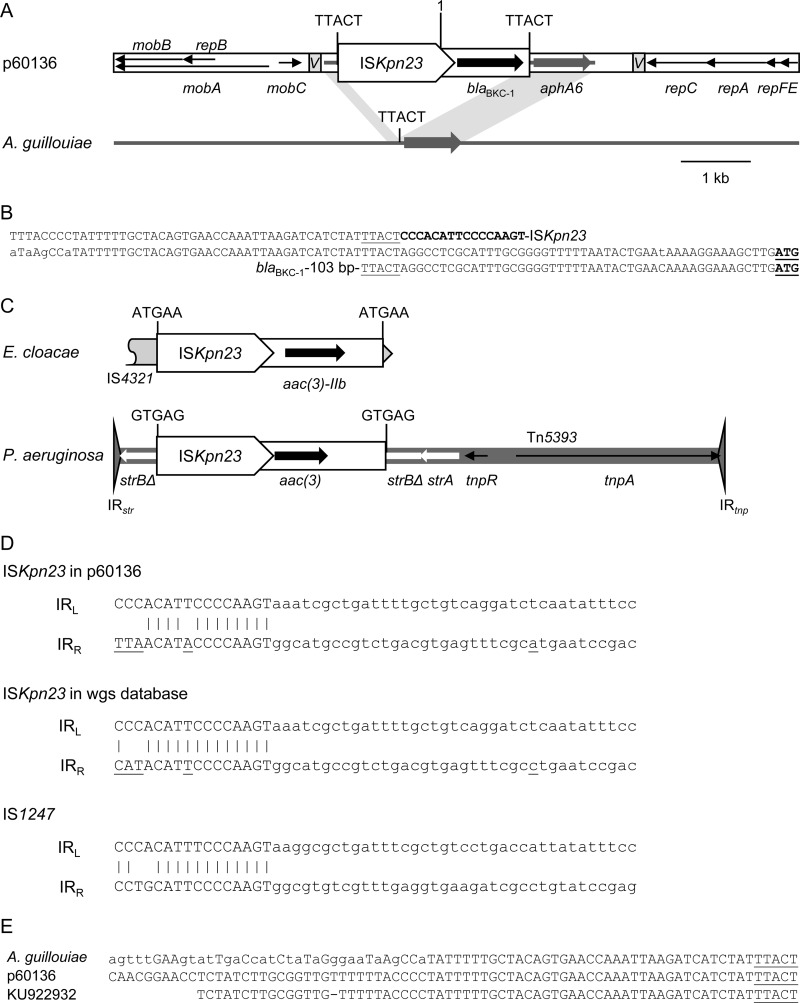
FIG 1.
(A) Diagram of the complete sequence of p60136 (GenBank accession no. KP689347) showing the proposed ISKpn23-blaBKC-1 TU, compared with aphA6 in the A. guillouiae chromosome (AP014630). “1” indicates position 1 in the p60136 sequence in KP689347, which was reversed to draw the diagram. ISKpn23 is shown as a box, with the pointed end indicating IRR. Different types of labeled arrows show the extents and directions of resistance and plasmid backbone genes. oriV regions are indicated by shaded boxes labeled “V.” The regions common to both sequences are indicated by shading, and the positions of the 5-bp DRs (TTACT) flanking the proposed TU in p60136 and the single copy of this sequence in A. guillouiae are shown. (B) Sequences around the insertion site in A. guillouiae (middle line) and flanking the proposed TU in p60136 (top and bottom lines). Differences are shown in lowercase letters in the A. guillouiae sequence. The proposed IRL of ISKpn23 is shown in bold typeface, the aphA6 start codon is bold and underlined, and the proposed DRs are underlined. (C) Potential ISKpn23-mediated TU in Enterobacter cloacae (JCKL01000012), carrying aac(3)-IIb inserted in a partial insertion sequence, IS4321 (gray), and in Pseudomonas aeruginosa (JTQL01000147) (15), carrying a putative aac(3) gene inserted in Tn5393. (D) Comparison of the ends of ISKpn23 in p60136, ISKpn23 in the NCBI WGS database (differences are underlined in each sequence), and IS1247 (as proposed in reference 8), with IRL and IRR in uppercase letters (16 bp, as listed in ISfinder). (E) Comparison of sequences upstream of the point of insertion of the proposed TU in p60136.
In addition to being similar to ISApr9 (1), ISKpn23, as defined in ISfinder (4; https://www-is.biotoul.fr/) (positions 1450 to 1 and 9786 to 9566 in GenBank accession no. KP689347), is ∼89% identical to IS1247. Like the better-known IS1380 family element ISEcp1 (5, 6), IS1247 is able to mobilize sequences adjacent to its right inverted repeat (IRR) by using alternative downstream sequences in conjunction with its left inverted repeat (IRL) (5, 7, 8). The inserted transposition unit (TU; or transposable module [TMo]) is flanked by 4- or 5-bp direct repeats (DRs), and IS1247 carries an outward-facing promoter (8).
A BLASTn search with the p60136 sequence downstream of ISKpn23 revealed two sequences with a single nucleotide difference: an Acinetobacter guillouiae chromosome (GenBank accession no. AP014630) (9), known to be the source of aphA6 genes (10), and a short sequence from K. pneumoniae (KU922932), where the aphA6 variant [12 nucleotide differences from aph(3′)-VIa (11)] was named aph(3′)-VIi. The match starts at TTACT 104 to 108 bp beyond the stop codon of blaBKC-1, and a potential DR of TTACT is also present immediately adjacent to the proposed IRL of ISKpn23 (Fig. 1A and B). The match to A. guillouiae and the KU922932 sequence continues upstream of ISKpn23, suggesting insertion of a 2.737-kb ISKpn23-blaBKC-1 TU. The aph(3′)-VIi coding sequence remains intact, and it is possible that both aph(3)-VIi and blaBKC-1 are expressed from a promoter in ISKpn23, as the original K. pneumoniae isolate KP60136 and an Escherichia coli transformant were resistant to amikacin and kanamycin in addition to β-lactams (1).
Searches of the NCBI whole-genome shotgun (WGS) contig database with ISKpn23 identified several examples of almost identical elements in different species, including one with 5-bp DRs, and several potential ISKpn23-mediated TUs flanked by DRs, in some cases with an uninterrupted version of the flanking sequence available, e.g., carrying different aac(3) genes (Fig. 1C). This ISKpn23 has five nucleotide differences from ISKpn23 in p61036 in the right-hand end, giving an IRR more similar to that of IS1247 (Fig. 1D).
The mechanism by which aph(3′)-VIi was acquired by an IncQ plasmid is currently unclear. The KU922932 sequence matches p60136 before the start of the match to A. guillouiae (Fig. 1E), but the available sequence ends before an identifiable IncQ (or other plasmid) backbone sequence. Resistance genes in IncQ plasmids are often associated with only, at most, remnants of mobile elements (2, 3), and acquisition of some genes (12) and/or subsequent fragmentation of mobile elements (13) may be due to recombination. Identification of additional sequences containing aph(3′)-VIi with context information may reveal more about the mobilization of this gene. blaBKC-1 has been reported in only six K. pneumoniae isolates from São Paulo, Brazil, to date (1, 14), and it will be interesting to see how successful this gene becomes, in comparison with bla genes associated with ISEcp1.
Footnotes
For the author reply, see doi:10.1128/AAC.01014-16.
REFERENCES
- 1.Nicoletti AG, Marcondes MF, Martins WM, Almeida LG, Nicolás MF, Vasconcelos AT, Oliveira V, Gales AC. 2015. Characterization of BKC-1 class A carbapenemase from Klebsiella pneumoniae clinical isolates in Brazil. Antimicrob Agents Chemother 59:5159–5164. doi: 10.1128/AAC.00158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loftie-Eaton W, Rawlings DE. 2012. Diversity, biology and evolution of IncQ-family plasmids. Plasmid 67:15–34. doi: 10.1016/j.plasmid.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Meyer R. 2009. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid 62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 35:820–855. doi: 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 6.Poirel L, Decousser JW, Nordmann P. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob Agents Chemother 47:2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Ploeg J, Willemsen M, van Hall G, Janssen DB. 1995. Adaptation of Xanthobacter autotrophicus GJ10 to bromoacetate due to activation and mobilization of the haloacetate dehalogenase gene by insertion element IS1247. J Bacteriol 177:1348–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szuplewska M, Bartosik D. 2009. Identification of a mosaic transposable element of Paracoccus marcusii composed of insertion sequence ISPmar4 (ISAs1 family) and an IS1247a-driven transposable module (TMo). FEMS Microbiol Lett 292:216–221. doi: 10.1111/j.1574-6968.2009.01495.x. [DOI] [PubMed] [Google Scholar]
- 9.Yee L, Hosoyama A, Ohji S, Tsuchikane K, Shimodaira J, Yamazoe A, Fujita N, Suzuki-Minakuchi C, Nojiri H. 2014. Complete genome sequence of a dimethyl sulfide-utilizing bacterium, Acinetobacter guillouiae strain 20B (NBRC 110550). Genome Announc 2:e01048-14. doi: 10.1128/genomeA.01048-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon EJ, Goussard S, Touchon M, Krizova L, Cerqueira G, Murphy C, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P. 2014. Origin in Acinetobacter guillouiae and dissemination of the aminoglycoside-modifying enzyme Aph(3′)-VI. mBio 5:e01972-14. doi: 10.1128/mBio.01972-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin P, Jullien E, Courvalin P. 1988. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol Microbiol 2:615–625. doi: 10.1111/j.1365-2958.1988.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 12.Kotsakis SD, Tzouvelekis LS, Lebessi E, Doudoulakakis A, Bouli T, Tzelepi E, Miriagou V. 2015. Characterization of a mobilizable IncQ plasmid encoding cephalosporinase CMY-4 in Escherichia coli. Antimicrob Agents Chemother 59:2964–2966. doi: 10.1128/AAC.05017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yau S, Liu X, Djordjevic SP, Hall RM. 2010. RSF1010-like plasmids in Australian Salmonella enterica serovar Typhimurium and origin of their sul2-strA-strB antibiotic resistance gene cluster. Microb Drug Resist 16:249–252. doi: 10.1089/mdr.2010.0033. [DOI] [PubMed] [Google Scholar]
- 14.Martins WM, Cordeiro-Moura JR, Ramos AC, Fehlberg LC, Nicoletti AG, Gales AC. 2016. Comparison of phenotypic tests for detecting BKC-1-producing Enterobacteriaceae isolates. Diagn Microbiol Infect Dis 84:246–248. doi: 10.1016/j.diagmicrobio.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Kos VN, Deraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm RA, Corbeil J, Gardner H. 2015. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 59:427–436. doi: 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]