Abstract
Emerging resistance to antimicrobials and the lack of new antibiotic drug candidates underscore the need for optimization of current diagnostics and therapies to diminish the evolution and spread of multidrug resistance. As the antibiotic resistance status of a bacterial pathogen is defined by its genome, resistance profiling by applying next-generation sequencing (NGS) technologies may in the future accomplish pathogen identification, prompt initiation of targeted individualized treatment, and the implementation of optimized infection control measures. In this study, qualitative RNA sequencing was used to identify key genetic determinants of antibiotic resistance in 135 clinical Pseudomonas aeruginosa isolates from diverse geographic and infection site origins. By applying transcriptome-wide association studies, adaptive variations associated with resistance to the antibiotic classes fluoroquinolones, aminoglycosides, and β-lactams were identified. Besides potential novel biomarkers with a direct correlation to resistance, global patterns of phenotype-associated gene expression and sequence variations were identified by predictive machine learning approaches. Our research serves to establish genotype-based molecular diagnostic tools for the identification of the current resistance profiles of bacterial pathogens and paves the way for faster diagnostics for more efficient, targeted treatment strategies to also mitigate the future potential for resistance evolution.
INTRODUCTION
The frequency and spectrum of infections with antibiotic-resistant bacteria are steadily increasing worldwide (1). The infections pose a serious threat to human health, as substantial mortality rates are reported in patients given ineffective empirical therapy, mainly due to resistance to the agents used (2). One exceedingly problematic Gram-negative pathogen is Pseudomonas aeruginosa. This opportunistic pathogen plays a dominant role as an infectious agent affecting the lungs of cystic fibrosis patients (3–5) and has emerged as one of the most important human pathogens involved in nosocomial infections (6). P. aeruginosa is known not only for its high intrinsic resistance to a broad spectrum of antimicrobial compounds (7, 8), but also for its remarkable ability to acquire new resistances via horizontal gene transfer and via the adoption of drug resistance-associated mutations during the course of infection (9–13). In particular, the accelerating development of multidrug-resistant P. aeruginosa strains represents a great diagnostic and therapeutic challenge in modern medicine (14, 15) and, with the lack of new antibiotic options, emphasizes the need for the optimization of current diagnostics, therapies, and prevention of the spread of these organisms.
Increasing numbers of whole-genome-sequencing (WGS) approaches that aim at the identification of genetic determinants that directly correlate with a particular bacterial phenotype are being reported (16, 17). The ongoing developments in DNA-sequencing technologies are likely to affect medical microbiological diagnostics and monitoring of pathogens. However, clinical adoption of WGS in resistance profiling is still challenging. While mutations that lead to modifications in drug targets, such as target mutations that confer quinolone resistance (18), represent direct and causal relationships to a resistance phenotype and thus are expected to be easily identified in phenotype-genotype association studies, the P. aeruginosa resistome is much more complex. It also includes expression changes of genes encoding efflux pumps or intrinsic β-lactamases, which can result from diverse mutations in regulatory genes (19–21). Furthermore, screenings of mutant libraries have revealed a large number of genes that seem to impact the resistance phenotype despite the lack of any direct link to a known resistance mechanism (22–25). Thus, many more, and probably also a combination of, specific mutations are expected to influence the bacterial resistance phenotype. This complicates the identification of causal relationships between genetic variations and resistance.
With the aim of shedding more light on the complex relationship between the evolution of antimicrobial resistance, genetic variations, and global phenotypic changes (26–29), we correlated resistance phenotypes, not only with genomic sequence variations, but also with gene expression profiles. Therefore, transcriptome-wide association studies of 135 phylogenetically broadly distributed P. aeruginosa isolates that had been RNA sequenced previously by our group (30, 31) were performed, and resistance to the three clinically most important classes of anti-pseudomonal antibiotics—fluoroquinolones, β-lactams, and aminoglycosides—was systematically explored. Indeed, transcriptional profiles of resistant isolates expressed distinct and resistance-specific sequence variation and gene expression patterns. We additionally applied machine learning algorithms to identify complex discriminatory markers for ciprofloxacin susceptibility versus resistance. Interestingly, the combined use of gene expression and sequence variation classifiers, which resulted from machine learning, enhanced the accuracy of resistance phenotype prediction in the clinical isolates. Thus, phenotype prediction on the basis of transcriptome profiles is a valuable and reliable approach and supports the identification of causal genotype-phenotype relationships.
MATERIALS AND METHODS
Strain collection and antibiotic resistance profile.
The 135 investigated clinical isolates were provided by different clinics or research institutions. In total, we included 73 isolates collected at the Hanover Medical School (MHH), 40 isolates from a strain collection provided by the University of Freiburg (sampled in numerous countries across Europe), 14 isolates from the Robert-Koch-Institute in Wernigerode, and 8 isolates provided by the National Reference Laboratory for Multidrug-Resistant Gram-Negative Bacteria in Bochum, Germany. The isolates were obtained from diverse infection sites (see Table S1 in the supplemental material).
Most of the isolates were categorized as multidrug resistant (resistant to three or more antimicrobial classes). The antibiotic susceptibility data were either hospital or institution derived or determined in house using the Vitek2 system (bioMérieux) or Etest strips (bioMérieux). The classification of MIC breakpoints was performed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.
As a reference for differential gene expression and sequence variation analysis, we chose the UCBPP-PA14 strain (32).
RNA sequencing.
Whole-transcriptome sequencing of the clinical isolates was previously performed by our group by the use of a custom-made protocol with barcoded RNA libraries to enable pooled sequencing of several samples (30, 31). The clinical isolates had been grown under standard conditions (LB broth; 37°C) and harvested in RNAprotect (Qiagen) at an optical density at 600 nm (OD600) of 2 before the transcriptome sequencing (RNA-seq) was performed (31).
The reads were mapped to the PA14 reference genome, which is available for download from the Pseudomonas Genome Database (http://v2.pseudomonas.com) (33). Mapping was performed using stampy, a short-read aligner that allows for gapped alignments (34), and SAMtools (35) was utilized for sequence variation calling. The reads per gene (rpg) values of all genes were calculated from the SAM output files. Testing for differential expression against the PA14 wild type (four biological replicates) was performed with DESeq (36), an R software package that uses a statistical model based on the negative binomial distribution.
To investigate the presence of horizontally acquired resistance genes in the accessory genomes (here defined as all genes that do not belong to the PA14 reference genome), all sequencing reads were mapped to a custom resistance gene collection as an artificial genome. This method allows the calculation of the relative sequence coverage of each resistance gene, providing reliable information on the presence or absence of certain genes. A threshold of ≥80% sequence coverage was used for positive detection of a gene. To confirm the bioinformatic prediction, PCR amplification and Sanger sequencing were carried out for some of the detected resistance genes or whole integron sequences using type 1 integron-specific primers binding to the 5′-conserved (intI1) and the 3′-conserved (ΔqacE1) segments.
Phylogeny.
The phylogenetic tree was created using a total of 148 genes that were ≥90% covered by sequencing reads in all clinical isolates and also had orthologs in all five P. aeruginosa reference strains included (PA14, PAO1, LESB58, PACS2, and PA7). The ortholog information was obtained from a precomputed Pseudomonas genome alignment with Mauve (http://v2.pseudomonas.com/mauve.jsp; 33). To extract the gene sequences of the clinical isolates, consensus sequences were created using the SAMtools package. From the consensus sequences and the reference genomes, the 148 gene sequences were extracted using the annotation information, resulting in one concatenated sequence per isolate. Phylogenetic distances between the strains were calculated using a k-mer approach, as described previously (37). The sequences were split into 17-mers, which were then compared between the isolates. The resulting distance matrix was used to build a neighbor-joining tree in R using the ape package (38), and supplemental information, like the antibiotic resistance phenotype or acquired resistance genes, was added and visualized using iTOL (http://itol.embl.de; 39).
MLST analysis.
For analysis of the multilocus sequence type (MLST) profiles based on RNA-seq data, a consensus sequence for each isolate was generated using SAMtools (35), from which the respective gene sequences (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) were extracted using a PERL script. Isolates with sufficient coverage in the respective genes were assigned a sequence type (ST) number according to the allelic profiles available in the MLST database (http://pubmlst.org/paeruginosa).
Storage of the comprehensive data set within a Web-based database.
Transcriptomes and phenotypes of all sequenced clinical isolates are stored within the Bactome (bacterial genome) database (https://bactome.helmholtz-hzi.de). The database further provides tools to extract gene expression or sequence variation information for all isolates on a genome-wide level and to perform phenotype-based group comparisons, e.g., antibiotic-resistant versus -susceptible isolates, to investigate an enrichment of sequence variations, nonsense mutations, or differentially expressed genes.
Computational modeling and statistical evaluation.
We used Pearson's correlation (r) to test for linear coherence of gene expression and MIC values and Kendall's rank correlation (τ) to analyze the dependence of MIC values on a single, distinct feature (e.g., occurrence of specific enzymes or mutations).
Linear mixed model analysis for SNP association studies.
Linear mixed models (LMMs) were calculated using the Python library LIMIX (https://github.com/PMBio/limix). Single nucleotide polymorphisms (SNPs) in coding regions that were covered by at least three reads and had a score of at least 50 were used as binary genotype information, with 1 marking the presence and 0 the absence of a SNP in an isolate. In cases where the read coverage was not sufficient, the missing values were replaced with 0.5. MIC values for the antibiotics (ceftazidime [CAZ], ciprofloxacin [CIP], and meropenem [MEM]) were used as phenotypic information and transformed using a rank-based transformation [the function “preprocess.rankStandardizeNormal()” in LIMIX] to achieve a normal distribution.
The different phenotypes were modeled as linear functions of the SNPs, with the sample relatedness as the covariate [the function “qtl.test_lmm()”], using the default likelihood ratio test to calculate the P values. The sample relatedness was also calculated using LIMIX [the function “getCovariance()”] and was normalized by dividing by the mean of the diagonal (covariance of the isolates with themselves). P values were regarded as significant when they were below the Bonferroni corrected significance threshold of 0.05.
Gene expression association analysis.
In order to measure how suitable expression values of single genes are to predict the resistance of an isolate to specific antibiotics, the normalized reads per gene were considered scores from which an area under the concentration-time curve (AUC) value was computed for every gene with respect to the two classes “resistant” and “susceptible.” Based on the work of Mason and Graham (40), P values for the AUC values were calculated, and a false-discovery rate correction was carried out. The AUC significance threshold was calculated based on a corrected maximal P value of 0.05 for each data set and resulted in 0.727 for CAZ, 0.730 for MEM, and 0.737 for CIP.
Machine learning with feature selection and the P-SVM algorithm.
The potential support vector machine (P-SVM) was employed for learning prediction models. P-SVM is a scale-invariant support vector machine that features two distinct regularization approaches (41). In the dyadic mode used for analyzing the data described in this paper, P-SVM essentially comes down to training a regularized linear model that tries to provide good generalization performance by choosing a compact set of markers, the combination of which is highly predictive.
The SNP data set was prefiltered for a minimal coverage of three reads, a Phred quality score of at least 50, and only nonsynonymous SNPs in coding regions, with 1 marking the presence and 0 the absence of an SNP in an isolate. For the SNP data set, the P-SVM was complemented by a feature selection step that was done beforehand: the position-dependent kernel association test (PODKAT) was used to detect genomic regions in which the SNPs are most significantly correlated with the phenotype to be predicted. PODKAT is a recent extension of the SNP set kernel association test (SKAT) (42), which uses a position-dependent kernel in order to better account for very rare and private SNPs (43, 44). Once the most significant regions are determined with PODKAT, the individual contributions of the SNPs to the test statistics are computed, and the 5,000 most promising SNPs are selected for further processing with P-SVM. For cross-validation with P-SVM, the data were divided into five nonoverlapping subsets, so-called folds, on which a 5-fold nested cross-validation approach was applied. While each time one fold was withheld in the outer loop as an independent test set (test fold) to compute unbiased estimates of prediction performance, parameter selection was carried out by 4-fold inner cross-validation on the remaining four folds (training folds). Once the best parameters had been found for the four training folds of the outer cross-validation loop, a model was trained on these four folds using the optimized parameters and applied to the withheld test fold to estimate the prediction performance on independent data (45, 46). This was applied to all five possible combinations of alternating splits (5-fold cross-validation), finally resulting in five sets of markers for any tested condition.
The gene expression data (normalized reads per kilobase of gene length) was analyzed by the P-SVM, with 5-fold double cross-validation (optimization criterion AUC) as described previously, though without data preprocessing and optional feature preselection.
Nucleotide sequence accession number.
All the short-read data are available at the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP034661.
RESULTS
Antimicrobial resistance profiles in clinical P. aeruginosa isolates with broad taxonomic distribution.
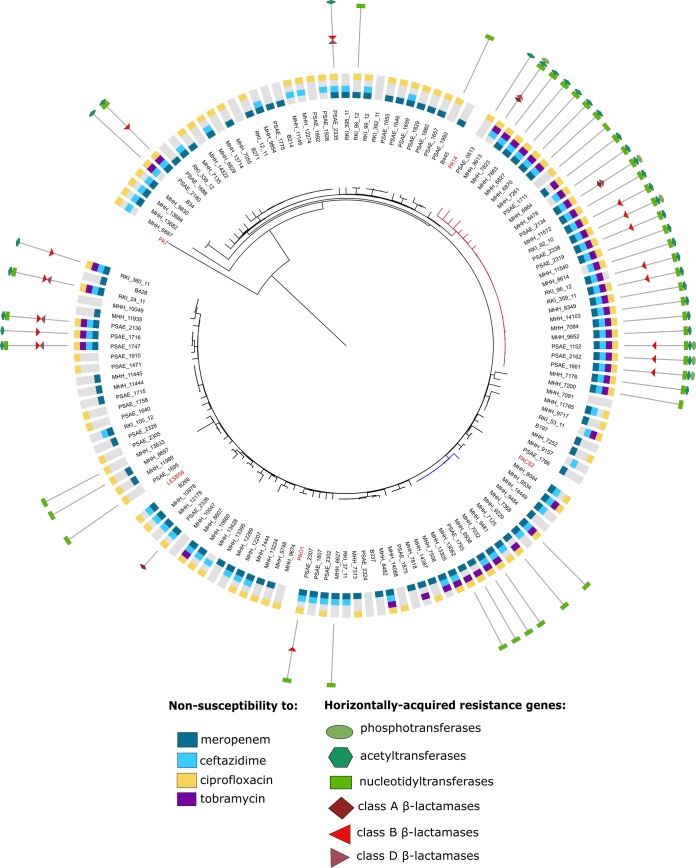
We determined the antimicrobial resistance profiles of 135 clinical P. aeruginosa isolates that had been sampled from different regions across Europe and from various infection sites. For all of these isolates, transcriptional profiles were recorded previously, and the phylogenetic relatedness was determined by using a k-mer approach (30). Figure 1 depicts a neighbor-joining tree based on the sequence similarity of 148 commonly expressed genes and includes five reference strains (PA14, PAO1, PA7, PACS2, and LESB58). In addition to the antibiotic resistance profile, the acquisition of horizontally acquired resistance genes is depicted in the phylogenetic tree. The isolates exhibit a broad taxonomic distribution and separate into two main lineages. One lineage contains the PA14 reference strain, as well as one large cluster including high-risk clone ST235 isolates (47); the other lineage contains PACS2, LESB58, and PAO1 and a cluster including high-risk clone ST175 isolates (47). There were also some taxonomically distant isolates (MHH6887, MHH13682, MHH13684, MHH9830, and B34), as well as the known outlier PA7 (48).
FIG 1.
Phylogenetic clustering and distribution of antibiotic resistance. The phylogenetic tree was constructed with a neighbor-joining algorithm based on a distance matrix calculated from k-mers of all genes with a coverage of at least 90% in all clinical isolates (total, 148 genes). Strain PA7 was defined as the outgroup. The data sets on antibiotic resistance and the horizontally acquired resistance genes were integrated using iTOL (39). The red tree branches indicate ST235, and the blue branches are ST175 isolates. Publicly available reference strains are indicated in red.
In general, the clinical isolates expressed diverging resistance phenotypes. Resistance against ciprofloxacin, tobramycin, ceftazidime, and meropenem was largely independent of phylogenetic relatedness (Fig. 1). However, the worldwide-disseminated lineages of sequence types 235 and 175 (47) were clear hot spots of resistance gene carriage. All ST235 isolates harbored aminoglycoside nucleotidyltransferases (mainly encoded by aadA1 or aadA6), and nearly all of them had an additional aminoglycoside acetyltransferase (mainly encoded by aacA4). Overall, 15 different aminoglycoside-modifying enzymes (AMEs) from all three enzymatic types were identified in 49 of the isolates (36%), with the majority of isolates harboring numerous different enzymes (between two and six). Furthermore, we identified nine different β-lactamases (OXA-2 and -4, VIM-1 and -2, GIM-2, IMP-1 and -7, PER-1, and CTX-M-3) in 19 isolates (14%), 6 of which contained two enzymes simultaneously. All but one isolate carried β-lactamases only in combination with at least one aminoglycoside-modifying enzyme, demonstrating their common association with integrons that harbor arrays of resistance gene cassettes.
We next correlated the presence and expression of acquired-resistance-conferring genes with the resistance profiles of the clinical isolates. We found that the great majority of isolates (41 out of 46 isolates [89%]) that exhibited resistance to tobramycin (MIC ≥ 8 μg/ml) carried at least one tobramycin-modifying enzyme (encoded by aacA4, aadB, aacA7, or aacA5). Only 14 out of 45 (31%) of the meropenem-resistant (MIC ≥ 16 μg/ml) and 17 out of 61 (28%) of the ceftazidime-resistant (MIC ≥ 32 μg/ml) isolates expressed a β-lactamase-encoding gene. Additionally, two isolates with Vim-2 carbapenemases showed a meropenem-intermediate resistance phenotype. Of note, the expression levels of resistance genes also correlated with the MIC level. As an example, two isolates (Psae1747 and Psae2136) were identified that harbored the same set of resistance genes but exhibited different levels of resistance (tobramycin MICs of >1,024 μg/ml and 128 μg/ml, respectively). This was in line with 2- to 3-fold-increased expression of the aacA7 and aacA5 genes in Psae1747, resulting in high-level tobramycin resistance.
More detailed information on the contribution of horizontally acquired-resistance-conferring genes versus other chromosomally acquired genetic determinants of resistance is provided in Fig. S1 in the supplemental material.
Correlation of genomic sequence variations with the expression of an antibiotic resistance phenotype.
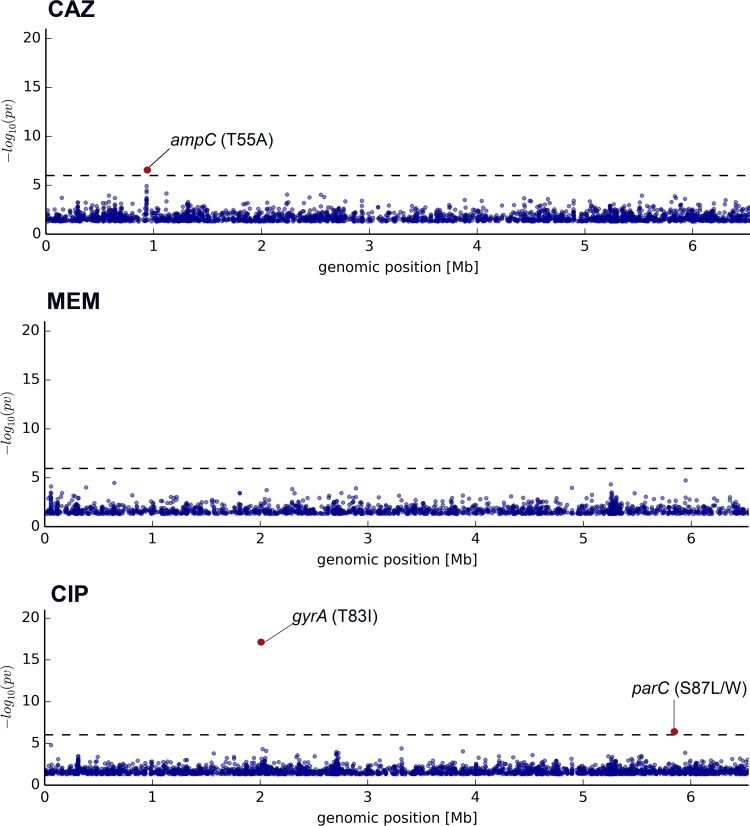
We systematically investigated the quantitative (transcriptional profiles) and qualitative (sequence information) RNA-seq data on the 135 bacterial isolates. Since an RNA sequence mirrors the sequence of the DNA from which it was transcribed, we first used the qualitative RNA-seq data to describe sequence variations on the single-nucleotide level within the transcribed genes, which constitute a high percentage of the overall number of genes in P. aeruginosa (30). In order to minimize false-positive associations that result from genetic relatedness and population structure (49), we applied LMMs to uncover the effect of nonsynonymous SNPs on the resistance phenotype. The genotype data set was combined with rank-transformed MIC values for the fluoroquinolone ciprofloxacin and the β-lactams ceftazidime and meropenem. Since we aimed at identifying markers of acquired chromosomally encoded resistance, only isolates that did not carry a horizontally acquired resistance gene were included in the association analysis (this is why tobramycin was not further analyzed here). Permutation tests with randomly shuffled MIC values were carried out to confirm the significance of true-positive associations. SNP associations for each of the tested antibiotics are shown in the Manhattan plots in Fig. 2 (see also Table S2 in the supplemental material). A clear association was found between the expression of ciprofloxacin resistance (77 isolates; MIC ≥ 4 μg/ml) and the presence of one SNP in gyrA encoding a DNA gyrase (T83I; P = 5.88 × 10−18) and one in parC encoding topoisomerase IV (S87L/W; P = 4.07 × 10−7). Both nucleotide positions are well-known targets for mutations involved in ciprofloxacin resistance (50, 51). The T83I SNP in gyrA was indeed present in 85 out of the 135 genomic sequences, 72 of which were ciprofloxacin-resistant isolates (MIC ≥ 4 μg/ml), and there was a clear overall correlation (τ = 0.637) between the presence of SNPs in gyrA and elevated ciprofloxacin MIC values. The S87L/W SNP in parC was present in 44 isolates, and there was also a clear correlation (τ = 0.727) between the presence of this particular SNP and ciprofloxacin MICs above 8 μg/ml.
FIG 2.
Transcriptome-wide mutation association with resistance phenotypes. Shown is a sequence variation association study for significant differences between clinical isolates resistant and susceptible to CAZ, MEM, and CIP. Each dot represents one nucleotide position, and only SNPs with an uncorrected P value of <0.05 were plotted. Hits above the significance threshold (corrected P value of <0.05) are indicated in red.
Manual inspection of the complete quinolone resistance-determining regions (QRDR) of the known target genes gyrA, gyrB, parC, and parE revealed additional mutations that had previously been associated with quinolone resistance. However, due to their overall low abundance, those mutations were not detected in the association studies. Eleven non-T83I SNPs in gyrA were found, and most of the mutations in gyrB, parC, and parE were detected in combination with the T83I mutation in gyrA.
No association of the presence of a particular SNP and resistance to meropenem (31 isolates, excluding the ones with acquired-resistance-conferring genes; MIC ≥ 16 μg/ml) was detected, and only one SNP in the chromosomally encoded β-lactamase AmpC was found to be associated with ceftazidime resistance (44 isolates, excluding the ones with acquired-resistance-conferring genes; MIC ≥ 32 μg/ml), whose impact still needs to be investigated.
Correlation of changes in gene expression with an antibiotic resistance phenotype.
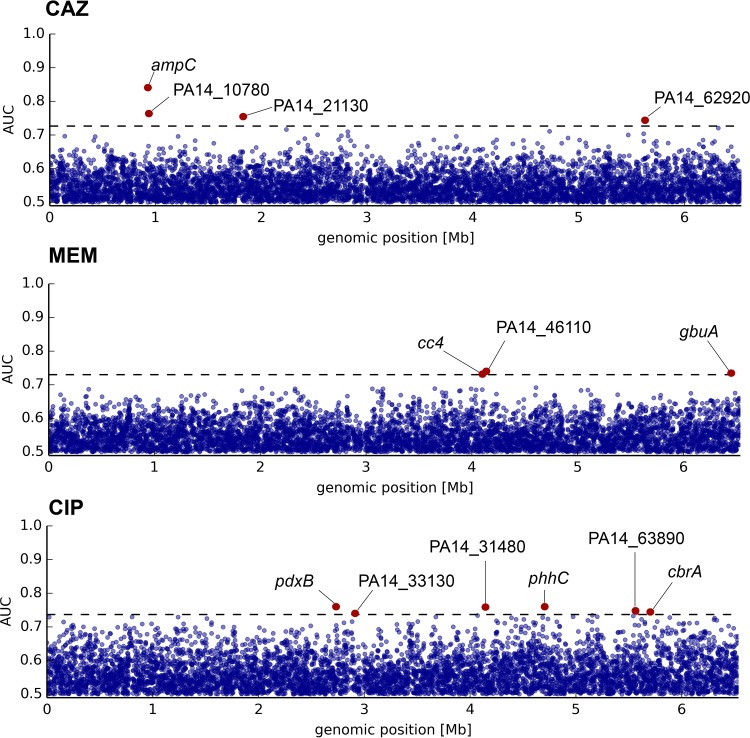
We next analyzed the quantitative RNA-seq data to uncover whether there are differentially expressed genes that can be associated with a resistance phenotype. To investigate resistance-associated single-gene expression patterns, receiver operating characteristic (ROC) analyses were performed. They resulted in an AUC-based probability for the association of the expression of a particular gene with resistance. Associations for each of the tested antibiotics are shown in the Manhattan plots in Fig. 3 (see Table S2 in the supplemental material). We found three genes with expression profiles significantly associated with MEM resistance, but all of them (PA14_46110, a predicted sodium-solute symporter; cc4, encoding the diheme protein cytochrome c4; and gbuA, a guanidinobutyrase involved in the arginine dehydrogenase pathway) only reached AUC values just above the defined significance threshold. Of note, just below the threshold (AUC = 0.692), the known carbapenem entry porin-encoding gene oprD was found to be downregulated in MEM-resistant isolates. However, a clear association of oprD was found when we looked exclusively for enrichment of nonsense mutations in the group of meropenem-nonsusceptible isolates (data not shown). A more detailed analysis of the distribution of premature translation termination mutations in oprD is shown in Fig. S2A in the supplemental material. These mutations occurred generally throughout the gene sequence. In contrast, single amino acid exchanges were mostly restricted to either the periplasmic hinge or the surface-associated loops (see Fig. S2B and C in the supplemental material). To evaluate how oprD gene inactivation was correlated with protein expression, immunoblot analyses with an OprD-specific antibody were performed, and we compared the actual protein levels of a total of 63 isolates with different MEM resistance phenotypes with the expression and sequence information obtained from RNA sequencing. All the tested nonsense mutations led to abolishment of efficient translation (data not shown). Interestingly, there was only partial correlation between the oprD mRNA levels and protein production. One reason could be that OprD is subject to frequent, possibly MexT-mediated posttranscriptional regulation, as has been described previously in PAO1 (52).
FIG 3.
Transcriptome-wide gene expression association with resistance phenotypes. Shown is a gene expression association study for significant differences between clinical isolates resistant and susceptible to CAZ, MEM, and CIP. Each dot represents one gene. Hits above the significance threshold (corrected P value of <0.05) are indicated in red.
Analysis of the CIP resistance phenotype revealed six genes whose expression was associated with resistance. Among them, we found the sensor kinase-encoding gene cbrA, which has been reported to be involved in resistance to polymyxins, tobramycin, and ciprofloxacin (53).
The most significant association was found for the expression of the chromosomally encoded β-lactamase AmpC (AUC = 0.84) with a CAZ-resistant phenotype. There was a clear correlation (r = 0.638) between increased ampC gene expression and elevated CAZ MICs (range, 0.5 to 64 μg/ml). Furthermore, the majority of CAZ-resistant (35/36) and intermediate-resistant (8/13) isolates exhibited high (>3 log2 fold change [FC]) ampC expression values, while most (49/56) susceptible variants in turn showed only slight or no constitutive overexpression. Interestingly, all seven CAZ-susceptible isolates that exhibited strongly increased ampC expression (>5 log2FC) harbored mutations in the ampC gene. Most of them were deletions within the first 10 nucleotides, as additionally confirmed by Sanger sequencing, which led to frameshifts and thus probably to a truncated signal peptide that is needed for β-lactamase maturation and secretion into the periplasmic space.
Of note, no efflux pump components appeared to be significantly associated with resistance to any of the investigated antibiotics. Manual inspection confirmed a high percentage of clinical isolates with efflux pump overexpression. This overexpression was correlated with an increased number of sequence variations in the respective regulatory genes (see Fig. S3 in the supplemental material). However, for none of the antibiotics a direct correlation to a resistance was observed. One reason could be the presence of other resistance mechanisms that masked the effects of antibiotic efflux. On the other hand, we also found a high number of susceptible isolates with strong efflux pump overexpression (e.g., 59 tobramycin-susceptible isolates strongly overexpressed mexXY with a change of ≥10-fold).
Identification of complex discriminatory markers for resistance by machine learning.
Standard phenotype-genotype association methods are suitable for the detection of directly associated single markers, as we have shown in this study, but they have limitations when it comes to identifying factors that contribute to more complex phenotypes. These phenotypes may result from different combinations of genetic variations, with the individual contributing factors being too rare to be detected in direct-association studies. Thus, complex machine learning classification algorithms are implemented more and more in clinical research to identify disease-causing biomarkers, especially in studies based on large-scale genomic approaches that result in a flood of sequence data (54, 55). Excessively high numbers of markers and typically low numbers of samples, plus the need to model interactions of multiple causal markers, call for advanced regularized linear models. The P-SVM (41 was used in this study to discriminate between two phenotypically opposed groups (here, resistant and susceptible). The P-SVM has been designed for exactly such purposes and has proven to be highly successful for analyzing high-dimensional molecular data (56). The training of the models, along with the selection of optimal hyperparameters, was done by nested cross-validation in order to facilitate model selection while still obtaining unbiased estimates of the generalization performance (45, 46).
In this study, the machine learning algorithms mentioned above were applied to the ciprofloxacin data set. The homogeneous distribution of molecular mechanisms (mainly target mutations) underlying ciprofloxacin resistance development seems to be particularly suited to the evaluation of whether resistance can be correlated, not only with distinct genetic variations, but also with the expression of a distinct and global transcriptional profile.
Genomic and transcriptomic ciprofloxacin resistance classifiers.
The predictive machine learning model based on the SNP data set revealed in total 247 unique mutations that were identified in at least one validation fold to be discriminatory for the ciprofloxacin resistance phenotype. A lower number of genes (167 in total) with changed expression were identified as transcriptomic classifiers in at least one validation fold. The vast majority of classifiers have been identified in only one of the five validation folds. However, eight mutation classifiers and six expression classifiers have been detected in at least three folds and thus can be considered strong discriminators (Tables 1 and 2).
TABLE 1.
Marker occurrence and classification reliability for ciprofloxacin resistance/susceptibility phenotype classification by machine learning
| Data set (CIPr vs. CIPs) | No. of markers per no. of folds |
AUCa | |||||
|---|---|---|---|---|---|---|---|
| 5 | 4 | 3 | 2 | 1 | Total | ||
| SNPs | 1 | 2 | 5 | 21 | 218 | 247 | 0.885 |
| Gene expression | 0 | 0 | 6 | 29 | 132 | 167 | 0.724 |
Area under the curve values calculated from an AUC-optimized ROC analysis for the PODKAT-PSVM double-cross-validation approach. The calculated AUC value (optimized for AUC) indicates the predictive power of the classifier combination.
TABLE 2.
Strong classification markers for the ciprofloxacin resistance data set identified by machine learning analysesa
| Data set (CIPr vs. CIPs) | Gene | SNP positionb | Product | Fold no. |
|---|---|---|---|---|
| SNPs | PA14_23260; gyrA | 2015001 | DNA gyrase subunit A | 5 |
| PA14_20440; phnN | 1758114 | Phosphonate transport ATP-binding protein | 4 | |
| PA14_60790 | 5418859 | Putative ABC transporter, ATP-binding protein | 4 | |
| PA14_25600 | 2238920 | Peptidase | 3 | |
| PA14_34600 | 3074288 | Putative glyceraldehyde-3-phosphate dehydrogenase | 3 | |
| PA14_62810; secG | 5604332 | Preprotein translocase subunit | 3 | |
| PA14_73360; gidB | 6529928 | Glucose-inhibited division protein B | 3 | |
| PA14_08060 | 693759 | Tail fiber assembly protein | 3 | |
| Gene expression | PA14_18480; algX | Alginate biosynthesis protein AlgX | 3 | |
| PA14_32290 | Hypothetical protein | 3 | ||
| PA14_48950 | Hypothetical protein | 3 | ||
| PA14_58410; opdP | Glycine-glutamate dipeptide porin | 3 | ||
| PA14_59390 | Hypothetical protein | 3 | ||
| PA14_62100; yedZ | Sulfide oxidase subunit | 3 |
The most frequently selected markers (a minimum of three outer training models [folds]) for differentiation between ciprofloxacin-susceptible and -resistant isolates are listed.
For the SNP data set, the genomic position of the mutation in the PA14 reference strain.
The overall reliability of the resistant versus susceptible phenotype classification based on the identified list of markers is demonstrated by the AUC score, calculated from the double-cross-validation P-SVM. The ciprofloxacin SNP-based phenotype classification seemed to be highly reliable, as it resulted in an AUC score of 0.885. While this might have been expected, strikingly, the gene expression data set also delivered a score of 0.724 (Table 1).
A closer look at the most frequently detected marker mutations confirmed the well-known gyrA T83I mutation to be the strongest classifier in the SNP data set, as it was detected in all five validation folds. Interestingly, the second most significant sequence variation that has been identified as directly associated with ciprofloxacin resistance (parC S87L/W) in the LMM was not among the 247 sequence variation classifiers. Instead, many other mutations without any previously known relation to antibiotic resistance contributed to the separation of resistant and susceptible isolates. For instance, mutations in two ATP-binding regions of transporters were found in four validation folds.
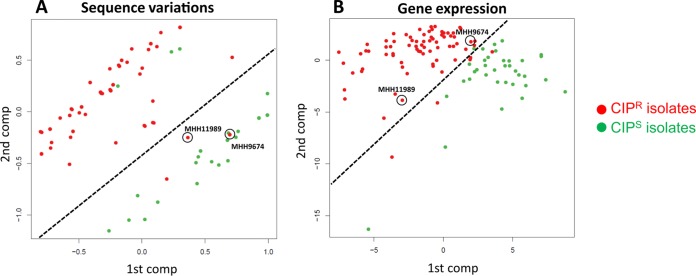
Also, the gene expression profile data set resulted in moderately accurate predictability (AUC > 0.7), although besides efflux pumps, no changes in the expression of single genes have been reported before to be associated with ciprofloxacin resistance. Among the strongest classifiers for this data set was the alginate biosynthesis gene algX, whose upregulation might contribute to the previously described protective effect of alginate against antibiotics, including ciprofloxacin (57) (Table 2). The strong discriminatory power of the machine learning classification is depicted in Fig. 4 in a principal-component analysis (PCA). The separation of resistant and susceptible isolates based on sequence variations (Fig. 4A), as well as on gene expression values (Fig. 4B), was much more precise when based on markers defined by machine learning than when based on random classifiers.
FIG 4.
PCA plots of all ciprofloxacin-resistant and -susceptible isolates. The PCAs on the left are based on the whole set of SNPs (A) or genes (B), and the PCAs on the right are based on only SNPs that occurred in a minimum of three validation folds (A) or on the whole set of phenotype-separating genes (B) identified by the P-SVM machine learning approach. The red and green dots represent ciprofloxacin-resistant and -susceptible isolates, respectively.
The transcriptional profile can predict ciprofloxacin resistance.
As we observed great classification power with machine learning, we next evaluated whether there is a characteristic transcriptional profile that can indeed be used to predict ciprofloxacin resistance. Interestingly, two ciprofloxacin-resistant clinical isolates that had acquired SNPs in the QRDR, albeit not the very frequent ones, were included in this study. Thus, since the dominant gyrA T83I mutation served as the only classifier mutation from the QRDR, both isolates were classified as ciprofloxacin susceptible when looking at the SNP-based classifiers identified by machine learning. However, when plotting the isolates based on the gene expression classifiers resulting from P-SVM, both of the isolates mentioned clearly clustered within the group of resistant isolates (Fig. 5). This demonstrates a distinct transcriptional profile expression of the resistant isolates, probably due to a direct effect of the resistance acquisition on the transcriptome or to mutational adaptations that have been acquired in order to compensate for the fitness burden of resistance-conferring mutations.
FIG 5.
Resistance phenotype prediction based on sequence variations or gene expression profiles. (A) Indicated are two isolates (circled) that are known to be resistant to ciprofloxacin but that clustered within the group of susceptible isolates (green dots) based on PCA analysis of the sequence variations that occurred in a minimum of three validation folds. (B) The same two isolates clearly clustered within the group of resistant isolates when the plot was based on the phenotype-separating genes identified by the gene expression P-SVM approach. Dashed lines indicate the separation of resistant and susceptible isolates.
DISCUSSION
The Achilles heel of global efforts to combat infectious diseases and accompanying antimicrobial resistance is early diagnosis (58). Consequently, there is a strong need for the introduction of improved diagnostic tools to enable more targeted and faster treatment and for implementation of effective infection control measures to diminish the development and spread of resistance (59, 60). To achieve this, there are expanding efforts worldwide to transform current diagnostics from the common culture-based methods to genomics-based tools. The application of next-generation sequencing (NGS) technologies has the potential to significantly accelerate bacterial species identification and resistance profiling (61) and could provide information, not only on the current drug susceptibility of a pathogen, but also on its potential to evolve resistance (62). However, precise prediction of antibiotic resistance profiles solely based on genotypes is still challenging due to the complexity of some resistance mechanisms (63). Although recent studies have demonstrated the potential of NGS as a tool for antibiotic resistance prediction (64–67), they were mainly restricted to the investigation of a limited set of well-known resistance-associated loci. Furthermore, all the previous studies relied exclusively on genome sequencing without considering the impact of multifactorial transcriptional regulation on the resistance phenotype.
This study is the first transcriptome-sequencing-based approach that has aimed at a systematic analysis of resistance markers using different methodologies and including both mutation and gene expression profiles. Our results demonstrate that, besides the acquisition of horizontally transferred resistance genes, dominant mutational variations (e.g., SNPs in target genes) or gene expression changes (which can be caused by diverse sequence changes), such as the upregulation of intrinsic β-lactamases, can be investigated in parallel by RNA-seq.
Many factors affect the accuracy of a prediction model, including the sample size, the numbers of variations underlying a particular phenotype (68), and, in the field of clinical microbiology, a high proportion of clinical samples without shared ancestry. Thus, the main objective of this study was to define the scale of information necessary for successful P. aeruginosa resistance phenotype-genotype correlations in terms of (i) the number and sufficient phylogenetic diversity of clinical isolates included in the study and (ii) the effectiveness of gene expression data to reduce the complexity of phenotypes that cannot be correlated with one directly associated marker.
We clearly demonstrated that RNA sequencing of 77 (ciprofloxacin), 61 (ceftazidime), and 45 (meropenem) β-lactam-resistant P. aeruginosa isolates with broad phylogenetic distribution was already sufficient to establish significant correlations and to identify the most dominant bacterial resistance traits in a combined gene expression and sequence variation approach. Thus, it can be expected that the application of unbiased transcriptome-wide association studies to a much larger number of resistant isolates (which do not express the most dominant resistance markers) has the potential to uncover novel mechanisms of resistance.
Most strikingly, the application of complex machine learning classification algorithms to our clinical isolates revealed a specific transcriptional profile within the group of ciprofloxacin-resistant isolates that was clearly distinct from that of the susceptible group. Since the cause of ciprofloxacin resistance in the clinical isolates could nearly always be attributed to target mutations (see Fig. S1 in the supplemental material), the observed subtle changes in the transcriptional profile are most likely a compensation for the perturbing effects of the target mutation rather than a contribution to the resistance phenotype. This is important, since it implies that there might be a general transcriptional fingerprint of resistance to be exploited for diagnostic purposes. Resistance phenotypes caused by rare genomic variations could be detected when screening for the phenotype-associated gene expression markers, which could thus significantly support the genomics-based identification of resistance.
In conclusion, our unbiased screening will undoubtedly have to be expanded in the future in order to identify more infrequent genetic resistance markers. Nevertheless, our results suggest that the availability of larger amounts of sequencing information from clinical-sample collections may accomplish the unraveling of the comprehensive resistome of P. aeruginosa and might also help to uncover how different treatment regimens affect the phenotypic and genotypic evolutionary paths to antibiotic resistance in the clinical setting. In the same vein, information about the presence of mutations that compensate for fitness costs or mutations that impact bacterial pathogenicity might influence the choice of antibacterial treatment. Combining transcriptional and mutational data sets and implementing them in predictive models will be essential to detect and understand resistance pathways and patterns in large, complex data sets. Especially in light of the steadily improving quality and quantity of sequencing data acquisition (69), it seems reasonable that faster and, in the future, also more economical genotype-based phenotype profiling has the potential to be used in routine medical microbiology diagnostics.
Supplementary Material
ACKNOWLEDGMENTS
We thank Iris F. Chaberny (Hanover Medical School, Hanover, Germany), Daniel Jonas (Freiburg University Medical Centre, Freiburg, Germany), Wolfgang Witte and Yvonne Pfeifer (Robert-Koch-Institute, Wernigerode, Germany), and Martin Kaase and Sören Gatermann (National Reference Laboratory for Multidrug-Resistant Gram-Negative Bacteria, Bochum, Germany) for providing us with clinical P. aeruginosa isolates. Furthermore, we are grateful to Ole Lund and Rolf Kaas (Technical University of Denmark, Lyngby, Denmark) for kindly providing us with a protocol describing their k-mer methodology for the calculation of phylogenetic distances prior to publication. We also thank Robert Geffers and the Genome Analytics Research Group (Helmholtz Centre for Infection Research, Braunschweig, Germany) for performing the Illumina sequencing and Klaus Hornischer (Helmholtz Centre for Infection Research, Braunschweig, Germany) for establishing the Bactome database.
TWINCORE is a joint venture between the Helmholtz Centre for Infection Research, Braunschweig, Germany, and the Hanover Medical School, Hanover, Germany.
S.H., A.K., and M.S. conceived and designed the experiments. A.K. and M.S. performed the experiments. D.E. and A.D. provided tools for data analysis. A.K., M.S., S.P., U.B., R.R., B.X., S.B., F.K., and M.P. analyzed the data. S.H., A.K., and M.S. wrote the paper.
Financial support from the European Research Council (http://erc.europa.eu/) (starter grant 260276) is gratefully acknowledged. M.S. was funded by the Ph.D. Program “Infection Biology” of the Hanover Biomedical Research School. A.K., S.P., and S.B. were supported by the Helmholtz International Graduate School for Infection Research under contract number VH-GS-202. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00075-16.
REFERENCES
- 1.World Health Organization. 2012. The evolving threat of antimicrobial resistance: options for action. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Fraser A, Paul M, Almanasreh N, Tacconelli E, Frank U, Cauda R, Borok S, Cohen M, Andreassen S, Nielsen AD, Leibovici L, TREAT Study Group. 2006. Benefit of appropriate empirical antibiotic treatment: thirty-day mortality and duration of hospital stay. Am J Med 119:970–976. doi: 10.1016/j.amjmed.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 3.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ratjen F, Doring G. 2003. Cystic fibrosis. Lancet 361:681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- 5.Tümmler B, Kiewitz C. 1999. Cystic fibrosis: an inherited susceptibility to bacterial respiratory infections. Mol Med Today 5:351–358. doi: 10.1016/S1357-4310(99)01506-3. [DOI] [PubMed] [Google Scholar]
- 6.Gaynes R, Edwards JR, National Nosocomial Infections Surveillance System. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis 41:848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 7.Schweizer HP. 2003. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet Mol Res 2:48–62. [PubMed] [Google Scholar]
- 8.Hancock RE, Brinkman FS. 2002. Function of pseudomonas porins in uptake and efflux. Annu Rev Microbiol 56:17–38. doi: 10.1146/annurev.micro.56.012302.160310. [DOI] [PubMed] [Google Scholar]
- 9.Breidenstein EB, de la Fuente-Nunez C, Hancock RE. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Bruchmann S, Dötsch A, Nouri B, Chaberny IF, Häussler S. 2013. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob Agents Chemother 57:1361–1368. doi: 10.1128/AAC.01581-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabot G, Ocampo-Sosa AA, Tubau F, Macia MD, Rodriguez C, Moya B, Zamorano L, Suarez C, Pena C, Martinez-Martinez L, Oliver A. 2011. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob Agents Chemother 55:1906–1911. doi: 10.1128/AAC.01645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehlberg LC, Xavier DE, Peraro PP, Marra AR, Edmond MB, Gales AC. 2012. Beta-lactam resistance mechanisms in Pseudomonas aeruginosa strains causing bloodstream infections: comparative results between Brazilian and American isolates. Microb Drug Resist 18:402–407. doi: 10.1089/mdr.2011.0174. [DOI] [PubMed] [Google Scholar]
- 13.Poole K. 2005. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:479–487. doi: 10.1128/AAC.49.2.479-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croughs PD, Li B, Hoogkamp-Korstanje JA, Stobberingh E, Antibiotic Resistance Surveillance Group . 2013. Thirteen years of antibiotic susceptibility surveillance of Pseudomonas aeruginosa from intensive care units and urology services in the Netherlands. Eur J Clin Microbiol Infect Dis 32:283–288. doi: 10.1007/s10096-012-1741-4. [DOI] [PubMed] [Google Scholar]
- 15.Obritsch MD, Fish DN, MacLaren R, Jung R. 2004. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother 48:4606–4610. doi: 10.1128/AAC.48.12.4606-4610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, Davis MR Jr, Skurnik D, Leiby N, LiPuma JJ, Goldberg JB, McAdam AJ, Priebe GP, Kishony R. 2011. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet 43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McAdam PR, Holmes A, Templeton KE, Fitzgerald JR. 2011. Adaptive evolution of Staphylococcus aureus during chronic endobronchial infection of a cystic fibrosis patient. PLoS One 6:e24301. doi: 10.1371/journal.pone.0024301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drlica K, Hiasa H, Kerns R, Malik M, Mustaev A, Zhao X. 2009. Quinolones: action and resistance updated. Curr Top Med Chem 9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagge N, Ciofu O, Hentzer M, Campbell JI, Givskov M, Hoiby N. 2002. Constitutive high expression of chromosomal beta-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob Agents Chemother 46:3406–3411. doi: 10.1128/AAC.46.11.3406-3411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidtke AJ, Hanson ND. 2008. Role of ampD homologs in overproduction of AmpC in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:3922–3927. doi: 10.1128/AAC.00341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuo Y, Eda S, Gotoh N, Yoshihara E, Nakae T. 2004. MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ-mexX intergenic DNA. FEMS Microbiol Lett 238:23–28. doi: 10.1111/j.1574-6968.2004.tb09732.x. [DOI] [PubMed] [Google Scholar]
- 22.Fajardo A, Martinez-Martin N, Mercadillo M, Galan JC, Ghysels B, Matthijs S, Cornelis P, Wiehlmann L, Tümmler B, Baquero F, Martinez JL. 2008. The neglected intrinsic resistome of bacterial pathogens. PLoS One 3:e1619. doi: 10.1371/journal.pone.0001619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breidenstein EB, Khaira BK, Wiegand I, Overhage J, Hancock RE. 2008. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob Agents Chemother 52:4486–4491. doi: 10.1128/AAC.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schurek KN, Marr AK, Taylor PK, Wiegand I, Semenec L, Khaira BK, Hancock RE. 2008. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:4213–4219. doi: 10.1128/AAC.00507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez-Ortega C, Wiegand I, Olivares J, Hancock RE, Martinez JL. 2010. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Antimicrob Agents Chemother 54:4159–4167. doi: 10.1128/AAC.00257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO. 2009. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol 27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoder-Himes DR, Chain PS, Zhu Y, Wurtzel O, Rubin EM, Tiedje JM, Sorek R. 2009. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc Natl Acad Sci U S A 106:3976–3981. doi: 10.1073/pnas.0813403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 29.Dötsch A, Eckweiler D, Schniederjans M, Zimmermann A, Jensen V, Scharfe M, Geffers R, Haussler S. 2012. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS One 7:e31092. doi: 10.1371/journal.pone.0031092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dotsch A, Schniederjans M, Khaledi A, Hornischer K, Schulz S, Bielecka A, Eckweiler D, Pohl S, Haussler S. 2015. The Pseudomonas aeruginosa transcriptional landscape is shaped by environmental heterogeneity and genetic variation. mBio 6:e00749. doi: 10.1128/mBio.00749-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pohl S, Klockgether J, Eckweiler D, Khaledi A, Schniederjans M, Chouvarine P, Tummler B, Haussler S. 2014. The extensive set of accessory Pseudomonas aeruginosa genomic components. FEMS Microbiol Lett 356:235–241. doi: 10.1111/1574-6968.12445. [DOI] [PubMed] [Google Scholar]
- 32.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunter G, Goodson M. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leekitcharoenphon PNE, Kaas RS, Lund O, Aarestrup FM. 2014. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paradis E, Claude J, Strimmer K. 2004. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 39.Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason SJ, Graham NE. 2002. Areas beneath the relative operating characteristics (ROC) and relative operating levels (ROL) curves. Q J R Meteorol Soc 128:2145–2166. doi: 10.1256/003590002320603584. [DOI] [Google Scholar]
- 41.Hochreiter S, Obermayer K. 2006. Support vector machines for dyadic data. Neural Comput 18:1472–1510. doi: 10.1162/neco.2006.18.6.1472. [DOI] [PubMed] [Google Scholar]
- 42.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. 2011. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bodenhofer U, Hochreiter S. 2013. PODKAT: a non-burden test for associating complex traits with rare and private variants, poster 1769W. 63rd Annu Meet Am Soc Hum Gene, Boston, MA. [Google Scholar]
- 44.Bodenhofer U. 2015. PODKAT: an R package for association testing involving rare and private variants, R package version 1.0.3 ed. http://www.bioinf.jku.at/software/podkat/.
- 45.Filzmoser P, Liebmann B, Varmuza K. 2009. Repeated double cross validation. J Chemometrics 23:160–171. [Google Scholar]
- 46.Salzberg SL. 1997. On comparing classifiers: pitfalls to avoid and a recommended approach. Data Min Knowl Discov 1:317–328. doi: 10.1023/A:1009752403260. [DOI] [Google Scholar]
- 47.Cholley P, Thouverez M, Hocquet D, van der Mee-Marquet N, Talon D, Bertrand X. 2011. Most multidrug-resistant Pseudomonas aeruginosa isolates from hospitals in eastern France belong to a few clonal types. J Clin Microbiol 49:2578–2583. doi: 10.1128/JCM.00102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy PH, Tetu SG, Larouche A, Elbourne L, Tremblay S, Ren Q, Dodson R, Harkins D, Shay R, Watkins K, Mahamoud Y, Paulsen IT. 2010. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One 5:e8842. doi: 10.1371/journal.pone.0008842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Listgarten J, Lippert C, Kadie CM, Davidson RI, Eskin E, Heckerman D. 2012. Improved linear mixed models for genome-wide association studies. Nat Methods 9:525–526. doi: 10.1038/nmeth.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruiz J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother 51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 51.Hooper DC, Wolfson JS. 1989. Bacterial resistance to the quinolone antimicrobial agents. Am J Med 87:17S–23S. [PubMed] [Google Scholar]
- 52.Köhler T, Epp SF, Curty LK, Pechere JC. 1999. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol 181:6300–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeung AT, Bains M, Hancock RE. 2011. The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J Bacteriol 193:918–931. doi: 10.1128/JB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Libbrecht MW, Noble WS. 2015. Machine learning applications in genetics and genomics. Nat Rev Genet 16:321–332. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sundaramurthy G, Eghbalnia HR. 2015. A probabilistic approach for automated discovery of perturbed genes using expression data from microarray or RNA-Seq. Comput Biol Med 67:29–40. doi: 10.1016/j.compbiomed.2015.07.029. [DOI] [PubMed] [Google Scholar]
- 56.Hochreiter S, Obermayer K. 2004. Gene selection for microarray data, p 319–355. In Schölkopf B, Tsuda K, Vert J-P (ed), Kernel methods for computational biology. MIT Press, Cambridge, MA. [Google Scholar]
- 57.Hodges NA, Gordon CA. 1991. Protection of Pseudomonas aeruginosa against ciprofloxacin and beta-lactams by homologous alginate. Antimicrob Agents Chemother 35:2450–2452. doi: 10.1128/AAC.35.11.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berkelman R, Cassell G, Specter S, Hamburg M, Klugman K. 2006. The “Achilles heel” of global efforts to combat infectious diseases. Clin Infect Dis 42:1503–1504. [DOI] [PubMed] [Google Scholar]
- 59.Barenfanger J, Drake C, Kacich G. 1999. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J Clin Microbiol 37:1415–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doern GV, Vautour R, Gaudet M, Levy B. 1994. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J Clin Microbiol 32:1757–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palmer AC, Kishony R. 2013. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat Rev Genet 14:243–248. doi: 10.1038/nrg3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woodford N, Sundsfjord A. 2005. Molecular detection of antibiotic resistance: when and where? J Antimicrob Chemother 56:259–261. doi: 10.1093/jac/dki195. [DOI] [PubMed] [Google Scholar]
- 64.Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, Kearns AM, Pichon B, Young B, Wilson DJ, Llewelyn MJ, Paul J, Peto TE, Crook DW, Walker AS, Golubchik T. 2014. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52:1182–1191. doi: 10.1128/JCM.03117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agerso Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 68:771–777. doi: 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
- 66.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TE, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244. doi: 10.1093/jac/dkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kos VN, Deraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm RA, Corbeil J, Gardner H. 2015. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 59:427–436. doi: 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Novianti PW, Jong VL, Roes KC, Eijkemans MJ. 2015. Factors affecting the accuracy of a class prediction model in gene expression data. BMC Bioinformatics 16:199. doi: 10.1186/s12859-015-0610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Minoche AE, Dohm JC, Himmelbauer H. 2011. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol 12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.