Abstract
Ribonucleoside analog inhibitors (rNAI) target the hepatitis C virus (HCV) RNA-dependent RNA polymerase nonstructural protein 5B (NS5B) and cause RNA chain termination. Here, we expand our studies on β-d-2′-C-methyl-2,6-diaminopurine-ribonucleotide (DAPN) phosphoramidate prodrug 1 (PD1) as a novel investigational inhibitor of HCV. DAPN-PD1 is metabolized intracellularly into two distinct bioactive nucleoside triphosphate (TP) analogs. The first metabolite, 2′-C-methyl-GTP, is a well-characterized inhibitor of NS5B polymerase, whereas the second metabolite, 2′-C-methyl-DAPN-TP, behaves as an adenosine base analog. In vitro assays suggest that both metabolites are inhibitors of NS5B-mediated RNA polymerization. Additional factors, such as rNAI-TP incorporation efficiencies, intracellular rNAI-TP levels, and competition with natural ribonucleotides, were examined in order to further characterize the potential role of each nucleotide metabolite in vivo. Finally, we found that although both 2′-C-methyl-GTP and 2′-C-methyl-DAPN-TP were weak substrates for human mitochondrial RNA (mtRNA) polymerase (POLRMT) in vitro, DAPN-PD1 did not cause off-target inhibition of mtRNA transcription in Huh-7 cells. In contrast, administration of BMS-986094, which also generates 2′-C-methyl-GTP and previously has been associated with toxicity in humans, caused detectable inhibition of mtRNA transcription. Metabolism of BMS-986094 in Huh-7 cells leads to 87-fold higher levels of intracellular 2′-C-methyl-GTP than DAPN-PD1. Collectively, our data characterize DAPN-PD1 as a novel and potent antiviral agent that combines the delivery of two active metabolites.
INTRODUCTION
Chronic infection with hepatitis C virus (HCV) represents a significant disease burden on global health. It is estimated that 3% of the human population carries a chronic HCV infection, which translates to roughly 170 million infections worldwide and 3.2 million infections in the United States (1). Although up to 25% of acutely infected individuals clear the virus spontaneously, chronic HCV infection develops in the remaining 75% of those infected and can lead to the development of liver cirrhosis and hepatocellular carcinoma (2–4).
HCV contains a 9.6-kb positive-strand RNA genome that codes for three structural and seven nonstructural proteins. The nonstructural protein 5B (NS5B) is the viral RNA-dependent RNA polymerase and is responsible for genome replication. Because of the high mutation rate of this enzyme, as well as the high virus replication rate, HCV propagation results in the generation of thousands of viral quasispecies (5, 6). Therefore, drug combination therapy is required in order to overcome development of resistance and to achieve successful viral clearance. HCV treatment comprised of ribavirin and pegylated interferon alpha, with or without protease inhibitors, can achieve sustained virologic responses in 40 to 80% of treated patients (7–9). However, curative outcomes historically have been suboptimal due to host polymorphisms, viral genotypic variability, and onset of adverse events.
In pursuit of new direct-acting antiviral agents with improved therapeutic outcome, several different virus replication steps, such as entry, replication complex formation, and NS5B-mediated viral RNA synthesis, are currently being investigated (reviewed in references 10–14). Members of a major class of anti-NS5B compounds, termed ribonucleoside analog inhibitors (rNAI), block HCV RNA replication inside the host cell cytoplasm through viral RNA chain termination. In December 2013, sofosbuvir became the first anti-HCV rNAI phosphoramidate prodrug to receive FDA approval (http://www.fda.gov/forpatients/illness/hepatitisbc/ucm377920.htm). Once inside the cell, host kinases must first phosphorylate rNAIs into the active ribonucleoside triphosphate (rNTP) form. Therefore, sufficient intracellular levels of triphosphorylated ribonucleoside analog (rNAI-TP) metabolites are critical for antiviral activity (14–16). The presence of a member of the phosphoramidate prodrug group on rNAI, such as sofosbuvir, allows the first, often rate-limiting, phosphorylation step to be bypassed, leading to increased intracellular rNAI-TP generation.
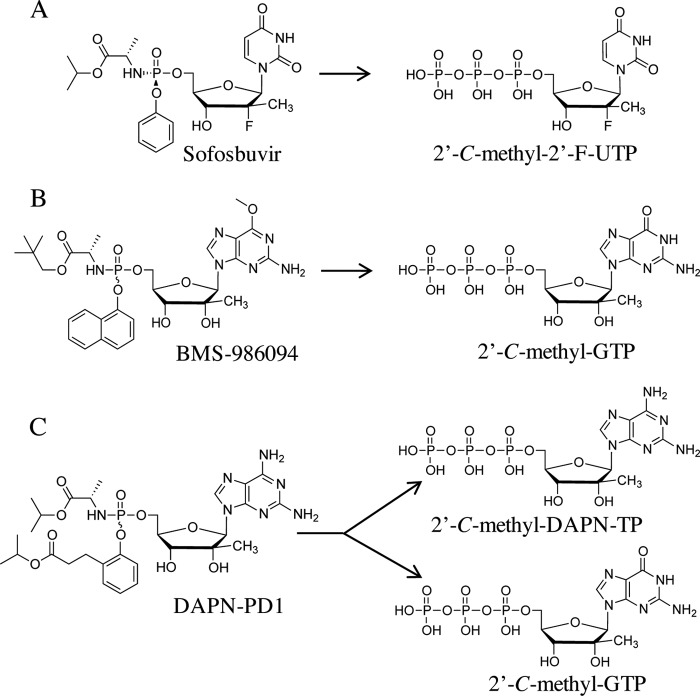
Despite the high mutation rate of HCV, clinical emergence of resistance toward sofosbuvir has rarely been observed. Although several resistance-associated substitutions and baseline polymorphisms recently have been associated with treatment, their phenotypic impact on conferring resistance to rNAI remains to be determined (17–19). For example, the infrequent emergence of resistance-conferring mutation S282T (20–22) is probably due to the unfit nature of the mutated virus (23). Sofosbuvir, which generates 2′-C-methyl-2′-F-UTP as its active metabolite in vivo (Fig. 1A), has demonstrated an exceedingly favorable safety profile in humans (24–26). Conversely, clinical development of many other anti-HCV rNAI has been terminated due to issues with toxicity (reviewed in reference 14). A recent important example pertains to phase II clinical trials with BMS-986094 (formerly known as INX-189), a nucleoside prodrug that is metabolized to generate 2′-C-methyl-GTP in vivo (Fig. 1B) (27, 28). Despite the high potency of this compound, clinical trials were halted after reports of severe adverse events and one death (29, 30). Although the exact cause of toxicity for BMS-986094 remains elusive (31), it is postulated that off-target inhibition of host cell nucleic acid synthesis at least in part accounts for the observed deleterious effects. In agreement with this hypothesis, recent studies have shown that human mtRNA polymerase (POLRMT) can indeed incorporate ribonucleoside 5′-triphosphate analogs in vitro. This is in turn correlated with inhibition of mtRNA transcription and interference with mitochondrial function in cell culture (32, 33). Thus, mechanisms associated with ribonucleotide analog toxicity are of significant interest as new rNAI are being developed for the treatment of infections with RNA viruses.
FIG 1.
Intracellular metabolism of anti-HCV phosphoramidate prodrugs. (A) Administration of sofosbuvir leads to intracellular generation of 2′-C-methyl-2′-F-UTP. (B) BMS-986094 prodrug group cleavage and removal of methyl group at position 6 of the guanine base leads to the generation of 2′-C-methyl-GTP. (C) Prodrug groups from β-d-2′-C-Me-2,6-diaminopurine ribonucleoside phosphoramidate prodrug 1 (DAPN-PD1) are cleaved, and 2′-C-methyl-DAPN-TP is generated after intracellular phosphorylation of the parent ribonucleoside. Parent 2′-C-methyl-DAPN monophosphate also can be a substrate for cellular adenosine deaminases that remove the amino group at position 6 of the purine ring. This leads to the generation of 2′-C-methyl-GTP.
While sofosbuvir has paved the way as the gold standard in HCV therapy, development of novel rNAI will be warranted for several reasons. Patients with genotype (GT) 3a infection treated with sofosbuvir-containing regimens generally attain lower sustained virologic response rates than other genotypes (34, 35). Additionally, the high cost of direct-acting antiviral agents will limit access to treatment globally, especially in developing countries (3, 36). Finally, considering the diversity of the chronically infected population, it is conceivable that niche populations require alternative therapies. In our search for innovative, potent, and safe anti-HCV therapeutic agents, we have recently described the chemical synthesis of β-d-2′-C-Me-2,6-diaminopurine ribonucleoside (DAPN) phosphoramidate prodrugs (PD) as novel inhibitors of HCV (37). We showed that cellular administration of DAPN-PD led to the generation of two chemically distinct bioactive ribonucleoside triphosphate analogs, namely, 2′-C-methyl-DAPN-TP and 2′-C-methyl-GTP (Fig. 1C). In this study, we examined the biochemical properties of each of the nucleoside 5′-triphosphate metabolites of prototype compound DAPN-PD1 (compound 14c [37]) and showed both metabolites were inhibitors of viral RNA polymerization. We also showed that although both active metabolites generated by DAPN-PD1 can be weak substrates for POLRMT in vitro, DAPN-PD1 did not inhibit mtRNA transcription. This is in contrast to BMS-986094, which showed significant untoward effects on the synthesis of mtRNA compared with DAPN-PD1. The relative abundance of intracellular 2′-C-methyl-GTP levels produced in off-target cells and differences in inhibition of mitochondrial transcription likely account for the distinct safety profiles of the two prodrugs. Work presented here characterizes DAPN-PD1 as a potent and differentiated antiviral agent that combines the delivery of two inhibitory metabolites with distinct incorporation profiles.
MATERIALS AND METHODS
Purified NS5B enzymes for GT 5a and 6a were provided by Cocrystal Pharma, Inc. Expression plasmids for the GT 1b NS5B wild type (WT) and S282T mutant were a gift from Matthias Götte (University of Alberta). Plasmid for the NS5B S96T mutant was generated by QuikChange site-directed mutagenesis. GpG primer (Trilink) and other RNA oligomer substrates (IDT) were 32P-radiolabeled as previously described (38) using [γ-32P]ATP (PerkinElmer Life Sciences). Reactions with T4 polynucleotide kinase (Fermentas) were allowed to proceed for 1 h at 37°C.
Chemical synthesis of rNAI-TP metabolites.
Nucleoside analog triphosphates were prepared as previously described (39–41). Briefly, high-performance liquid chromatography (HPLC) purification was performed using a Dionex NucleoPac PA-200 (9 by 250 mm) column, eluting with a gradient of 0 to 100% of 0.5 M triethylammonium bicarbonate (TEAB) over 25 min. The appropriate fractions were collected, solvents were evaporated by lyophilization, and the products were freeze-dried with deionized water several times to remove excess buffer. Nucleoside analog triphosphates were isolated as trimethylammonium salts with >95% purity as measured by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Expression and purification of HCV NS5B.
NS5B purification was performed essentially as previously described (42). Briefly, HCV GT 1b NS5B (accession number AJ238799.1), in which the 21 amino acids at the C terminus were replaced by a His tag, was inserted into the expression vector pET-21b (Novagen). WT, S282T, or S96T mutant proteins were expressed in 3 liters of transformed BL21(DE3) cells grown to an optical density (OD) of 0.6. After induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), cultures were shaken for 16 h at 25°C. Protein was purified using a Talon Ni2+ column (Clontech) and eluted with increasing concentrations of imidazole. Following additional purification with a DEAE anion exchange column, samples were eluted from the cation exchange SP column using increasing concentrations of NaCl. Purified protein was stored in 10 mM HEPES buffer, pH 7.5, with 50% glycerol, 1 mM dithiothreitol (DTT), and 600 mM NaCl. GT 2a JFH1 (accession number AB114136), GT 3a (accession number EF523597), and GT 4a (accession number EF523599) were purified using the same protocol.
In vitro NS5B-mediated RNA polymerization inhibition assay.
C-terminal His-tagged NS5BΔ21 enzyme was incubated at 30°C with a synthetic 20mer RNA template (T20; 5′-AACCGUAUCCAAAACAGUCC-3′) and 1 μM 32P-radiolabeled GpG primer in a buffer containing 40 mM Tris, pH 7.5, 6 mM NaCl, and 2 mM MgCl2. Reactions were initiated with the addition of 10 μM nucleotide triphosphate (NTP) mix, 1 μM competing NTP, and various concentrations of inhibitor. Reactions were allowed to proceed for 90 min and subsequently stopped with the addition of 10 mM EDTA and formamide. Samples were visualized on 20% denaturing polyacrylamide gel and quantified using Quantity One software. Fifty percent inhibitory concentrations (IC50s) were calculated using KaleidaGraph software.
Rapid single-ribonucleotide incorporation assay.
NS5BΔ21 (3 μM) was incubated with 1 μM RNA template (T20G10; 5′-AAUGUAUAAGCAUUAUAUCC-3′) (43) and 1 μM 32P-radiolabeled GpG primer. ATP and UTP (20 μM) were added to the mix, and the elongation complex was allowed to form for 1 h at 30°C and paused at position +10. Various amounts of GTP or GTP analog subsequently were added, and synthesis of +11 RNA product was measured over time. A similar experimental setup was used when assessing ATP or ATP analog incorporation where the RNA template (T20A16; 5′-AACCUGAGAAGGAGAAAGCC-3′) (38) was preincubated with 20 μM CTP and UTP. Reactions were stopped with the addition of 10 mM EDTA and formamide. Samples were subsequently denatured at 95°C for 3 min and resolved in a 20% denaturing polyacrylamide gel. A single-exponential equation, y = ymax(1 − exp(−kobs[S])), was used to obtain rates of incorporation (kobs) in KaleidaGraph software, where S represents substrate concentration and y represents amount of product formed. Obtained rates were fitted to a nonlinear regression using the hyperbolic equation y = (kpol[S])/(Kd,app+ [S]) in order to obtain Kd,app and kpol values. Statistical significance (unpaired t test) was calculated using GraphPad Prism software.
In vitro IC50 assay with host DNA polymerases.
Increasing concentrations of 2′-C-methyl-DAPN-TP or 2′-C-methyl-GTP (up to 200 μM) were incubated with recombinant human DNA polymerase α (catalog number 1075; CHIMERx), recombinant human polymerase β (catalog number 1077; CHIMERx), or recombinant human polymerase γ (catalog number 1076; CHIMERx). Each enzyme was incubated with a DNA/DNA hybrid, and DNA synthesis was performed essentially according to the manufacturer's protocol. Briefly, nucleotide incorporation was allowed to proceed for 5 min at 37°C for human DNA polymerase α and β and for 200 min at 37°C for human DNA polymerase γ.
In vitro POLRMT assay.
In vitro RNA synthesis assays with POLRMT (Indigo Biosciences) were performed as previously described (32). Briefly, 32P-radiolabeled RNA primer (5′-UUUUGCCGCGCC) was hybridized to a 3 M excess of the appropriate DNA template (5′-GGGAATGCANGGCGCGGC, where position N was replaced by A, T, or C). POLRMT (125 nM) was incubated with 500 nM 5′-radiolabeled RNA/DNA hybrid, 10 mM MgCl2, and 100 μM corresponding NTP or rNAI-TP. Incorporation was allowed to proceed for 2 h at 30°C, and reactions were stopped by the addition of 10 mM EDTA and formamide. Samples were visualized on 20% denaturing polyacrylamide gel. Data were analyzed by normalizing the product fraction for each rNAI-TP to that of the corresponding natural NTP.
Measurement of intracellular rNAI-TP levels.
Huh-7 cells were exposed to medium containing 50 μM the nucleotide prodrugs for 4 h at 37°C. Cells were washed with 3× PBS to remove extracellular compounds. Intracellular prodrugs and metabolites were extracted from 1 × 106 cells using 1 ml 70% ice-cold methanol (containing 20 nM internal standard ddATP). Samples were dried and resuspended in HPLC during mobile phase before analysis by LC-MS/MS (44). The calibration curves were generated from standards of parent nucleotides, prodrugs, 2′-C-methyl-DAPN-TP, and 2′-C-methyl-GTP. Similar protocols were repeated for primary human hepatocytes incubated with 50 μM nucleotide prodrugs for 24 h at 37°C.
Mitochondrial transcription inhibition assay.
Huh-7 cells were maintained in growth medium containing advanced Dulbecco's modified Eagle medium (DMEM) (Gibco, Life Sciences) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 2 nM l-glutamine, and 5 mM HEPES. Huh-7 cells were cultured in a 24-well plate at a density of 0.5 × 106 cells. After attachment of the cells, medium with 50 ng/ml ethidium bromide (EtBr) was added to each well and placed at 37°C in a 5% CO2 incubator (32). Following EtBr treatment for 24 h, medium was removed and cells were washed with PBS two times. Fresh medium containing 10 μM different ribonucleoside analog prodrugs was added to each well in duplicates. The plate was placed at 37°C in a 5% CO2 incubator for 48 h with medium/compound replenishment every 24 h. At 48 h, cells were washed with PBS and harvested to perform reverse transcription-PCR (RT-PCR). Total RNA was isolated using an RNeasy minikit (Qiagen), and RNA samples were treated with DNase RQ1 (Promega). Extracted RNA was converted to cDNA using SuperScript-VILO (Invitrogen). RT-PCR was performed using a LightCycler 480 probe master (Roche) and 900 nM forward and reverse primers for mitochondrial ND1 transcript (forward, 5′-AACCTCTCCACCCTTATCACAA-3′; reverse, 5′-TCATATTATGGCCAAGGGTCA-3′) and nuclear glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript (forward, 5′-TCCACTGGCGTCTTCACC-3′; reverse, 5′-GGCAGAGATGATGACCCTTTT-3′). Amplicons were detected using Roche FastStart universal probe master numbers 45 and 60 for ND1 and GAPDH, respectively. RT-PCR was conducted at 95°C for 10 min, followed by 95°C for 15 s and 60°C for 1 min for 40 cycles. The threshold cycle value (CT) of each amplicon was normalized to the internal control (GAPDH). Data were analyzed by the 2−ΔΔCT relative quantification method (45). Statistical significance (unpaired t test) was calculated using GraphPad Prism software.
RESULTS
DAPN-PD1 generates two nucleoside 5′-triphosphate metabolites that are active against HCV NS5B polymerase.
We have previously described the chemical synthesis of DAPN-PD1 as a novel ribonucleoside analog with anti-HCV activity (median effective concentration [EC50] of 0.7 μM in HCV GT 1b replicon) (37). The intracellular metabolism of 2,6-diaminopurine prodrug resulted in the generation of two nucleoside metabolites, namely, 2′-C-methyl-GTP and 2′-C-methyl-DAPN-TP, in both primary human hepatocytes and Huh-7 cells (37). Upon prodrug group cleavage, 2′-C-methyl-DAPN monophosphate was subjected to intracellular phosphorylation and 2′-C-methyl-DAPN-TP was generated (Fig. 1C). In parallel, 2′-C-methyl-DAPN nucleosides also can be a substrate for intracellular deamination at position 6 of the purine ring, which in turn leads to the generation of 2′-C-methyl-GTP (Fig. 1C). Prior studies have described an inhibitory role for 2′-C-methyl-GTP in both HCV replicon-based and cell-free NS5B polymerase assays (20, 28, 46). However, 2′-C-methyl-DAPN-TP has not previously been examined as an inhibitor of HCV NS5B-mediated RNA polymerization. In order to assess chain termination, we chemically synthesized 2′-C-methyl-DAPN-TP and performed cell-free RNA polymerization assays with purified NS5BΔ21 enzyme. 2′-C-methyl-DAPN-TP inhibited RNA synthesis at an IC50 of 3.4 ± 1.1 μM for HCV GT 1b (Table 1), while the IC50 for 2′-C-methyl-GTP was 5.6 ± 1.6 μM. Similar results were obtained for NS5B enzymes from HCV GT 2a, 3a, 4a, 5a, and 6a. The IC50s obtained for each metabolite of DAPN-PD1 were comparable to that of 2′-C-methyl-2′-F-UTP, the active metabolite of sofosbuvir (Table 1).
TABLE 1.
In vitro inhibition of NS5B-mediated RNA synthesis by nucleoside 5′-triphosphate analogs
| NS5B GT | IC50a (μM) |
||
|---|---|---|---|
| 2′-C-Methyl-DAPN-TP | 2′-C-Methyl-GTP | 2′-C-Methyl-2′-F-UTP | |
| 1b | 3.4 ± 1.1 | 5.6 ± 1.6 | 2.9 ± 0.6 |
| 2a | 3.6 ± 2.1 | 5.1 ± 1.2 | 3.0 ± 0.9 |
| 3a | 1.6 ± 0.1 | 3.6 ± 0.8 | 7.2 ± 2.1 |
| 4a | 1.8 ± 1.4 | 7.4 ± 4.2 | 12.8 ± 10.7 |
| 5a | 4.7 ± 3.5 | 3.5 ± 1.0 | ND b |
| 6a | 5.1 ± 5.1 | 2.1 ± 0.4 | ND |
In vitro IC50s are the averages from two to four replicates ± standard deviations.
ND, not determined.
Incorporation profiles for active nucleoside 5′-triphosphate metabolites of DAPN-PD1.
In vitro nucleotide incorporation assays preformed on NS5B/RNA elongation complexes (described in Materials and Methods) were conducted in order to confirm the incorporation of 2′-C-methyl-DAPN-TP as an A analog. The RNA templates used allowed for ATP or GTP analog incorporation at position +16 or +11, respectively (Fig. 2A and C). Our biochemical data confirmed that as expected, 2′-C-methyl-DAPN-TP behaves as an ATP analog, while 2′-C-methyl-GTP maintained a GTP analog incorporation profile. No incorporation was observed with up to 250 μM 2′-C-methyl-DAPN-TP when cytidine was present in the RNA template at position +11, indicating that 2′-C-methyl-DAPN-TP is not a GTP analog (Fig. 2B and D). Overall, these data suggest that DAPN-PD1 can deliver two rNAI-TP metabolites with distinct incorporation profiles.
FIG 2.
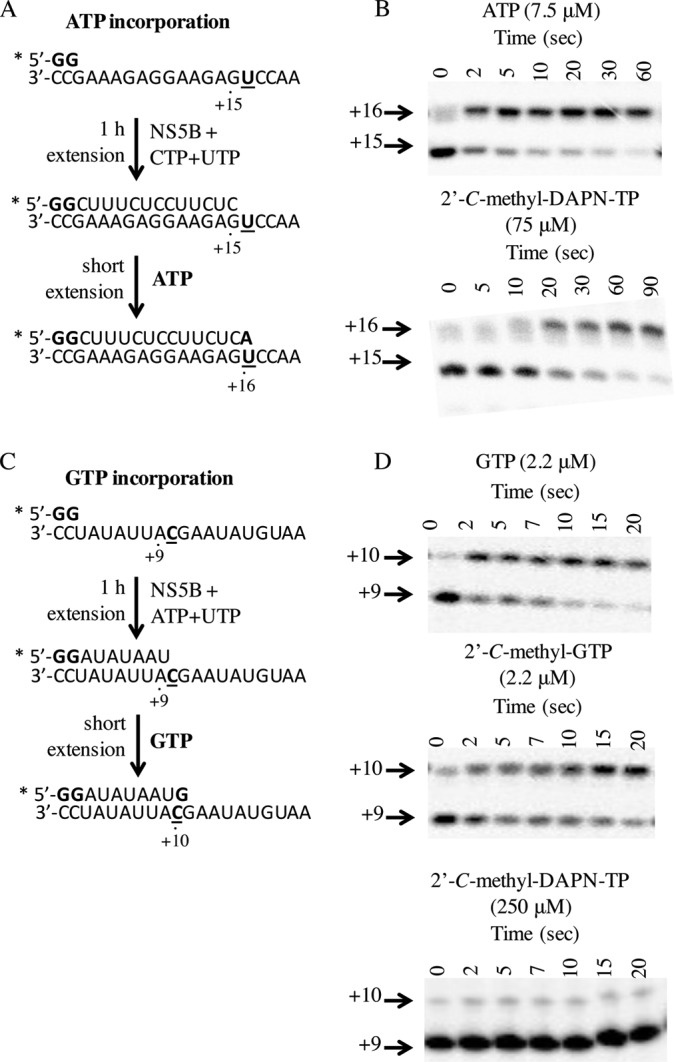
NS5B elongation complex single-nucleotide extension. (A) Radiolabeled GpG primer is extended to position +15 with the addition of CTP and UTP. Incorporation of ATP or ATP analog subsequently is measured at position +16. (B) Nucleoside triphosphate incorporation was resolved on a denaturing polyacrylamide gel where incubation with ATP (7.5 μM) or 2′-C-methyl-DAPN-TP (75 μM) results in extension by one position over time. (C) Radiolabeled GpG primer is extended to position +9 with the addition of ATP and UTP. Incorporation of GTP or GTP analog subsequently was measured at position +10. (D) Nucleoside 5′-triphosphate incorporation was resolved on a denaturing polyacrylamide gel where GTP (2.2 μM) and 2′-C-methyl-GTP (2.2 μM), but not 2′-C-methyl-DAPN-TP (250 μM), show extension by one position over time. Assays were repeated at various NTP concentrations in order to obtain Kd,app and kpol values for each inhibitor (summarized in Table 2).
Kinetics of incorporation for rNAI-TPs generated by DAPN-PD1.
Rapid nucleotide incorporation by NS5B in the elongation complex was assessed in order to determine the apparent nucleotide dissociation equilibrium constant (Kd,app) and maximal incorporation rate (kpol). Increasing concentrations of nucleoside triphosphate analogs were incubated with the appropriate NS5B/RNA elongation complex, and incorporation was measured over a short time period (Fig. 2). The amount of product formed was fitted to a single-turnover nucleotide incorporation plot where productively bound NS5B/RNA complexes could be extended by one nucleotide (Fig. 3). Kd,app and kpol values are summarized in Table 2, with incorporation efficiency reported as a kpol/Kd,app ratio. 2′-C-methyl-GTP appears to have over 13-fold lower catalytic efficiency than GTP substrate, while 2′-C-methyl-DAPN-TP appears to be more severely compromised with regard to both binding affinity and incorporation rate (over 900-fold) (Table 2). Although earlier results suggest that both 2′-C-methyl-DAPN-TP and 2′-C-methyl-GTP are inhibitors of NS5B-mediated RNA synthesis, the kinetic parameters measured here suggest that 2′-C-methyl-GTP is more readily incorporated by NS5B polymerase (see Discussion).
FIG 3.
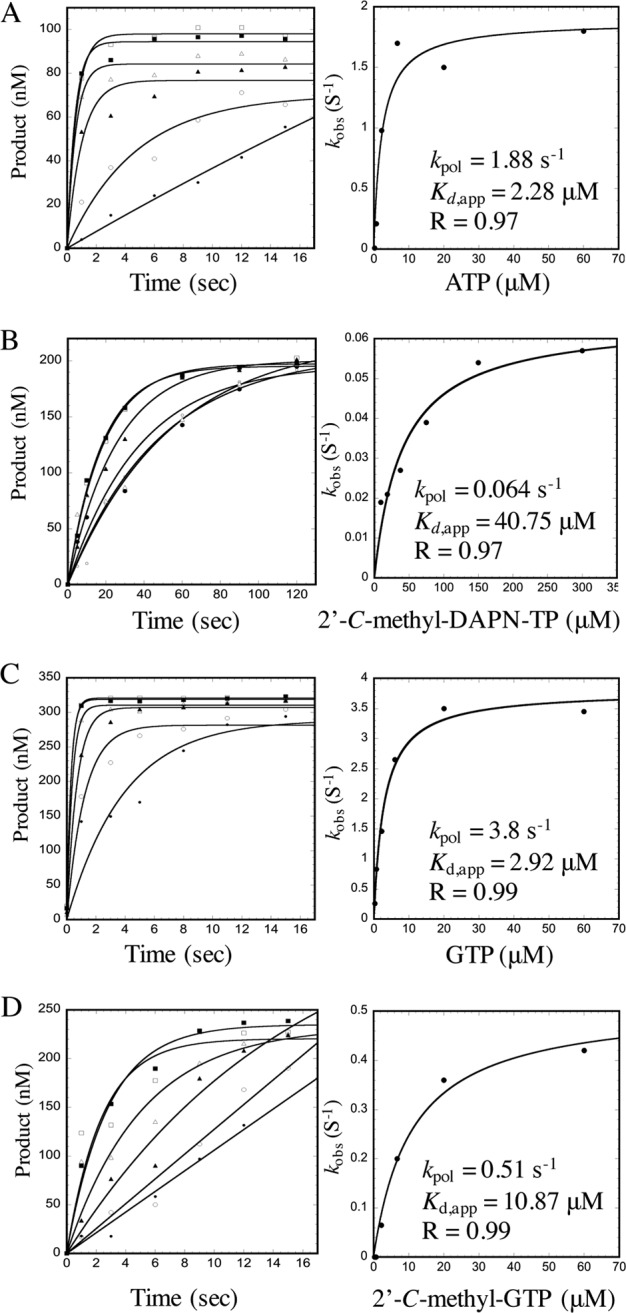
Kinetic parameters for nucleoside 5′-triphosphate incorporation by NS5B polymerase. Single representative plots are shown for kinetic parameters reported in Table 2. Incorporation of increasing amounts of rNTP coincubated with preformed NS5B/RNA elongation complexes was monitored over time. The amount of product formed was quantified and fitted to a single exponential equation, y = ymax(1 − exp(−kobs[S]), in order to obtain rates of incorporation. Obtained kobs(S − 1) values were plotted as a function of NTP concentration and fitted to a nonlinear regression using the hyperbolic equation y = (kpol[S])/(Kd,app+ [S]). kpol and Kd,app values are indicated on each plot for ATP (A), 2′-C-methyl-DAPN-TP (B), GTP (C), and 2′-C-methyl-GTP (D).
TABLE 2.
Kinetic parameters for nucleoside 5′-triphosphate incorporation by HCV NS5B polymerase
| NTP substrate | Kd,appa (μM) | kpola (s−1) | Incorporation efficiencyb | Selectivityc |
|---|---|---|---|---|
| ATP | 1.4 ± 0.9 | 1.3 ± 0.6 | 0.9 | |
| 2′-C-Methyl-DAPN-TP | 37 ± 1.0 | 0.05 ± 0.02 | 0.001 | 900 |
| GTP | 4.1 ± 1.6 | 4.1 ± 0.9 | 1.0 | |
| 2′-C-Methyl-GTP | 9.6 ± 1.9 | 0.7 ± 0.3 | 0.07 | 13.3 |
Kinetic parameters were calculated based on at least two separate replicates ± standard deviations (see Fig. 3 for additional information).
Incorporation efficiency was determined as kpol/Kd,app for each NTP.
Selectivity was determined as (kpol/Kd,app)NTP/(kpol/Kd,app)rNAI-TP.
In addition to enzymatic measurements of incorporation, cytoplasmic rNAI-TP levels are another important factor to take into consideration when estimating in vivo incorporation rates of nucleoside analogs. The rate constant for incorporation (keff, NS5B [per second]) values can be calculated through experimentally determined kinetic parameters and measurement of intracellular rNAI-TP levels according to the equation Keff, NS5B (per second) = (kpol × [NTP])/(Kd,app + [NTP]) (Table 3) (32). Intracellular metabolism of DAPN-PD1 was monitored in primary human hepatocyte cells, which represent the primary host cell environment where inhibition of HCV viral replication occurs. By taking into account intracellular levels of natural NTPs and rNAI-TPs, we were able to determine keff, NS5B values for each nucleoside species. Based on these variables, we estimated that fold differences between rate constants for incorporation of 2′-C-methyl-DAPN-TP to be ∼43-fold lower than those for ATP. A difference of 8-fold was observed for 2′-C-methyl-GTP compared to GTP (Table 3).
TABLE 3.
Intracellular metabolism and NS5B-catalyzed incorporation rates for nucleoside triphosphate analogs
| NTP substrate | Intracellular NTP (pmol/106 cells) | Intracellular NTP (μM) | keff, NS5Bc (s−1) | FDd |
|---|---|---|---|---|
| ATP | 8,700.0a | 4,400.0 | 1.3 | |
| 2′-C-Methyl-DAPN-TP | 96.0b | 48.0 | 0.03 | 43 |
| GTP | 1,600.0a | 790.0 | 4.1 | |
| 2′-C-Methyl-GTP | 47.0b | 24.0 | 0.5 | 8.0 |
Intracellular ATP and GTP concentrations were determined in Huh-7 cell lines as described in Materials and Methods. Primary human hepatocytes were estimated to contain amounts of natural NTPs comparable to those of Huh-7 cells. Nucleotide levels measured by LC-MS/MS were converted to micromolar concentrations, estimating a cell volume of 2 pl/cell (33).
Intracellular 2′-C-methyl-DAPN-TP and 2′-C-methyl-GTP levels measured in primary human hepatocytes after treatment with 50 μM DAPN-PD1 for 24 h.
Rate constant for incorporation was determined with the equation Keff, NS5B (per second) = (kpol × [NTP])/(Kd,app + [NTP]). kpol and Kd,app values were determined experimentally as reported in Table 2.
Fold difference (FD) was calculated by dividing the Keff, NS5B of natural NTP substrate by that of the corresponding rNAI-TP.
Resistance profiles of 2′-C-methyl-DAPN-TP and 2′-C-methyl-GTP metabolites.
In order to assess resistance to active rNAI-TP metabolites of DAPN-PD1, cell-free IC50 assays were performed with purified NS5BΔ21 containing a mutation either at position 96 or 282, each of which has been associated with drug resistance. The presence of S96T mutation did not affect susceptibility to either 2′-C-methyl-DAPN-TP or 2′-C-methyl-GTP (IC50 of 2.5 μM and 8.8 μM, respectively). However, compared to WT enzyme, addition of S282T mutation resulted in IC50s of above 50 μM, indicating at least 15-fold and 9-fold resistance to 2′-C-methyl-DAPN-TP and 2′-C-methyl-GTP, respectively (Table 4). Similar trends were observed for 2′-C-methyl-2′-F-UTP.
TABLE 4.
Effect of resistance-conferring mutations on nucleoside analog potency in cell-free assays
| NS5B | 2′-C-Methyl-DAPN-TP |
2′-C-Methyl-GTP |
2′-C-Methyl-2′-F-UTP |
|||
|---|---|---|---|---|---|---|
| IC50a (μM) | FDb | IC50 (μM) | FD | IC50 (μM) | FD | |
| GT 1b WT | 3.4 ± 1.1 | 1 | 5.6 ± 1.6 | 1 | 2.9 ± 0.6 | 1 |
| GT 1b S96T | 2.5 ± 0.6 | 0.7 | 8.8 ± 2.9 | 1.6 | 3.5 ± 0.9 | 1.2 |
| GT 1b S282T | >50 | >15 | >50 | >9 | >50 | >19 |
In vitro IC50s were determined with cell-free assays using purified recombinant NS5BΔ21. Each value is the average from two to three replicates (± standard deviations).
FD, fold difference.
Incorporation of 2′-C-methyl-DAPN-TP by human host DNA polymerases.
In order to examine potential off-target inhibition of host cellular polymerases by 2′-C-methyl-DAPN-TP, increasing amounts of the rNAI-TP were incubated with human DNA polymerases α, β, and γ. No inhibition of DNA synthesis was observed for up to 200 μM 2′-C-methyl-DAPN-TP or 2′-C-methyl-GTP (Table 5), suggesting these nucleoside analog triphosphates are not substrates for host DNA polymerases. The active metabolite of sofosbuvir, 2′-C-methyl-2′-F-UTP, did not show any inhibitory effect up to 100 μM (Table 5).
TABLE 5.
In vitro inhibition of host DNA polymerase-mediated DNA synthesis
| Inhibitor | IC50 (μM) |
||
|---|---|---|---|
| DNA Pol α | DNA Pol β | DNA Pol γ | |
| 2′-C-Methyl-DAPN-TP | >200 | >200 | >200 |
| 2′-C-Methyl-GTP | >200 | >200 | >200 |
| 2′-C-Methyl-2′-F-UTP | >100 | >100 | >100 |
| 3′-dTTPa | NDb | 17.7 | 41.7 |
| Aphidicolinc | 5.4 | ND | ND |
3′-dTTP was used as a positive control for inhibition of DNA synthesis by host DNA polymerase (Pol) β and γ.
ND, not determined.
Aphidicolin was used as a positive control for inhibition of DNA synthesis by host DNA polymerase α.
Incorporation of 2′-C-methyl-DAPN-TP by human host mitochondrial RNA polymerase POLRMT.
We next examined whether nucleoside 5′-triphosphate metabolites of DAPN-PD1 were substrates for host mitochondrial RNA polymerase (POLRMT). When purified POLRMT was incubated with annealed 32P-radiolabeled RNA/DNA hybrids containing an A, C, or T on the DNA template at position 9 (Fig. 4A), we found that the addition of 100 μM 2′-C-methyl-DAPN-TP resulted in 34% incorporation when normalized to incorporation of natural ATP substrate (Fig. 4B). Consistent with previous findings (32), 39% incorporation was observed for 2′-C-methyl-GTP normalized to natural GTP substrate (Fig. 4B), while the active metabolite of sofosbuvir, 2′-C-methyl-2′-F-UTP, was not found to be a substrate for POLRMT when tested up to 100 μM.
FIG 4.
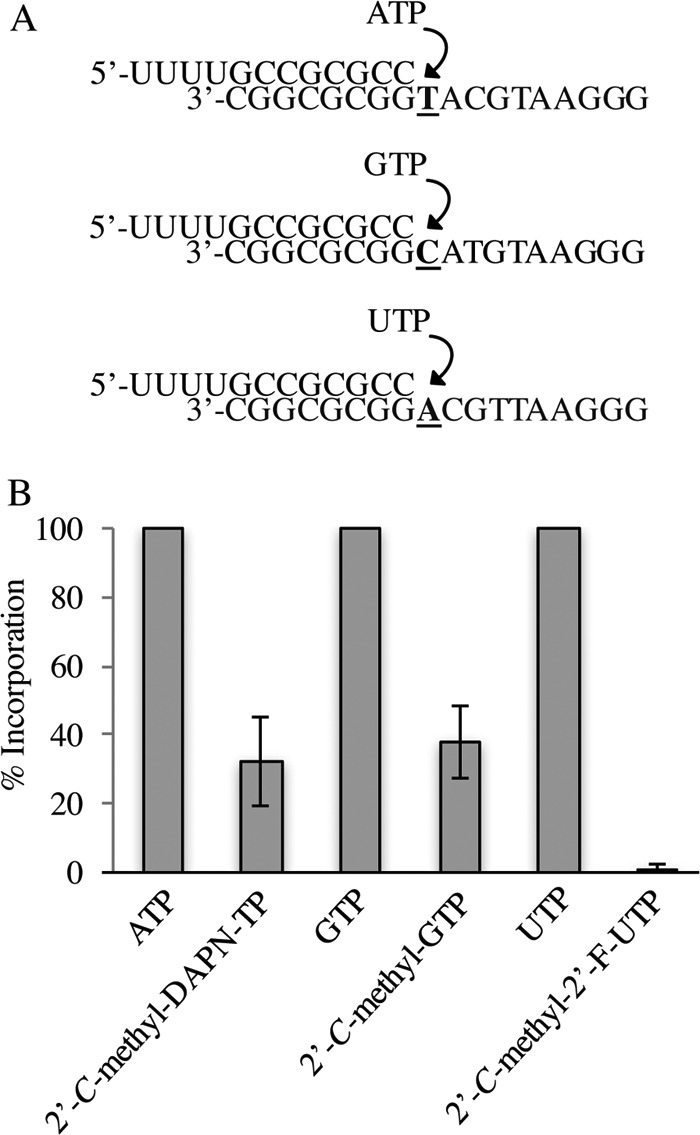
Ribonucleoside 5′-triphosphate analog incorporation by POLRMT. (A) Schematic representation of RNA/DNA primer/template substrates used for incorporation of each nucleoside triphosphate. (B) Nucleotide incorporation by POLRMT was allowed to proceed for 2 h in the presence of 100 μM each NTP or rNAI-TP. The percentage of RNA product at n + 1 for each rNAI-TP was normalized to that of natural NTP substrates. Error bars represent standard deviation (SD) values from three separate experiments.
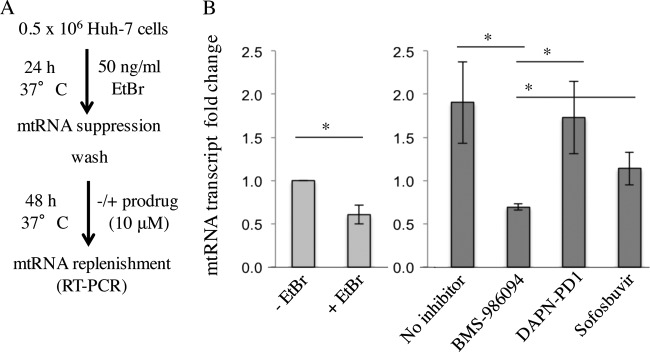
Effect of DAPN-PD1 on mitochondrial transcription in Huh-7 cells.
A phenotypic cell-based assay that measures the impact of DAPN-PD1 on mtRNA transcription was performed. Considering the observed clinical safety of sofosbuvir, and conversely the clinical toxicity of BMS-986094, these compounds were used as negative and positive controls, respectively. When Huh-7 cells were coincubated with EtBr (Fig. 5A), which selectively reduces mtRNA synthesis, we observed a 2-fold reduction in mtRNA transcript levels. mtRNA replenishment subsequently was observed after EtBr removal (Fig. 5B). Interestingly, we found that cells previously treated with EtBr exhibited increased mitochondrial transcription as normalized to nuclear GAPDH (thus accounting for increased cell growth). This suggests that cells overcompensate for EtBr-induced mtRNA suppression by selectively upregulating mtRNA synthesis. Conversely, treatment with 10 μM BMS-986094 (which generates 2′-C-methyl-GTP intracellularly) was sufficient to fully inhibit mtRNA replenishment after EtBr removal (Fig. 5B). Treatment with DAPN-PD1 (which generates 2′-C-methyl-GTP and 2′-C-methyl-DAPN-TP intracellularly) did not impede mtRNA replenishment. Treatment with the comparator sofosbuvir did not inhibit mtRNA replenishment. Our data suggest that although both DAPN-PD1 and BMS-986094 generate rNAI-TPs that can be substrates for POLRMT, only BMS-986094 demonstrates a deleterious effect with regard to replenishment of mitochondrial transcription. The phenotypic impact of the observed rNAI-mediated mtRNA suppression on mitochondrial function is not yet fully understood.
FIG 5.
Effect of ribonucleotide prodrugs on mitochondrial RNA transcription. (A) Huh-7 cells were incubated with ethidium bromide (EtBr) for 24 h. EtBr was subsequently removed by washing the cells with PBS buffer in the absence or presence of 10 μM each prodrug. RT-PCR was employed to measure mtRNA transcript after 48 h of replenishment. (B) Representative plot of mtRNA transcript levels after treatment with EtBr and nucleoside prodrugs as normalized to no-EtBr treatment controls (−EtBr). “+EtBr” sample represents mtRNA levels at 24 h (light gray bars) after EtBr treatment. “No inhibitor” sample represents the amount of mtRNA replenishment at 48 h (dark gray bars) after EtBr removal in the absence of inhibitors. All mtRNA measurements are normalized to nuclear GAPDH RNA, which also serves as an internal control for cell growth. Error bars represent SD values from two to four separate replicates. The differences between the −EtBr control sample, +EtBr sample, and no inhibitor control were significant (P < 0.01). The differences between BMS-986094-treated samples compared to either DAPN-PD1-treated or sofosbuvir-treated samples were significant (P < 0.05). Differences between no inhibitor, DAPN-PD1-treated samples, and sofosbuvir-treated samples were not statistically significant. An unpaired t test (GraphPad Prism software) was used to determine significance for all samples.
Intracellular concentrations of nucleoside 5′-triphosphate metabolites.
In order to address the underlying cause of selective mtRNA depletion with BMS-986094 but not DAPN-PD1, we examined the intracellular metabolism of each inhibitor. Huh-7 cells were incubated with 50 μM either BMS-986094 or DAPN-PD1 for 4 h at 37°C, and intracellular 2′-C-methyl-GTP levels were measured using quantitative LC-MS/MS. We found that metabolism of BMS-986094 generated 87-fold higher intracellular 2′-C-methyl-GTP levels than DAPN-PD1 (680 pmol/106 cells and 7.8 pmol/106 cells, respectively) (Table 6). Taking into account previously reported kinetic parameters of incorporation of 2′-C-methyl-GTP by POLRMT (32), we calculated the rate constant for the incorporation value keff, POLRMT (also known as the mitovir score [32]) for this inhibitor. When the intracellular 2′-C-methyl-GTP levels generated by BMS-986094 or DAPN-PD1 were compared, mitovir scores of 0.04 s−1 and 0.0007 s−1, respectively, were obtained (Table 6). The mitovir score for 2′-C-methyl-GTP generated by BMS-986094 was determined to be 57-fold higher than the mitovir score for 2′-C-methyl-GTP generated by DAPN-PD1.
TABLE 6.
Generation of 2′-C-methyl-GTP from DAPN-PD1 and BMS-986094 in Huh-7 cells
| Prodrug | 2′-C-Methyl-GTPa (pmol/106 cells) | 2′-C-Methyl-GTPb (μM) | keff, POLRMTc (s−1) | Selectivityd |
|---|---|---|---|---|
| DAPN-PD1 | 7.8 ± 0.2 | 3.9 | 0.0007 | |
| BMS-986094 | 680 ± 40 | 340 | 0.04 | 57.0 |
rNAI prodrug (50 μM) was incubated with Huh-7 cells for 4 h at 37°C. Intracellular 2′-C-methyl-GTP levels generated from each prodrug were determined using LC-MS/MS. Each value is the average from two separate replicates (± standard deviations).
Amount of nucleoside 5′-triphosphate metabolites was converted from picomoles per million cells to molarity using an estimated cellular volume of 2 pl per cell (33).
Rate constant for incorporation was determined with the equation Keff, POLRMT (per second) = (kpol × [NTP])/(Kd,app + [NTP]). Keff, POLRMT was calculated based on reported kinetic properties of 2′-C-methyl-GTP incorporation by POLRMT enzyme (33).
Selectivity was determined as (Keff, POLRMT)2′-C-methyl-GTP from DAPN-PD1/(Keff, POLRMT) 2′-Cmethyl-GTP from BMS-986094.
DISCUSSION
β-d-2′-C-Methyl-2,6-diaminopurine ribonucleoside is a new phosphoramidate prodrug with selective antiviral activity against HCV in vitro. This prodrug can produce two nonobligate chain-terminator rNAI-TPs. While 2′-C-methyl-GTP behaves as a GTP analog, we observed that 2′-C-methyl-DAPN-TP is an ATP analog. This is consistent with previous reports that modified 2,6-diaminopurine nucleosides form base pairs opposite thymidine or uridine residues (47, 48). We found that both 2′-C-methyl-DAPN-TP and 2′-C-methyl-GTP inhibited NS5B-mediated RNA synthesis in cell-free assays against all NS5B genotypes tested (Table 1). These findings suggest that both nucleoside 5′-triphosphate metabolites have the potential to contribute to anti-HCV inhibition in vivo. In order to address this hypothesis, we took into consideration the kinetic parameters of nucleotide incorporation by NS5B, as well as intracellular rNAI-TP levels for each active metabolite. Rapid single-nucleotide incorporation assays demonstrated that incorporation efficiency was reduced for both rNAI-TPs, albeit to different degrees. The incorporation efficiency of 2′-C-methyl-GTP was reduced by 13-fold compared to that of GTP, largely due to changes in the binding affinity (P = 0.0005). On the other hand, 2′-C-methyl-DAPN-TP was 900-fold less efficiently incorporated than ATP. This effect is largely attributed to reductions in the rate of polymerization (kpol) of this inhibitor (Table 2).
The incorporation rate constants (keff, NS5B) for rNAI-TP metabolites of DAPN-PD1 were calculated based on their intracellular metabolism in primary human hepatocytes. We estimated a 43-fold difference in keff, NS5B values between 2′-C-methyl-DAPN-TP and ATP, while the difference in keff, NS5B values is 8-fold for 2′-C-methyl-GTP and GTP (Table 3). It is worth noting that the 9.6-kb genome of HCV GT 1b is comprised of 58% G·C content, indicating that slightly more opportunities exist for GTP or CTP analog incorporation. Conversely, the 3′-untranslated poly(U/UC) region of the viral genome (81% U·A content) can represent a hot spot for chain termination by ATP analogs, such as 2′-C-methyl-DAPN-TP. Overall, calculations described above suggest that under the cellular conditions tested, 2′-C-methyl-GTP has a higher probability for incorporation by NS5B enzyme. At the same time, considering that a single incorporation event is sufficient to abrogate virus replication, the contribution of 2′-C-methyl-DAPN-TP to chain termination of viral RNA synthesis in vivo should not be ruled out.
Cell culture selection of resistance-conferring mutations such as S282T or S96T has been associated with rNAIs containing a 2′-C-methyl or 4′-azido group, respectively, on the ribose ring (15, 23, 49, 50). Here, we report a low level of resistance conferred by recombinant S282T mutant NS5B enzyme when each rNAI-TP was tested separately. However, we have previously reported that the GT 1b replicon harboring the S282T mutation does not show resistance to DAPN-PD1 (37). We hypothesize that the combined delivery of both metabolites in cell culture can overcome resistance development. Additional studies are under way to address the distinct resistance profiles seen in cell-based and cell-free systems.
In addition to antiviral activity, a thorough examination of the safety profile of rNAI is critical for preclinical development of investigational compounds. As highlighted recently, phase II clinical trials with BMS-986094 were halted after reports of severe toxicity and one death (51). Because 2′-C-methyl-GTP is an intracellular metabolite common to both BMS-986094 and DAPN phosphoramidate prodrugs, we previously reported head-to-head comparative assays wherein the cytotoxicity of each prodrug was monitored in various cell lines. We were unable to detect cytotoxicity with DAPN phosphoramidate prodrugs in Vero, Huh-7, HepG2, CEM, peripheral blood mononuclear cells (PBMC), or PC3 cell lines up to 100 μM. Similarly, no bone marrow toxicity or any changes in lactic acid production were observed with DAPN prodrug treatment. On the other hand, BMS-986094 treatment led to mitochondrial DNA toxicity, increased lactic acid production, and Huh-7 cell death (37). In this study, we aimed to look more closely at correlates of cytotoxicity with BMS-986094 treatment in order to better understand the lack of cytotoxicity observed with DAPN phosphoramidate prodrugs. It was recently reported that human mitochondrial RNA polymerase (POLRMT) incorporates 2′-C-methyl-GTP as well as a number of other anti-HCV ribonucleoside analogs (32, 33). Here, we show that 2′-C-methyl-DAPN-TP is also a substrate for POLRMT (Fig. 4), suggesting that similar to BMS-986094, metabolites of DAPN-PD1 also have the potential to interfere with mitochondrial transcription. However, the biochemical data alone do not fully explain the distinct cell-based toxicity profiles of BMS-986094 and DAPN phosphoramidate prodrugs. When changes in mtRNA levels were measured after treatment with inhibitors, we observed that treatment with BMS-986094 prevented RNA replenishment while treatment with either DAPN-PD1 or sofosbuvir did not have an effect in this regard (Fig. 5). These data were consistent with the observation that BMS-986094, but not DAPN-PD1, causes mitochondrial toxicity in Huh-7 cells in a 14-day assay (37).
In order to address the underlying cause of selective mtRNA inhibition by BMS-986094, we monitored the intracellular metabolism of each prodrug in Huh-7 cells. BMS-986094 prodrug generated ∼87 times higher 2′-C-methyl-GTP levels than DAPN-PD1 (Table 6). This finding is consistent with the low-nanomolar median effective concentration (EC50) value reported for BMS-986094 (28). As proposed previously (32), the data support the hypothesis that inhibition of mtRNA transcription by rNAIs is dependent not only on POLRMT substrate specificity but also on intracellular concentrations of rNAI-TP generated by each prodrug.
As a caveat, it is worth noting that all rNAI-TP measurements reported in this study are cytoplasmic, and the calculated mitovir score values are based on the previously reported assumption that intramitochondrial rNAI-TP levels correlate with rNAI-TP levels detected in the cytoplasm (32). Therefore, the obtained mitovir scores may change with more accurate measurements of intramitochondrial rNAI-TP levels. It is also possible that prodrug group choice may differently affect entry or accumulation of rNAI-TPs in the mitochondria, partially accounting for the distinct inhibitory profile of mtRNA transcription observed with DAPN-PD1 and BMS-986094 treatment. For example, BMS-986094 may be particularly well suited for targeting the mitochondria. Conversely, the DAPN prodrug group may modulate mitochondrial entry, or the mitochondria may not have the necessary adenosine deaminases for 2,6-diaminopurine-to-guanine base conversion. Further studies are under way to examine the dynamics of rNAI-TP generation inside the mitochondria relative to prodrug choice.
Although we and others (32) have reported on a correlation between cytotoxicity and mtRNA suppression, it is worth highlighting that little is known about the phenotypic effects of rNAI-mediated mtRNA inhibition on mitochondrial protein production and function. Mitochondrial damage caused by inhibition of DNA polymerase γ by anti-HIV 3′-deoxynucleoside inhibitors has been shown to gradually accumulate over time (52). It is not clear whether inhibition of mtRNA transcription with ribonucleoside analog inhibitors would show similar kinetics with regard to cytotoxicity or whether deleterious effects would appear in a more immediate fashion. As more antiviral rNAIs are developed against RNA viruses, a better understanding of the dynamics of transcription-mediated mitochondrial toxicity will be warranted. Furthermore, it is conceivable that cells that require higher ATP consumption are more sensitive to changes in mtRNA transcript levels. Considering that cardiotoxicity was observed in BMS-986094 phase II clinical trials (31), high 2′-C-methyl-GTP may accumulate in cardiac tissue in a somewhat selective matter. This accumulation may in turn have more immediate consequences because of increased energy requirements for cardiac tissue. In agreement with this hypothesis, we have observed that the metabolism of BMS-986094 in cardiomyocytes leads to high intracellular 2′-C-methyl-GTP accumulation (S. Tao, personal communication).
Finally, we cannot exclude the possibility that other non-rNAI-TP components of ribonucleoside prodrugs contribute to cytotoxicity. Indeed, the intracellular metabolism of BMS-986094 leads to the generation of potentially hazardous by-products, such as 1-naphtol, neopentanol, and methanol. To address this issue, we synthesized each of the aforementioned metabolites separately and tested their effect on cytotoxicity in VERO, CEM, PBM, and Huh-7 cells. We did not observe toxicity with up to 100 μM each metabolite in any of the cell lines tested (data not shown). These data suggest that the observed toxicity involves the antiviral molecule as a whole. Importantly, it is worth noting that cellular metabolism of the prodrug moiety of DAPN-PD1 results in the generation of dihydrocoumarin metabolites, a nontoxic food additive that has been in use for human consumption for over 40 years (53, 54).
In conclusion, this study describes the biochemical properties of nucleoside metabolites of DAPN-PD1. Our cell-based (37) and cell-free assays demonstrate that the potency and resistance profiles of DAPN-PD1 metabolites are comparable to that of the sofosbuvir metabolite 2′-C-methyl-2′-F-UTP. Assessment of DAPN-PD1 and sofosbuvir metabolism also shows that comparable levels of intracellular rNAI-TP are achieved with each prodrug in primary human hepatocytes (S. Tao, personal communication). Direct comparisons between DAPN-PD1, BMS-986094, and sofosbuvir highlight the importance of prodrug group choice in maintaining antiviral activity while minimizing off-target inhibition of host polymerases. These findings have important implications for addressing cytotoxicity with ribonucleoside analogs in development. Finally, DAPN-PD1 can deliver intracellularly two ribonucleotide analog chain terminators with distinct incorporation profiles, making it an attractive prodrug that needs to be further preclinically developed.
ACKNOWLEDGMENTS
Emmanuela Anandarajah and Dana Ditje are acknowledged for technical assistance with regard to NS5B protein purification. Emily Hammond is acknowledged for experimental assistance. Joseph Hollenbaugh and Judy Mathew are acknowledged for careful reading of the manuscript.
S.S.L. and R.F.S. are founders and shareholders of Cocrystal Pharma, Inc. Emory received no funding from Cocrystal Pharma, Inc., to perform this work and vice versa.
REFERENCES
- 1.Hutin HKM, Dore GJ, Perz JF, Armstrong GL, Dusheiko G. 2004. Global burden of disease (GBD) for hepatitis C. J Clin Pharmacol 44:20–29. doi: 10.1177/0091270003258669. [DOI] [PubMed] [Google Scholar]
- 2.Rein DB, Wittenborn JS, Weinbaum CM, Sabin M, Smith BD, Lesesne SB. 2011. Forecasting the morbidity and mortality associated with prevalent cases of pre-cirrhotic chronic hepatitis C in the United States. Dig Liver Dis 43:66–72. doi: 10.1016/j.dld.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Hagan LM, Schinazi RF. 2013. Best strategies for global HCV eradication. Liver Int 33(Suppl 1):S68–S79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong JB, McQuillan GM, McHutchison JG, Poynard T. 2000. Estimating future hepatitis C morbidity, mortality, and costs in the United States. Am J Public Health 90:1562–1569. doi: 10.2105/AJPH.90.10.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogata N, Alter HJ, Miller RH, Purcell RH. 1991. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci U S A 88:3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuevas JM, Gonzalez-Candelas F, Moya A, Sanjuan R. 2009. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J Virol 83:5760–5764. doi: 10.1128/JVI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaefer EA, Chung RT. 2011. The impact of human gene polymorphisms on HCV infection and disease outcome. Semin Liver Dis 31:375–386. doi: 10.1055/s-0031-1297926. [DOI] [PubMed] [Google Scholar]
- 8.Zeuzem S, Berg T, Moeller B, Hinrichsen H, Mauss S, Wedemeyer H, Sarrazin C, Hueppe D, Zehnter E, Manns MP. 2009. Expert opinion on the treatment of patients with chronic hepatitis C. J Viral Hepat 16:75–90. doi: 10.1111/j.1365-2893.2008.01012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoulim F, Liang TJ, Gerbes AL, Aghemo A, Deuffic-Burban S, Dusheiko G, Fried MW, Pol S, Rockstroh JK, Terrault NA, Wiktor S. 2015. Hepatitis C virus treatment in the real world: optimising treatment and access to therapies. Gut 64:1824–1833. doi: 10.1136/gutjnl-2015-310421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartenschlager R, Lohmann V, Penin F. 2013. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol 11:482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- 11.Scheel TK, Rice CM. 2013. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med 19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wendt A, Adhoute X, Castellani P, Oules V, Ansaldi C, Benali S, Bourliere M. 2014. Chronic hepatitis C: future treatment. Clin Pharmacol 6:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler JJ, Nettles JH, Amblard F, Hurwitz SJ, Bassit L, Stanton RA, Ehteshami M, Schinazi RF. 2014. Approaches to hepatitis C treatment and cure using NS5A inhibitors. Infect Drug Resist 7:41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coats SJ, Garnier-Amblard EC, Amblard F, Ehteshami M, Amiralaei S, Zhang H, Zhou L, Boucle SR, Lu X, Bondada L, Shelton JR, Li H, Liu P, Li C, Cho JH, Chavre SN, Zhou S, Mathew J, Schinazi RF. 2014. Chutes and ladders in hepatitis C nucleoside drug development. Antiviral Res 102:119–147. doi: 10.1016/j.antiviral.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sofia MJ. 2013. Nucleotide prodrugs for the treatment of HCV infection. Adv Pharmacol 67:39–73. doi: 10.1016/B978-0-12-405880-4.00002-0. [DOI] [PubMed] [Google Scholar]
- 16.Schinazi RF, Shi J, Whitaker T. 2015. Sofosbuvir (Sovaldi): the first-in-class HCV NS5B nucleotide polymerase inhibitor, p 61–80. Innovative drug synthesis. John Wiley & Sons, Inc, Hoboken, NJ. [Google Scholar]
- 17.Donaldson EF, Harrington PR, O'Rear JJ, Naeger LK. 2015. Clinical evidence and bioinformatics characterization of potential hepatitis C virus resistance pathways for Sofosbuvir. Hepatology 61:56–65. doi: 10.1002/hep.27375. [DOI] [PubMed] [Google Scholar]
- 18.Tong X, Le Pogam S, Li L, Haines K, Piso K, Baronas V, Yan JM, So SS, Klumpp K, Najera I. 2014. In vivo emergence of a novel mutant L159F/L320F in the NS5B polymerase confers low-level resistance to the HCV polymerase inhibitors mericitabine and sofosbuvir. J Infect Dis 209:668–675. doi: 10.1093/infdis/jit562. [DOI] [PubMed] [Google Scholar]
- 19.Svarovskaia ES, Dvory-Sobol H, Parkin N, Hebner C, Gontcharova V, Martin R, Ouyang W, Han B, Xu S, Ku K, Chiu S, Gane E, Jacobson IM, Nelson DR, Lawitz E, Wyles DL, Bekele N, Brainard D, Symonds WT, McHutchison JG, Miller MD, Mo H. 2014. Infrequent development of resistance in genotype 1-6 hepatitis C virus-infected subjects treated with sofosbuvir in phase 2 and 3 clinical trials. Clin Infect Dis 59:1666–1674. doi: 10.1093/cid/ciu697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, Bartholomew L, Bosserman MR, Ceccacci A, Colwell LF, Cortese R, De Francesco R, Eldrup AB, Getty KL, Hou XS, LaFemina RL, Ludmerer SW, MacCoss M, McMasters DR, Stahlhut MW, Olsen DB, Hazuda DJ, Flores OA. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem 278:49164–49170. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- 21.Olsen DB, Eldrup AB, Bartholomew L, Bhat B, Bosserman MR, Ceccacci A, Colwell LF, Fay JF, Flores OA, Getty KL, Grobler JA, LaFemina RL, Markel EJ, Migliaccio G, Prhavc M, Stahlhut MW, Tomassini JE, MacCoss M, Hazuda DJ, Carroll SS. 2004. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob Agents Chemother 48:3944–3953. doi: 10.1128/AAC.48.10.3944-3953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Pogam S, Jiang WR, Leveque V, Rajyaguru S, Ma H, Kang H, Jiang S, Singer M, Ali S, Klumpp K, Smith D, Symons J, Cammack N, Najera I. 2006. In vitro selected Con1 subgenomic replicons resistant to 2′-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology 351:349–359. doi: 10.1016/j.virol.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 23.Ludmerer SW, Graham DJ, Boots E, Murray EM, Simcoe A, Markel EJ, Grobler JA, Flores OA, Olsen DB, Hazuda DJ, LaFemina RL. 2005. Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob Agents Chemother 49:2059–2069. doi: 10.1128/AAC.49.5.2059-2069.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asselah T. 2014. Sofosbuvir for the treatment of hepatitis C virus. Expert Opin Pharmacother 15:121–130. doi: 10.1517/14656566.2014.857656. [DOI] [PubMed] [Google Scholar]
- 25.Jacobson IM, Gordon SC, Kowdley KV, Yoshida EM, Rodriguez-Torres M, Sulkowski MS, Shiffman ML, Lawitz E, Everson G, Bennett M, Schiff E, Al-Assi MT, Subramanian GM, An D, Lin M, McNally J, Brainard D, Symonds WT, McHutchison JG, Patel K, Feld J, Pianko S, Nelson DR. 2013. Sofosbuvir for hepatitis C genotype 2 or 3 in patients without treatment options. N Engl J Med 368:1867–1877. doi: 10.1056/NEJMoa1214854. [DOI] [PubMed] [Google Scholar]
- 26.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR, Jacobson IM, Kowdley KV, Nyberg L, Subramanian GM, Hyland RH, Arterburn S, Jiang D, McNally J, Brainard D, Symonds WT, McHutchison JG, Sheikh AM, Younossi Z, Gane EJ. 2013. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med 368:1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 27.McGuigan C, Madela K, Aljarah M, Gilles A, Brancale A, Zonta N, Chamberlain S, Vernachio J, Hutchins J, Hall A, Ames B, Gorovits E, Ganguly B, Kolykhalov A, Wang J, Muhammad J, Patti JM, Henson G. 2010. Design, synthesis and evaluation of a novel double pro-drug: INX-08189. A new clinical candidate for hepatitis C virus. Bioorg Med Chem Lett 20:4850–4854. [DOI] [PubMed] [Google Scholar]
- 28.Vernachio JH, Bleiman B, Bryant KD, Chamberlain S, Hunley D, Hutchins J, Ames B, Gorovits E, Ganguly B, Hall A, Kolykhalov A, Liu Y, Muhammad J, Raja N, Walters CR, Wang J, Williams K, Patti JM, Henson G, Madela K, Aljarah M, Gilles A, McGuigan C. 2011. INX-08189, a phosphoramidate prodrug of 6-O-methyl-2′-C-methyl guanosine, is a potent inhibitor of hepatitis C virus replication with excellent pharmacokinetic and pharmacodynamic properties. Antimicrob Agents Chemother 55:1843–1851. doi: 10.1128/AAC.01335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheridan C. 2012. Calamitous HCV trial casts shadow over nucleoside drugs. Nat Biotechnol 30:1015–1016. doi: 10.1038/nbt1112-1015. [DOI] [PubMed] [Google Scholar]
- 30.Pollack A, Gale J. 2 August 2012. BMS suspends study of nucleotide BMS094 formerly INX189. http://www.natap.org/2012/HCV/080312_01.htm.
- 31.Ahmad T, Yin P, Saffitz J, Pockros PJ, Lalezari J, Shiffman M, Freilich B, Zamparo J, Brown K, Dimitrova D, Kumar M, Manion D, Heath-Chiozzi M, Wolf R, Hughes E, Muir AJ, Hernandez AF. 2014. Cardiac dysfunction associated with a nucleotide polymerase inhibitor for treatment of hepatitis C. Hepatology 62:409–416. [DOI] [PubMed] [Google Scholar]
- 32.Arnold JJ, Sharma SD, Feng JY, Ray AS, Smidansky ED, Kireeva ML, Cho A, Perry J, Vela JE, Park Y, Xu Y, Tian Y, Babusis D, Barauskus O, Peterson BR, Gnatt A, Kashlev M, Zhong W, Cameron CE. 2012. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog 8:e1003030. doi: 10.1371/journal.ppat.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng JY, Xu Y, Barauskas O, Perry JK, Ahmadyar S, Stepan G, Yu H, Babusis D, Park Y, McCutcheon K, Perron M, Schultz BE, Sakowicz R, Ray AS. 2015. Role of the mitochondrial RNA polymerase in the toxicity of nucleotide inhibitors of the hepatitis C virus. Antimicrob Agents Chemother 60:806–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sofia MJ. 2014. Beyond sofosbuvir: what opportunity exists for a better nucleoside/nucleotide to treat hepatitis C? Antiviral Res 107:119–124. doi: 10.1016/j.antiviral.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 35.McQuaid T, Savini C, Seyedkazemi S. 2015. Sofosbuvir, a significant paradigm change in HCV treatment. J Clin Transl Hepatol 3:27–35. doi: 10.14218/JCTH.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagan LM, Wolpe PR, Schinazi RF. 2013. Treatment as prevention and cure towards global eradication of hepatitis C virus. Trends Microbiol 21:625–633. doi: 10.1016/j.tim.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou L, Zhang HW, Tao S, Bassit L, Whitaker T, McBrayer TR, Ehteshami M, Amiralaei S, Pradere U, Cho JH, Amblard F, Bobeck D, Detorio M, Coats SJ, Schinazi RF. 2015. β-D-2′-C-methyl-2,6-diaminopurine ribonucleoside phosphoramidates are potent and selective inhibitors of hepatitis C virus (HCV) and are bioconverted intracellularly to bioactive 2,6-diaminopurine and guanosine 5′-triphosphate forms. J Med Chem 58:3445–3458. doi: 10.1021/jm501874e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powdrill MH, Tchesnokov EP, Kozak RA, Russell RS, Martin R, Svarovskaia ES, Mo H, Kouyos RD, Gotte M. 2011. Contribution of a mutational bias in hepatitis C virus replication to the genetic barrier in the development of drug resistance. Proc Natl Acad Sci U S A 108:20509–20513. doi: 10.1073/pnas.1105797108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludwig W, Follmann H. 1978. The specificity of ribonucleoside triphosphate reductase. Multiple induced activity changes and implications for deoxyribonucleotide formation. Eur J Biochem 82:393–403. [DOI] [PubMed] [Google Scholar]
- 40.Ludwig J, Eckstein F. 1989. Rapid and efficient synthesis of nucleoside 5′-0-(1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J Org Chem 54:631–635. doi: 10.1021/jo00264a024. [DOI] [Google Scholar]
- 41.Kovács T, Ötvös L. 1988. Simple synthesis of 5-vinyl- and 5-ethynyl-2′-deoxyuridine-5′-triphosphates. Tetrahedron Lett 29:4525–4528. doi: 10.1016/S0040-4039(00)80537-7. [DOI] [Google Scholar]
- 42.Powdrill MH, Deval J, Narjes F, De Francesco R, Gotte M. 2010. Mechanism of hepatitis C virus RNA polymerase inhibition with dihydroxypyrimidines. Antimicrob Agents Chemother 54:977–983. doi: 10.1128/AAC.01216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin Z, Leveque V, Ma H, Johnson KA, Klumpp K. 2012. Assembly, purification, and pre-steady-state kinetic analysis of active RNA-dependent RNA polymerase elongation complex. J Biol Chem 287:10674–10683. doi: 10.1074/jbc.M111.325530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang HW, Detorio M, Herman BD, Solomon S, Bassit L, Nettles JH, Obikhod A, Tao SJ, Mellors JW, Sluis-Cremer N, Coats SJ, Schinazi RF. 2011. Synthesis, antiviral activity, cytotoxicity and cellular pharmacology of l-3′-azido-2′,3′-dideoxypurine nucleosides. Eur J Med Chem 46:3832–3844. doi: 10.1016/j.ejmech.2011.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Eldrup AB, Allerson CR, Bennett CF, Bera S, Bhat B, Bhat N, Bosserman MR, Brooks J, Burlein C, Carroll SS, Cook PD, Getty KL, MacCoss M, McMasters DR, Olsen DB, Prakash TP, Prhavc M, Song Q, Tomassini JE, Xia J. 2004. Structure-activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J Med Chem 47:2283–2295. doi: 10.1021/jm030424e. [DOI] [PubMed] [Google Scholar]
- 47.Cheong C, Tinoco I Jr, Chollet A. 1988. Thermodynamic studies of base pairing involving 2,6-diaminopurine. Nucleic Acids Res 16:5115–5122. doi: 10.1093/nar/16.11.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strobel SA, Cech TR, Usman N, Beigelman L. 1994. The 2,6-diaminopurine riboside.5-methylisocytidine wobble base pair: an isoenergetic substitution for the study of GU pairs in RNA. Biochemistry 33:13824–13835. doi: 10.1021/bi00250a037. [DOI] [PubMed] [Google Scholar]
- 49.Le Pogam S, Kang H, Harris SF, Leveque V, Giannetti AM, Ali S, Jiang WR, Rajyaguru S, Tavares G, Oshiro C, Hendricks T, Klumpp K, Symons J, Browner MF, Cammack N, Najera I. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J Virol 80:6146–6154. doi: 10.1128/JVI.02628-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klumpp K, Leveque V, Le Pogam S, Ma H, Jiang WR, Kang H, Granycome C, Singer M, Laxton C, Hang JQ, Sarma K, Smith DB, Heindl D, Hobbs CJ, Merrett JH, Symons J, Cammack N, Martin JA, Devos R, Najera I. 2006. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J Biol Chem 281:3793–3799. doi: 10.1074/jbc.M510195200. [DOI] [PubMed] [Google Scholar]
- 51.Pockros P. 2013. Lessons learned from failed clinical trials in hepatitis C drug development. HepDART, Big Island, HI. [Google Scholar]
- 52.Kohler JJ, Lewis W. 2007. A brief overview of mechanisms of mitochondrial toxicity from NRTIs. Environ Mol Mutagen 48:166–172. doi: 10.1002/em.20223. [DOI] [PubMed] [Google Scholar]
- 53.Rastogi SC, Johansen JD, Menne T. 1996. Natural ingredients based cosmetics. Content of selected fragrance sensitizers. Contact Dermat 34:423–426. [DOI] [PubMed] [Google Scholar]
- 54.Fenaroli G. 2010. Fenaroli's handbook of flavor ingredients, 6th ed CRC Press, Boca Raton, FL. [Google Scholar]