Abstract
The blaIMP-14 carbapenem resistance gene has largely previously been observed in Pseudomonas aeruginosa and Acinetobacter spp. As part of global surveillance and sequencing of carbapenem-resistant Escherichia coli, we identified a sequence type 131 strain harboring blaIMP-14 within a class 1 integron, itself nested within an ∼54-kb multidrug resistance region on an epidemic IncA/C2 plasmid. The emergence of blaIMP-14 in this context in the ST131 lineage is of potential clinical concern.
TEXT
The emergence of carbapenemases in clinically prevalent Escherichia coli lineages such as sequence type 131 (ST131) is a major problem for the management of patients infected with these strains (1, 2). Globally, five major transmissible carbapenemase enzymes predominate, represented by the KPC, OXA-48-like, NDM, VIM, and IMP families (2, 3).
An IMP metallo-beta-lactamase enzyme (IMP-1) was first detected in Japan in Pseudomonas aeruginosa in the late 1980s (4); since then, 52 genetically diverse blaIMP gene variants 738 to 747 bp in length have been identified (5). Most blaIMP variants have been isolated from either Pseudomonas or Acinetobacter spp. and demonstrate a degree of geographic structuring (6); however, some, such as blaIMP-4 and blaIMP-8, have emerged successfully in members of the family Enterobacteriaceae and are distributed over wider geographic regions (6, 7). Associations of blaIMP with E. coli ST131 have, to date, been restricted to blaIMP-4 and blaIMP-8 in Taiwan, China, and Australia (8–11). As part of the Merck Study for Monitoring Antimicrobial Resistance Trends (SMART) (1), we identified an IMP-14-producing ST131 E. coli isolate, Ecol_732, that was isolated in Bangkok, Thailand, in 2012 and was sequenced in order to ascertain the genetic structures associated with this IMP variant in ST131.
Ecoli_732 was obtained from the urine of a hospitalized elderly male with a lower urinary tract infection. The MICs of ampicillin-sulbactam, piperacillin-tazobactam, cefoxitin, ceftriaxone, ceftazidime, cefepime, ertapenem, imipenem, amikacin, and ciprofloxacin were determined with microdilution panels prepared at International Health Management Associates, Inc. (Schaumburg, IL, USA), in accordance with 2015 CLSI guidelines. It tested nonsusceptible (i.e., either intermediate or resistant) to the above-mentioned agents. The MICs of colistin and tigecycline (determined by E tests) were 0.12 and 1 mg/liter, respectively.
DNA (chromosomal plus plasmid) was extracted from pure overnight subcultures of the isolate for both PacBio (long-read) sequencing and Illumina MiSeq (short-read) sequencing with the Qiagen Genomic-tip 100/G kit and the QIAamp DNA minikit (catalogue numbers 10243 and 51304; Qiagen, Valencia, CA), respectively. Preliminary de novo assembly of PacBio reads with HGAP3 was performed; resulting contigs were annotated with Prokka (12) and then trimmed on the basis of sequence/annotation overlaps in Geneious (version 9.04) (13). One-hundred-fifty-base paired-end MiSeq reads for each of the isolates were trimmed with Trimmomatic (version 0.35) (14) and then mapped to the corresponding PacBio assemblies with BWA mem (version 0.7.9a-r786) (15). Read pileups were inspected to confirm the structural integrity of the contigs and correct any small errors in the assembled contigs. Unmapped MiSeq reads were assembled with A5MiSeq (16) in order to identify any small plasmids (<7 kb) that may have been filtered out during the size selection process implemented as part of PacBio library preparation. Additional annotation focused on resistance genes and insertion sequences was performed with reference to the ResFinder (17), PlasmidFinder (18), and ISFinder (19) databases.
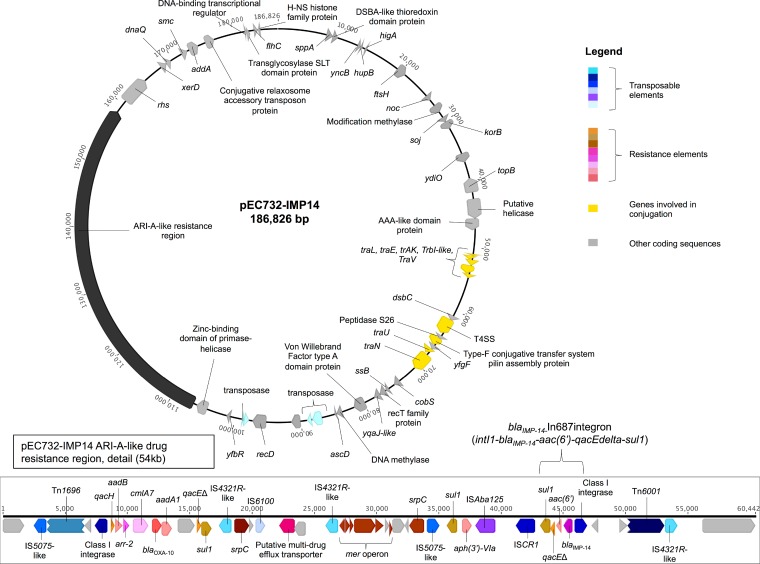
The Ecol_732 genome consists of a 5,009,900-bp chromosome and six plasmids, five of which could be fully resolved. These included pEC732-IMP14 (186,826 bp, IncA/C2), pEC732_2 (129,154 bp, IncFII/FIA/FIB/col), pEC732_3 (82,588 bp, IncB/O/K/Z), pEC732_4 (4,072 bp, untyped), and pEC732_5 (1,549 bp, untyped). A partial sixth mob plasmid fragment was also present (4,204 bp, untyped). The blaIMP-14 sequence in pEC732-IMP14 differed from the reference AY553332 by a single synonymous substitution (A249C), resulting in the same amino acid sequence. It was located within a 3,791-bp class 1 integron, In687 [intI1-blaIMP-14-aac(6′)-qacEdelta-sul1]. This integron is almost identical (single nucleotide difference, A925G) to that in Achromobacter xylosoxidans strain R4, which was cultured from a urine sample, also in Thailand (GenBank accession number KJ406505).
The backbone of pEC732-IMP14 was highly similar to that of prototype type 1 IncA/C2 plasmid pRMH760 (RefSeq database accession no. NC_023898; from a Klebsiella pneumoniae strain) and other type 1 IncA/C2 plasmids (recently reviewed in reference 20), almost all of which also include a specific region designated ARI-A that contains a variable array of resistance genes and is located 1,711 bp upstream of rhs (20). Similarly, in pEC732-IMP14, the blaIMP-14-harboring integron was part of a much larger, 54,454-bp ARI-A-like region containing antimicrobial, heavy metal, and biocide resistance genes (Fig. 1), including those encoding resistance to beta-lactams (blaOXA-10, blaIMP-14), macrolides (drug efflux), rifampin (arr-2), sulfonamides (sul1), aminoglycosides [aadA1, aadB, aph(3′)-IVa, aac(6′)] chloramphenicol (cmlA7), chromate (srpC), mercury (mer operon), and quaternary ammonium compounds (qac). Some of these were part of a second, novel, integron designated In1286 (intI1-qacH-aadB-arr-2-cmlA7-blaOXA-10-aadA1).
FIG 1.
Plasmid pEC732-IMP14 with a detailed view of the ARI-A-like 54-kb resistance region (bottom).
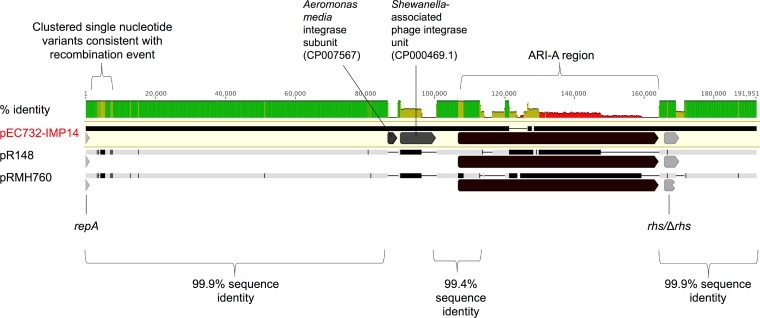
An alignment of pEC732-IMP14, prototype IncA/C2 type 1 plasmid pRMH760, and the only publicly available type 1 IncA/C2 sequence from Thailand, pR148 (RefSeq database accession no. NC_019380, from Aeromonas hydrophila [21]), demonstrates the genetic similarity of these plasmids (Fig. 2). All three sequences were >99% similar in the 1- to 86,573-bp region and in the ∼27.5-kb region downstream of ARI-A (Fig. 2). Differences in pEC732-IMP14 include a region of clustered single-nucleotide variants suggestive of a recombination event (region, 3,100 to 8,000 kb) and the acquisition of two integrase subunits (regions, 86,573 to 89,203 and 90,138 to 200,167 bp; Fig. 2). Interestingly, the pR148-containing A. hydrophila strain was identified on a Thai tilapia fish farm that had successively used several antimicrobial classes (21).
FIG 2.
Alignment of pEC732-IMP14, pR148, and pRMH760 demonstrating mean percent pairwise identity over alignment columns (green, 100%; olive, 30 to 70%; red <30%; no color, 0%). Vertical black lines in the sequence bars represent differences between the nucleotide sequences with respect to the pEC732-IMP14 reference; thin horizontal black lines represent deleted regions. The repA gene, variable ARI-A resistance region, and rhs gene are indicated, as are the region of clustered single-nucleotide variants downstream of the repA region and two additional mobile genetic elements present in pEC732-IMP14 and absent from the other two sequences.
To date, blaIMP-14 has not been described in E. coli, to our knowledge, and has largely previously been reported in P. aeruginosa and Acinetobacter baumannii strains by several hospital centers in Thailand, in some cases as part of clonal outbreaks (22–25). Although blaIMP-14 is similarly associated with class 1 integrons in these cases, as in pEC732-IMP14, the wider plasmid contexts and sequences of these integrons in P. aeruginosa and A. baumannii strains have not been investigated. It is, however, conceivable that the blaIMP-14-harboring integron observed in pEC732-IMP14 and A. xylosoxidans strain R4 has been exchanged more widely with Pseudomonas and Acinetobacter spp. in Thailand. Class 1 integrons have been linked with the recent successful spread and expansion of another metallo-beta-lactamase, blaVIM, in IncA/C2 plasmids in members of the family Enterobacteriaceae in Greece (26) and blaVIM and blaIMP in Spain (27).
The presence of the extensively drug-resistant region observed here on an epidemic IncA/C2 plasmid in an E. coli ST131 strain from Thailand is therefore of concern and may represent wider, regional, horizontal dissemination of blaIMP-14 mediated by mobile genetic elements across bacterial families. The homology of pEC732-IMP14 with an A. hydrophila plasmid found on a fish farm and the presence of blaIMP-harboring plasmids in E. coli in other environmental (28) and animal sampling frames (29) suggest that the transmission network for IMP-positive E. coli may extend beyond the health care setting. Broad surveillance and control measures that are targeted at both community and health care contexts may be required to monitor and limit blaIMP dissemination.
Nucleotide sequence accession numbers.
Complete sequence data for Ecol_732 have been deposited in GenBank under BioProject number PRJNA316786. The accession numbers of the sequences are CP015138 (chromosome), CP015139 (pEC732-IMP14), CP015140 (pEC732_2), CP015141 (pEC732_3), CP015142 (pEC732_4), CP015143 (pEC732_5), and CP015144 (pEC732_6 [partial sequence only]).
ACKNOWLEDGMENTS
We are grateful for the support of the Modernizing Medical Microbiology Informatics Group. We also thank Daryl Hoban and the Merck SMART surveillance team.
N.S. is currently funded through a Public Health England/University of Oxford clinical lectureship; the sequencing work was also partly funded through a previous Wellcome Trust doctoral research fellowship (099423/Z/12/Z). Additional funding support was provided by a research grant from Calgary Laboratory Services (10006465) and by the Health Innovation Challenge Fund (a parallel funding partnership between the Wellcome Trust [WT098615/Z/12/Z] and the Department of Health [grant HICF-T5-358]). This research was supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre Programme.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The views expressed are those of the authors and not necessarily those of the National Health Service, the NIHR, or the Department of Health.
REFERENCES
- 1.Peirano G, Bradford PA, Kazmierczak KM, Badal RE, Hackel M, Hoban DJ, Pitout JD. 2014. Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg Infect Dis 20:1928–1931. doi: 10.3201/eid2011.141388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 35:147–151. doi: 10.1128/AAC.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K, Palzkill T, Jacoby G. 2015. β-Lactamase classification and amino acid sequences for TEM, SHV and OXA extended-spectrum and inhibitor resistant enzymes. http://www.lahey.org/studies/ Accessed 1 March 2016.
- 6.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect Dis 11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 7.Sidjabat HE, Townell N, Nimmo GR, George NM, Robson J, Vohra R, Davis L, Heney C, Paterson DL. 2015. Dominance of IMP-4-producing Enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob Agents Chemother 59:4059–4066. doi: 10.1128/AAC.04378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho PL, Cheung YY, Wang Y, Lo WU, Lai EL, Chow KH, Cheng VC. 2016. Characterization of carbapenem-resistant Escherichia coli and Klebsiella pneumoniae from a healthcare region in Hong Kong. Eur J Clin Microbiol Infect Dis 35:379–385. doi: 10.1007/s10096-015-2550-3. [DOI] [PubMed] [Google Scholar]
- 9.Yan JJ, Tsai LH, Wu JJ. 2012. Emergence of the IMP-8 metallo-beta-lactamase in the Escherichia coli ST131 clone in Taiwan. Int J Antimicrob Agents 40:281–282. doi: 10.1016/j.ijantimicag.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Zhang F, Zhu D, Xie L, Guo X, Ni Y, Sun J. 2015. Molecular epidemiology of carbapenemase-producing Escherichia coli and the prevalence of ST131 subclone H30 in Shanghai, China. Eur J Clin Microbiol Infect Dis 34:1263–1269. doi: 10.1007/s10096-015-2356-3. [DOI] [PubMed] [Google Scholar]
- 11.Sidjabat HE, Robson J, Paterson DL. 2015. Draft genome sequences of two IMP-4-producing Escherichia coli sequence type 131 isolates in Australia. Genome Announc 3:e00983-15. doi: 10.1128/genomeA.00983-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 13.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton A, Markowitz S, Duran C, Thierer T, Ashton B, Metjies P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 17.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32-36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmer CJ, Hall RM. 2014. pRMH760, a precursor of A/C(2) plasmids carrying blaCMY and blaNDM genes. Microb Drug Resist 20:416–423. doi: 10.1089/mdr.2014.0012. [DOI] [PubMed] [Google Scholar]
- 21.Del Castillo CS, Hikima J, Jang HB, Nho SW, Jung TS, Wongtavatchai J, Kondo H, Hirono I, Takeyama H, Aoki T. 2013. Comparative sequence analysis of a multidrug-resistant plasmid from Aeromonas hydrophila. Antimicrob Agents Chemother 57:120–129. doi: 10.1128/AAC.01239-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khuntayaporn P, Montakantikul P, Santanirand P, Kiratisin P, Chomnawang MT. 2013. Molecular investigation of carbapenem resistance among multidrug-resistant Pseudomonas aeruginosa isolated clinically in Thailand. Microbiol Immunol 57:170–178. doi: 10.1111/1348-0421.12021. [DOI] [PubMed] [Google Scholar]
- 23.Piyakul C, Tiyawisutsri R, Boonbumrung K. 2012. Emergence of metallo-beta-lactamase IMP-14 and VIM-2 in Pseudomonas aeruginosa clinical isolates from a tertiary-level hospital in Thailand. Epidemiol Infect 140:539–541. doi: 10.1017/S0950268811001294. [DOI] [PubMed] [Google Scholar]
- 24.Kansakar P, Dorji D, Chongtrakool P, Mingmongkolchai S, Mokmake B, Dubbs P. 2011. Local dissemination of multidrug-resistant Acinetobacter baumannii clones in a Thai hospital. Microb Drug Resist 17:109–119. doi: 10.1089/mdr.2010.0062. [DOI] [PubMed] [Google Scholar]
- 25.Samuelsen O, Toleman MA, Sundsfjord A, Rydberg J, Leegaard TM, Walder M, Lia A, Ranheim TE, Rajendra Y, Hermansen NO, Walsh TR, Giske CG. 2010. Molecular epidemiology of metallo-beta-lactamase-producing Pseudomonas aeruginosa isolates from Norway and Sweden shows import of international clones and local clonal expansion. Antimicrob Agents Chemother 54:346–352. doi: 10.1128/AAC.00824-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papagiannitsis CC, Dolejska M, Izdebski R, Giakkoupi P, Skalova A, Chudejova K, Dobiasova H, Vatopoulos AC, Derde LP, Bonten MJ, Gniadkowski M, Hrabak J. 2016. Characterisation of IncA/C2 plasmids carrying an In416-like integron with the blaVIM-19 gene from Klebsiella pneumoniae ST383 of Greek origin. Int J Antimicrob Agents 47:158–162. doi: 10.1016/j.ijantimicag.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Zamorano L, Miro E, Juan C, Gomez L, Bou G, Gonzalez-Lopez JJ, Martinez-Martinez L, Aracil B, Conejo MC, Oliver A, Navarro F. 2015. Mobile genetic elements related to the diffusion of plasmid-mediated AmpC beta-lactamases or carbapenemases from Enterobacteriaceae: findings from a multicenter study in Spain. Antimicrob Agents Chemother 59:5260–5266. doi: 10.1128/AAC.00562-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieffer N, Poirel L, Bessa LJ, Barbosa-Vasconcelos A, da Costa PM, Nordmann P. 2016. VIM-1, VIM-34, and IMP-8 carbapenemase-producing Escherichia coli strains recovered from a Portuguese river. Antimicrob Agents Chemother 60:2585–2586. doi: 10.1128/AAC.02632-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolejska M, Masarikova M, Dobiasova H, Jamborova I, Karpiskova R, Havlicek M, Carlile N, Priddel D, Cizek A, Literak I. 2016. High prevalence of Salmonella and IMP-4-producing Enterobacteriaceae in the silver gull on Five Islands, Australia. J Antimicrob Chemother 71:63–70. doi: 10.1093/jac/dkv306. [DOI] [PMC free article] [PubMed] [Google Scholar]