Abstract
Emerging multidrug-resistant (MDR) Gram-negative bacilli (GNB), including Escherichia coli sequence type 131 (ST131) and its resistance-associated H30 subclone, constitute an ever-growing public health threat. Their reservoirs and transmission pathways are incompletely defined. To assess diarrheal stools as a potential reservoir for ST131-H30 and other MDR GNB, we cultured 100 clinical stool samples from a Veterans Affairs Medical Center clinical laboratory (October to December 2011) for fluoroquinolone- and extended-spectrum cephalosporin (ESC)-resistant E. coli and other GNB, plus total E. coli. We then characterized selected resistant and susceptible E. coli isolates by clonal group, phylogenetic group, virulence genotype, and pulsotype and screened all isolates for antimicrobial resistance. Overall, 79 of 100 stool samples yielded GNB (52 E. coli; 48 other GNB). Fifteen samples yielded fluoroquinolone-resistant E. coli (10 were ST131, of which 9 were H30), 6 yielded ESC-resistant E. coli (2 were ST131, both non-H30), and 31 yielded susceptible E. coli (1 was ST131, non-H30), for 13 total ST131-positive samples. Fourteen non-E. coli GNB were ESC resistant, and three were fluoroquinolone resistant. Regardless of species, almost half (46%) of the fluoroquinolone-resistant and/or ESC-resistant non-E. coli GNB were resistant to at least three drug classes. Fecal ST131 isolates closely resembled reference clinical ST131 isolates according to virulence genotypes and pulsed-field gel electrophoresis (PFGE) profiles. Thus, a substantial minority (30%) of veterans with diarrhea who undergo stool testing excrete antibiotic-resistant GNB, including E. coli ST131. Consequently, diarrhea may pose transmission risks for more than just diarrheal pathogens and may help disseminate clinically relevant ST131 strains and other MDR GNB within hospitals and the community.
Multidrug-resistant (MDR) Gram-negative bacilli (GNB) are important extraintestinal pathogens that pose an ever-growing public health threat. Among the various clinically important GNB, Escherichia coli is a leading source of illness, death, and health care costs (1). Over the past 2 decades E. coli clinical isolates have shown an increasing prevalence of resistance to first-line antibiotics, notably fluoroquinolones (FQs) and extended-spectrum cephalosporins (ESCs) (2–4). The main contributor to this worsening resistance trend is the H30 subclone within E. coli sequence type 131 (ST131), which is a newly emerged, globally disseminated MDR extraintestinal pathogen (5).
One suspected route of transmission of MDR GNB is human-to-human spread via the fecal-oral route. Supporting this hypothesis for ST131 is the evidence of fecal colonization with ST131 among patients with an ST131 infection and their healthy close contacts in patterns suggesting within-household transmission (6–8). Compared with formed stools, diarrhea presumably would provide even more opportunities for fecal shedding, environmental contamination, and person-to-person transmission. To our knowledge, however, diarrheal stools have not been studied as a vehicle for MDR GNB, especially ST131 and its H30 subclone.
Accordingly, we surveyed 100 diarrheal fecal samples submitted to the clinical microbiology laboratory at the Minneapolis Veteran Affairs Medical Center (MVAMC) for fluoroquinolone-resistant and ESC-resistant E. coli and other GNB, screened the samples and E. coli isolates by PCR for ST131 and H30 subclone status, and determined extended antimicrobial susceptibility profiles for relevant isolates. Additionally, to better assess the likely pathogenic potential of the drug-resistant and susceptible E. coli isolates, we used multiple molecular typing modalities to further characterize these isolates and to compare them with archived clinical E. coli isolates.
MATERIALS AND METHODS
Fecal samples.
From October through December 2011, 100 residual fecal samples were obtained from the clinical microbiology laboratory at the MVAMC. The samples had been submitted as loose or liquid stool for diarrheal pathogen testing and, after removal of a portion for the requested clinical assays, had been held at 4°C without preservative. Study personnel retrieved the samples from the clinical microbiology laboratory within 24 to 48 h of submission. The samples represented all available fecal samples in the clinical laboratory on the days that study personnel visited the laboratory to retrieve specimens, and the samples were retrieved without knowledge of which tests had been requested, indication for testing, source of sample (i.e., inpatient versus outpatient, unit, or clinic), and patient identity or other characteristics.
Sample processing.
In the research laboratory, fecal samples were processed the same day or the day after they were retrieved from the clinical laboratory. They were streaked for isolation on Tergitol 7 (T7) agar plates (Sigma-Aldrich, St. Louis, MO, USA), either plain or supplemented with (separately) ciprofloxacin (4 mg/liter), ceftazidime (2 mg/liter), cefotaxime (8 mg/liter), or imipenem (2 mg/liter). Plates were incubated overnight at 37°C. Bacterial isolates recovered from the samples were stored at −80 C° for further analysis.
Isolation of E. coli and determination of ST131 status.
For each sample that yielded GNB, mixed Gram-negative growth harvested from the inoculum area of the plate underwent PCR-based testing for ST131-specific single nucleotide polymorphisms (SNPs) in gyrB and mdh (9). If population DNA PCR screening detected ST131, up to 10 morphologically distinct colonies of presumptive E. coli (i.e., lactose- and indole-positive, citrate-negative GNB with a characteristic E. coli colonial morphology), as available, underwent single-colony ST131 PCR testing. Furthermore, if the ciprofloxacin-supplemented T7 plate grew E. coli, up to 10 E. coli colonies from the nonsupplemented T7 plate (as available) were replica plated to ciprofloxacin-supplemented T7 agar. Results from this testing were used to define the presence and within-sample prevalence of ST131 and other fluoroquinolone-resistant E. coli isolates. From each sample that yielded E. coli, one resistant isolate (i.e., resistant to fluoroquinolones and/or ESCs) and one susceptible isolate (i.e., susceptible to both fluoroquinolones and ESCs), as available, were selected for further molecular testing.
Isolation of non-E. coli antibiotic-resistant GNB.
The antibiotic-supplemented T7 agar plates from the primary stool cultures were also scrutinized for non-E. coli Gram-negative bacilli (GNB). One representative of each morphologically distinct colony type, as available, was identified using the API-20E system (bioMérieux, Marcy l'Etoile, France).
Virulence genotyping, phylotyping, and clonal group typing.
To assess the extraintestinal virulence potential of E. coli isolates, boiled lysates were prepared in duplicate from single colonies and tested for 51 virulence genes (VGs) using established multiplex PCR-based assays (10, 11). The VG score was calculated as the total number of VGs detected, adjusted for multiple detection of the pap (P fimbriae), sfa and foc (S and F1C fimbriae), and kps (group 2 capsule) operons.
E. coli isolates were assigned to phylogenetic groups (phylogroups) A, B1, B2, C, D, E, F, and cryptic clade I by a multiplex PCR-based assay (12). ST131 isolates were tested by allele-specific primers for allele 30 of fimH (encoding a variant of the type 1 fimbrial adhesin), corresponding with the H30 subclone, the main fluoroquinolone resistance-associated subset within ST131 (5, 13). For non-ST131 isolates from phylogroups B2 and D, clonal lineage was identified where possible by using established PCR-based assays specific for clonal complexes ST12, ST31, ST69, ST73, ST81, ST95, ST127, ST141, ST144, ST372, and ST405 (14–16).
Antimicrobial susceptibility testing.
Susceptibility to 23 antimicrobial agents was determined by disk diffusion, using Clinical and Laboratory Standards Institute (CLSI)-recommended methods, reference strains, and interpretive criteria (17). The agents included amikacin, amoxicillin-clavulanate, ampicillin, aztreonam, cefepime, cefoxitin, ceftazidime, ceftriaxone, cephatholin, chloramphenicol, ciprofloxacin, gentamicin, imipenem, levofloxacin, nalidixic acid, nitrofurantoin, piperacillin, piperacillin-tazobactam, streptomycin, tetracycline, sulfamethoxazole, trimethoprim, and trimethoprim-sulfamethoxazole. Intermediate results were analyzed as resistant. The resistance score was the number of agents to which an isolate exhibited resistance. Multidrug resistance was defined as resistance to at least one representative each of at least three drug classes, counting penicillins and cephalosporins separately (18).
PFGE analysis.
XbaI pulsed-field gel electrophoresis (PFGE) analysis was used to assign ST131 isolates to pulsotypes based on 94% profile similarity to reference strains (19). Dendrograms were inferred within BioNumerics, version 6.6 (Applied Maths, Austin, TX), according to the unweighted pair group method based on Dice coefficients. Profiles also were compared with a large private PFGE profile reference library containing 1,925 pulsotypes and representing 4,916 E. coli isolates, as collected from diverse locales, specimen types, hosts, clinical syndromes, and time periods (20).
Comparator clinical isolates.
For internal and external comparisons based on VGs and antimicrobial resistance, the fecal isolates were divided into three groups based on resistance phenotype and ST131 status, as follows: (i) ST131 (regardless of resistance phenotype), (ii) non-ST131, resistant to fluoroquinolones and/or ESCs, and (iii) non-ST131, susceptible to both fluoroquinolones and ESCs. For comparison with the present fecal E. coli isolates, an equal number of isolates were selected from an archived set of 595 deidentified human clinical E. coli isolates, as obtained systematically during 2011 from 24 nationally distributed VAMCs (5). These isolates had been assessed previously for ST131 status and virulence genotype using the same methods as used for the present fecal isolates (5). The comparison clinical isolates were selected randomly in appropriate numbers for each of the above three groups, as defined based on ST131 status and fluoroquinolone and ESC phenotype.
Statistical analysis.
Comparisons of proportions were tested by using Fisher's exact test. Comparisons of VG scores were tested by using a Mann-Whitney U test (two-tailed). Pairwise similarity relationships according to extended VG profiles and, separately, PFGE profiles were used to construct dendrograms according to the unweighted pair group method with arithmetic averages (21). Dendrograms were inferred within BioNumerics, version 6.6 (Applied Maths, Austin, TX). PERMANOVA, as implemented using PAST (PAlaeontological STatistics), package 3.10 (Øyvind Hammer, Natural History Museum, University of Oslo), was used to compare statistically the various isolate groups (as defined based on resistance phenotype, ST131 status, and fecal versus clinical source) for dissimilarity according to aggregate VG profiles. The criterion for statistical significance throughout was a P value of <0.05.
RESULTS
Prevalence of E. coli and non-E. coli GNB.
Of the 100 clinical fecal specimens from the MVAMC clinical microbiology laboratory, 79 (79%) yielded GNB, including 52 (52%) with E. coli (of which 21 also had other GNB) and 48 (48%) with other GNB (of which 21 also had E. coli). Of the 52 samples with E. coli, 15 (29%; 15% overall) yielded fluoroquinolone-resistant E. coli isolates and 6 (12%; 6% overall) yielded ESC-resistant E. coli isolates. In contrast, only 3 (6%; 3% overall) of the 48 samples with non-E. coli GNB yielded fluoroquinolone-resistant non-E. coli GNB, whereas 14 (29%; 14% overall) yielded ESC-resistant non-E. coli GNB. Overall, 30 samples (30%) yielded GNB (including E. coli and non-E. coli) that were resistant to fluoroquinolones and/or ESCs.
E. coli sequence types and subclones.
From the 52 E. coli-positive fecal samples, 58 E. coli isolates (including all 19 that were resistant to FQs and/or ESCs, plus 39 that were susceptible to both drug classes) were chosen for molecular analysis. Selective clonal typing showed that six familiar human-associated ST complexes that are known for causing endemic or epidemic extraintestinal E. coli infections, with or without associated antimicrobial resistance (22, 23), accounted for 24 (40%) of the 58 E. coli isolates. These ST complexes, in order of descending frequency (number of isolates), included ST131 (12), ST95 (6), ST73 (2), ST127 (2), ST69 (1), and ST405 (1). Additionally, one fecal sample screened positive for ST131 using population DNA PCR, but the responsible organism could not be isolated, suggesting the presence of a low-frequency ST131 strain that was not among the 10 colonies picked for ST131 PCR testing. Thus, 13 total samples (13%) contained ST131.
Of the 12 ST131 isolates, 10 (83%) were fluoroquinolone resistant (1 of these was also ESC resistant) and 2 (17%) were fluoroquinolone susceptible (1 of these was also ESC resistant), giving 2 (17%) ESC-resistant isolates and 10 (83%) ESC-susceptible isolates. Subclonal typing of the 12 ST131 isolates showed that the H30 subclone accounted for 9 (90%) of 10 fluoroquinolone-resistant ST131 isolates but neither (0%) of the 2 fluoroquinolone-susceptible ST131 isolates (P = 0.04) or the 2 ESC-resistant ST131 isolates. Overall, ST131 accounted for 10 (67%) of the 15 total fluoroquinolone-resistant E. coli isolates but only 2 (33%) of the 6 total ESC-resistant E. coli isolates.
In contrast to the ST131 isolates, none of the 12 E. coli isolates from defined STs other than ST131 were resistant to either fluoroquinolones or ESCs (versus ST131, P < 0.001 and P = 0.50, respectively). Similarly, of the 46 total non-ST131 E. coli isolates, only 5 (11%) were resistant to fluoroquinolones, and 4 (9%) were resistant to ESCs (versus ST131, P < 0.001 and P = 0.60, respectively).
Within-sample prevalence of fluoroquinolone-resistant and ST131 E. coli.
For 14 (93%) of the 15 fecal samples that yielded fluoroquinolone-resistant E. coli, the local E. coli population was dominated by fluoroquinolone-resistant E. coli, which accounted for 80 to 100% of the 10 E. coli colonies per sample that were picked arbitrarily from the nonselective primary plate for fluoroquinolone resistance screening. This was true regardless of ST131 status; that is, fluoroquinolone-resistant E. coli predominated in 10/10 samples for ST131 versus 4/5 for non-ST131 isolates.
Phylogroup distribution.
Of the eight recognized E. coli phylogroups, five (A, B1, B2, D, and F) were detected among the E. coli study isolates, each accounting for from 7% (group F) to 41% (group B2) of isolates (Table 1). Phylogroup distribution varied in relation to fluoroquinolone resistance status, with resistant isolates being predominantly from group B2 (67%, versus 5% for FQ-susceptible E. coli; P < 0.001) and never from group B1 (0%, versus 23% for fluoroquinolone-susceptible E. coli; P = 0.03). This association of phylogroup B2 with fluoroquinolone resistance resulted mainly from the predominance among resistant isolates of ST131 (phylogroup B2) since among non-ST131 isolates phylogroup B2 was similarly prevalent among resistant and susceptible isolates, accounting for only a minority of either group (Table 1). In contrast, among the non-ST131 isolates, phylogroup F (instead of B2) was associated with resistance (Table 1).
TABLE 1.
Bacterial traits by ST131 status and antimicrobial drug resistance in 58 Escherichia coli isolates from clinical stool samples from veterans
| Phylogroup or genea | Prevalence by isolate group (no. of isolates [%])e |
P valued | |||
|---|---|---|---|---|---|
| Total (n = 58) | ST131 (n = 12) | Non-ST131, resistant (n = 8)b | Non-ST131, susceptible (n = 38)c | ||
| Group A | 12 (21) | 0 (0) | 1 (12.5) | 11 (29) | 0.66 |
| Group B1 | 12 (21) | 0 (0) | 2 (25) | 10 (26) | 1.0 |
| Group B2 | 24 (41) | 12 (100) | 1 (12.5) | 11 (29) | 0.66 |
| Group D | 6 (10) | 0 (0) | 1 (12.5) | 5 (13) | 1.0 |
| Group F | 4 (7) | 0 (0) | 3 (37.5) | 1 (3) | 0.01 |
| iha | 16 (28) | 9 (75) A | 2 (25) B | 5 (13) B | <0.001 |
| iutA | 20 (35) | 8 (67) A | 5 (63) A | 7 (18) B | 0.002 |
| malX | 26 (45) | 11 (92) A | 4 (50) B | 11 (29) B | <0.001 |
| kfiC | 4 (7) | 3 (25) A | 1 (13) A | 0 (0) B | 0.007 |
| sat | 13 (22) | 8 (67) A | 2 (25) AB | 3 (8) B | <0.001 |
| usp | 22 (38) | 11 (92) A | 1 (13) B | 10 (26) B | <0.001 |
Groups A, B1, B2, D, and F are E. coli phylogenetic groups; virulence genes are those which had a P value of <0.05 for at least one comparison each. iha, adhesin-siderophore receptor; sat, secreted autotransporter toxin; iutA, aerobactin system; usp, uropathogenic-specific protein; malX, pathogenicity island marker; kfiC, subgroup of group II.
Resistant, resistant to at least one of the following: ciprofloxacin or levofloxacin (fluoroquinolones) and ceftriaxone or ceftazidime (extended-spectrum cephalosporins).
Susceptible, susceptible to both ciprofloxacin and levofloxacin (fluoroquinolones) and to both ceftriaxone and ceftazidime (extended-spectrum cephalosporins), regardless of other possible drug resistance.
P values (by Fisher's exact test) are for two-way comparisons of the prevalence of individual phylogroups between resistant and susceptible non-ST131 isolates and three-way comparisons for the prevalence of individual virulence genes across subgroups.
Uppercase letters identify proportions that, within the given row, do not differ significantly at a P value of 0.05.
Extended virulence genotypes.
Of the 51 studied VGs, 41 (80%) were detected in 2% to 100% of the 58 E. coli isolates each. Multiple VGs were detected within each functional category. Compared with 46 non-ST131 E. coli isolates, the 12 ST131 E. coli isolates had a significantly higher prevalence of six VGs (Table 1), i.e., iha (iron-regulated gene homologue adhesin), iutA (aerobactin receptor), kfiC (capsule gene), malX (pathogenicity island marker), sat (secreted autotransporter toxin), and usp (uropathogenic-specific protein), and no significantly lower prevalence of any VG.
Overall, VG scores among the E. coli isolates ranged from 0 to 15 (median, 5). Collectively, the 12 ST131 isolates had numerically (but not statistically significantly) higher VG scores (median 9, range 0 to 10) than the non-ST131 resistant isolates (median 4.5, range 2 to 13) or the non-ST131 susceptible isolates (median 4, range 0 to 15) (P = 0.44, overall).
Extended virulence profiles for the 58 present fecal E. coli isolates and 58 archival clinical E. coli isolates, which were matched to the fecal isolates by ST131 and resistance status, were used to construct a similarity dendrogram (Fig. 1). The dendrogram contained three major zones, which corresponded broadly with the constituent strains' clonal backgrounds, clinical versus fecal source, and resistance status. That is, the dendrogram's upper zone was dominated by a mixture of resistant non-ST131 isolates (whether fecal or clinical) and susceptible non-ST131 fecal isolates, the middle zone was dominated by resistant ST131 isolates (with fecal and clinical ST131 isolates intermingled extensively), and the lower zone was dominated by susceptible non-ST131 clinical isolates.
Fig 1.
Dendrogram of virulence genotypes among 58 fecal and 58 matched clinical Escherichia coli isolates from veterans (2011). The dendrogram (by the unweighted-pair group method with arithmetic averages) was based on pairwise similarity relationships according to the presence or absence of 51 individual virulence genes. Only the 40 virulence genes detected in at least 1 isolate are shown. Trait definitions: afaBC, Dr antigen-specific adhesion operon; astA, enteroaggregative E. coli heat-stable cytotoxin; bmaE, blood group M-specific adhesion; cdtB, cytolethal distending toxin; clbBN, hybrid peptide-polyketide; cnf1, cytotoxic necrotizing factor 1; cvaC, microcin (colicin) V; fimH, d-mannose-specific adhesion, type-1 fimbriae; fimH30, allele 30 of fimH; focG, F1C fimbriae (sialic acid-specific); fyuA, Yersinia siderophore receptor; H7, flagellin variant; hlyD, alpha-hemolysin; hlyF, hemolysin F; cnf1, cytotoxic necrotizing factor 1; hra, heat-resistant agglutinin; ibeA, invasion of brain endothelium; iha, iron-regulated gene homologue adhesin; ire, iron-regulated element; iroN, catecholate siderophore receptor; iss, increased serum survival (outer membrane protein); iutA, ferric aerobactin receptor; K2, group II kpsM; K5, kfiC (subgroup of group II); K15, specific kpsM protein (polysialic acid transport); kpsM II, group 2 capsule polysaccharide synthesis (e.g., K1, K5, and K12); kpsM K1 and K2/K100, group 2 capsule variants; malX, pathogenicity-associated island marker; ompT, outer membrane protein T (protease); papAH, P fimbrial structural subunit (note: papC, papEF, papG, and papG allele II gave the same result as papA); pic, protein involved in intestinal colonization (serine protease); sat, secreted autotransporter toxin; sfaDE, S and F1C fimbriae; sfaS, S fimbriae (sialic acid specific); traT, surface exclusion, serum survival-associated; tsh, temperature-sensitive hemagglutinin; usp, uropathogenic-specific protein (bacteriocin); vat, vacuolating autotransporter (serine protease). Virulence genes (with colored squares indicating presence) are color-coded by functional category as follows: red, adhesins; green, toxins; blue, siderophores; yellow, protectins; purple, miscellaneous; orange, fimH30. Data to the right of the dendrogram include sample source (only fecal source indicated; blank, clinical), FQ/ESC (fluoroquinolone and extended-spectrum cephalosporin) phenotype (RES, resistant to one or both classes; blank, susceptible to both), and ST131, sequence type 131 status (POS, positive; blank, negative).
This visual impression of segregation by ST131/resistance/source group was supported statistically by PERMANOVA (Table 2). That is, according to aggregate VG profiles, the ST131 clinical and fecal isolates (middle zone in the dendrogram) did not differ from one another but were highly significantly different from all other strain categories. Similarly, the resistant non-ST131 isolates (whether clinical or fecal) and the susceptible non-ST131 fecal isolates (upper zone in the dendrogram) did not differ significantly from one another but were significantly different from all other strain categories, except for a significant difference (P < 0.001) between the fecal versus clinical non-ST131 susceptible isolates. Finally, the susceptible non-ST131 clinical isolates (lower zone in the dendrogram) were highly significantly different from all other strain categories apart from resistant non-ST131 clinical isolates, with a comparison yielding a P value of 0.08 (Table 2).
TABLE 2.
P values for pairwise comparisons between strain groups according to similarity of extended virulence genotypes
| Isolate source and group (n)a | Descriptionb |
P value for difference between groupsc |
||||
|---|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | Group 5d | ||
| Fecal | ||||||
| Group 0 (38) | Non-ST131, susceptible | 0.30 | <0.001 | <0.001 | <0.001 | 0.55 |
| Group 1 (8) | Non-ST131, resistant | 0.008 | <0.001 | 0.006 | 0.92 | |
| Group 2 (12) | ST131 | 0.45 | <0.001 | 0.001 | ||
| Clinical | ||||||
| Group 3 (12) | ST131 | <0.001 | <0.001 | |||
| Group 4 (38) | Non-ST131, susceptible | 0.08 | ||||
Fecal, stool isolate from this study; clinical, clinical isolate from 2011 national VA survey (5); n, number of isolates.
Susceptible, susceptible to fluoroquinolones and extended-spectrum cephalosporins (ESCs); resistant, resistant to fluoroquinolones and/or ESCs.
P values were determined by PERMANOVA for pairwise comparisons between isolate groups based on Dice similarity.
Group 5, clinical, non-ST131 resistant isolates (n = 8).
Antimicrobial resistance phenotype: E. coli.
Resistance to each of the 23 study antibiotics but 3 (amikacin, imipenem, and nitrofurantoin) was detected in one or more of the 58 fecal E. coli isolates, ranging in prevalence from 5% (chloramphenicol, gentamicin, and piperacillin-tazobactam) to 80% (ampicillin and nalidixic acid) (see Table S1 in the supplemental material). Overall, 48 (83%) of the 58 isolates exhibited resistance to at least one antibiotic. Resistance scores, which overall ranged from 0 to 13 (median, 2), were similar among ST131 isolates (median 5.5, range 1 to 11) and non-ST131 resistant isolates (median 7.5, range 3 to 13) (P = 0.62) but were significantly higher in each of these two groups than among non-ST131 susceptible isolates (median 1, range 0 to 7) (P < 0.001 for both comparisons). Similarly, of the ST131 and non-ST131 resistant isolates, 8 (67%) and 7 (88%) qualified as MDR (P = 0.60), compared to only 5 (13%) of the non-ST131 susceptible isolates (P < 0.001 for both comparisons).
PFGE analysis.
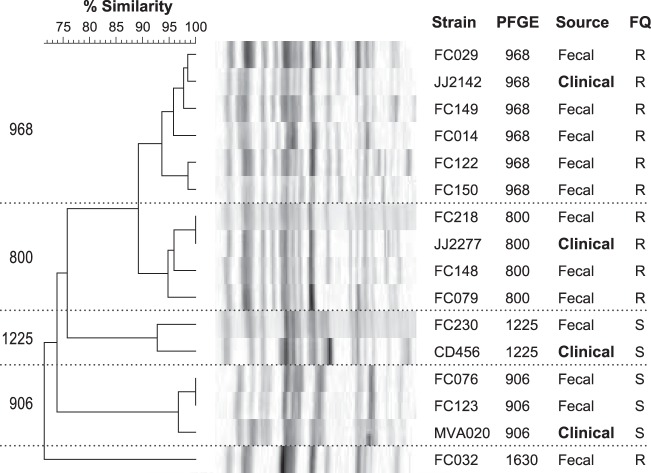
The PFGE profiles of the 12 fecal ST131 E. coli isolates fell into five pulsotypes (Fig. 2). Of these, the four that were represented by multiple isolates each also occurred among ST131 clinical isolates in the external PFGE reference library. Notably, pulsotypes 968 and 800, the two most prevalent pulsotypes among the present ST131 fecal isolates, were also the most common ST131-associated pulsotypes in the reference library (20).
FIG 2.
Dendrogram of pulsed-field gel electrophoresis (PFGE) profiles of 12 fecal ST131 isolates from veterans and 4 reference ST131 clinical isolates. The dendrogram (as inferred according to the unweighted-pair group method with arithmetic averages) was based on Dice similarity coefficients from XbaI PFGE profiles. The four clinical isolates are reference isolates for the respective pulsotypes from a large private (mainly clinical) PFGE database (20). The 94% similarity threshold resolved five pulsotypes among the ST131 isolates (968, 800, 1225, 906, and 1630), four of which included ≥2 fecal isolates each and corresponded with pulsotypes from the clinical database, leaving only 1 fecal isolate (FC032) unmatched to a clinical isolate. FQ, fluoroquinolone phenotype (R, resistant; S, susceptible).
Prevalence and characteristics of antimicrobial-resistant non-E. coli GNB.
The 15 fecal samples that yielded fluoroquinolone- or ESC-resistant non-E. coli GNB yielded one to three distinct such organisms each, for 20 isolates total. These included (number of isolates) Acinetobacter spp. (n = 2), Citrobacter freundii (2), Citrobacter koseri (1), Enterobacter cloacae (5), Flavimonas oryzihabitans (1), Hafnia alvei (2), Klebsiella pneumoniae (3), Proteus mirabilis (2), and Pseudomonas fluorescens (1). Among these 20 resistant isolates, coresistance to agents other than fluoroquinolones and ESCs was detected for all 23 study antibiotics except imipenem and amikacin, ranging in prevalence from 5% (gentamicin) to 95% (ampicillin) (see Table S2 in the supplemental material). Resistance scores among the resistant non-E. coli GNB ranged from 5 to 18 (median, 9), with 16 isolates (80%) qualifying as MDR.
DISCUSSION
In this cross-sectional survey, we screened 100 clinical fecal samples at the MVAMC for the presence of E. coli and other resistant GNB; we then characterized selected E. coli isolates as to ST, phylogenetic group, virulence genotype, and pulsotype and tested both the E. coli and other resistant GNB for resistance to a broad panel of antibiotics. We found that nearly one-third (30%) of samples contained fluoroquinolone- or ESC-resistant GNB, including E. coli ST131 (overall prevalence, 13%), and that most such isolates qualified as MDR. Additionally, we found that among the GNB overall, most fluoroquinolone-resistant isolates were E. coli (75%), but most ESC-resistant isolates were non-E. coli (75%). Thus, at least among veterans, diarrhea may contribute to the dissemination of ST131 E. coli and other MDR GNB within the hospital and/or community.
We also found that, in these diarrheal fecal samples, E. coli ST131 accounted for most fluoroquinolone-resistant GNB overall (67%) and most fluoroquinolone-resistant E. coli (80%). This demonstrates a significant fecal reservoir of ST131 that likely contributes importantly to the current clinical epidemic of fluoroquinolone resistance, which is driven mainly by ST131 (24, 25). Notably, our findings provide a minimum estimate of the prevalence of resistant GNB because the antibiotic concentrations used in the selective agar were chosen based on pre-2011 CLSI breakpoints for cephalosporin and carbapenem resistance, with the goal of detecting primarily fully resistant isolates. As such, they provide a stringent, albeit incompletely sensitive, screen for the presence of resistant organisms.
Supporting the potential clinical relevance of the present fecal ST131 isolates was the finding that their predominant subclone (H30) and pulsotypes (especially 968 and 800) matched those that predominate globally among ST131 clinical isolates (20, 26). Likewise, their extended virulence genotypes were indistinguishable statistically from those of contemporaneous clinical ST131 isolates from veterans but highly distinct from those of non-ST131 E. coli, regardless of source (fecal or clinical) or resistance phenotype. This argues against the possible alternate hypothesis that fecal ST131 isolates represent a distinct population from clinical ST131 isolates and, therefore, are of no public health importance.
In comparison to the non-ST131 fecal E. coli isolates, the present ST131 fecal isolates (whether susceptible or resistant to ESCs or fluoroquinolones) exhibited numerically more VGs and had distinctive combinations of VGs. To the extent that traits regarded conventionally as associated with extraintestinal VGs also promote gut colonization (27), the abundance and distinctive combinations of VGs present in ST131 may confer enhanced fitness in the intestinal niche, thereby potentially contributing to ST131's epidemiological success (28, 29).
At least 11 of the non-ST131 E. coli isolates represented familiar human-associated ST complexes, such as ST95, ST73, ST127, ST69, and ST405, that are known for causing endemic or epidemic extraintestinal E. coli infections (23). However, none of these non-ST131 STs contributed more than 6% to the total sample population. Therefore, with an estimated 13% overall by-sample prevalence, ST131 was by far the most prevalent defined E. coli clonal group among these clinical fecal samples, outnumbering the next most prevalent defined ST by more than 2-fold. This large fecal prevalence gap between ST131 and traditional high-prevalence extraintestinal pathogenic E. coli (ExPEC) clonal groups mirrors the findings of a recent study of clinical isolates from veterans (5), suggesting that clonal prevalence in the fecal reservoir may influence clonal prevalence in the pathogenic niche.
Regarding phylogroup distribution, the present fluoroquinolone-resistant isolates were predominantly from group B2, reflecting the dominance of (group B2-derived) ST131. In contrast, although phylogroup F was rare overall (7%), among non-ST131 isolates it was associated significantly with resistance to fluoroquinolones and/or ESCs. Since both phylogroups B2 and F are associated with extraintestinal virulence (30–32), their associations here with fluoroquinolone resistance underscore the potential risk of acquiring antimicrobial-resistant pathogenic E. coli from diarrheal feces.
When a fluoroquinolone-resistant strain was present in the fecal samples (15% of samples), it was almost always the only E. coli strain detected among the 10 tested colonies, with no significant difference between ST131 and non-ST131 isolates. Thus, fluoroquinolone-resistant E. coli were abundant in these samples, suggesting a substantial potential risk for transmission to exposed individuals.
Apart from their E. coli content, 17 (17%) of the fecal samples also contained fluoroquinolone- and/or ESC-resistant non-E. coli GNB, which accounted for most of the study's ESC-resistant isolates. Most of these genera/species, including Acinetobacter, Citrobacter, Enterobacter, Klebsiella, Proteus, and Pseudomonas, commonly cause human disease. Coresistance was frequent among these isolates, 80% of which qualified as MDR. The presence of such potentially pathogenic MDR non-E. coli GNB in diarrheal feces may pose an added risk to exposed individuals.
The study has several limitations. Because it was conducted at a single institution and using clinical fecal samples from unidentified patients, the generalizability of results is unknown. In that regard, due to a possible bias toward patients receiving antimicrobial therapy (and, therefore, having antibiotic-associated or Clostridium difficile-associated diarrhea, which could have triggered stool testing), the results may overestimate the prevalence of resistant fecal GNB and ST131 among veterans generally. Additionally, the small sample size in certain subgroups reduced the power for finding associations. As such, a new multicenter study with a larger sample size would be desirable. Study strengths include the close matching of fecal and clinical E. coli isolates from veterans, extensive molecular typing using virulence-relevant markers, use of multiple analytical modalities, and attention to clinically relevant resistance phenotypes.
In summary, our findings establish that, at the MVAMC, diarrheal feces submitted for clinical testing in 2011 frequently contained ST131 and other MDR GNB. The presence of such organisms may place exposed health care workers, patients, and household members at risk for acquiring MDR extraintestinal pathogens, thereby potentially contributing to the dissemination of such organisms within the host population.
Supplementary Material
ACKNOWLEDGMENT
We thank the MVAMC clinical microbiology laboratory for assistance with sample collection.
This material is based upon work supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, grants I01 CX000192 01 and I01 CX000920-01A1 (J.R.J.).
The opinions expressed are strictly those of the authors and do not necessarily reflect those of the MVAMC or Department of Veterans Affairs.
J.R.J. has received research grants or consultancies from Actavis, Crucell/Jannsen, ICET, Merck, Syntiron, and Tetraphase and has patent applications for tests to detect specific E. coli clones. The other authors report no financial conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00383-16.
REFERENCES
- 1.Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect 5:449–456. doi: 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 2.Gupta K, Scholes D, Stamm WE. 1999. Increasing prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in women. JAMA 281:736–738. doi: 10.1001/jama.281.8.736. [DOI] [PubMed] [Google Scholar]
- 3.Gupta K, Hooton TM, Stamm WE. 2001. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann Intern Med 135:41–50. doi: 10.7326/0003-4819-135-1-200107030-00012. [DOI] [PubMed] [Google Scholar]
- 4.Madeiros AA. 1993. Nosocomial outbreaks of multiresistant bacteria: extended-spectrum beta-lactamases have arrived in North America. Ann Intern Med 119:428–430. doi: 10.7326/0003-4819-119-5-199309010-00015. [DOI] [PubMed] [Google Scholar]
- 5.Colpan AJB, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR, VICTORY (Veterans Influence of Clonal Types on Resistance: Year 2011) investigators. 2013. Escherichia coli sequence type 131 (ST131) as an emergent multidrug-resistant pathogen among U.S. veterans. Clin Infect Dis 57:1256–1265. doi: 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ender PT, Gajanana D, Johnston B, Clabots C, Tamarkin FJ, Johnson JR. 2009. Transmission of an extended-spectrum-beta-lactamase-producing Escherichia coli (sequence type ST131) strain between a father and daughter resulting in septic shock and emphysematous pyelonephritis. J Clin Microbiol 47:3780–3782. doi: 10.1128/JCM.01361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Cerero L, Navarro MD, Bellido M, Martin-Pena A, Vinas L, Cisneros JM, Gomez-Langley SL, Sanchez-Monteseirin H, Morales I, Pascual A, Rodriguez-Bano J. 2014. Escherichia coli belonging to the worldwide emerging epidemic clonal group O25b/ST131: risk factors and clinical implications. J Antimicrob Chemother 69:809–814. doi: 10.1093/jac/dkt405. [DOI] [PubMed] [Google Scholar]
- 9.Denisuik AJ, Lagace-Wiens PR, Pitout JD, Mulvey MR, Simner PJ, Tailor F, Karlowsky JA, Hoban DJ, Adam HJ, Zhanel GG, Canadian Antimicrobial Resistance Alliance. 2013. Molecular epidemiology of extended-spectrum beta-lactamase-, AmpC beta-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolated from Canadian hospitals over a 5 year period: CANWARD 2007–11. J Antimicrob Chemother 68(Suppl 1):i57–i65. doi: 10.1093/jac/dkt027. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JR, Kuskowski MA, Owens K, Soto S, Horcajada JP, Jimenez de Anta MT, Vila J. 2005. Extended virulence genotypes of Escherichia coli isolates from patients with cystitis, pyelonephritis, or prostatitis. J Infect Dis 191:46–50. doi: 10.1086/426450. [DOI] [PubMed] [Google Scholar]
- 12.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 13.Johnson JR, Johnston B, Kuskowski MA, Sokurenko EV, Tchesnokova V. 2015. Intensity and mechanisms of fluoroquinolone resistance within the H30 and H30Rx subclones of Escherichia coli sequence type 131 compared with other fluoroquinolone-resistant E. coli. Antimicrob Agents Chemother 59:4471–4480. doi: 10.1128/AAC.00673-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002–2004. Antimicrob Agents Chemother 53:2733–2739. doi: 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clermont O, Christenson JK, Daubie AS, Gordon DM, Denamur E. 2014. Development of an allele-specific PCR for Escherichia coli B2 sub-typing, a rapid and easy to perform substitute of multilocus sequence typing. J Microbiol Methods 101:24–27. doi: 10.1016/j.mimet.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Matsumura Y, Yamamoto M, Nagao M, Hotta G, Matsushima A, Ito Y, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group. 2012. Emergence and spread of B2-ST131-O25b, B2-ST131-O16 and D-ST405 clonal groups among extended-spectrum-beta-lactamase-producing Escherichia coli in Japan. J Antimicrob Chemother 67:2612–2620. doi: 10.1093/jac/dks278. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2011. Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. Document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Sahm DF, Thornsberry C, Mayfield DC, Jones ME, Karlowsky JA. 2001. Multidrug-resistant urinary tract isolates of Escherichia coli: prevalence and patient demographics in the United States in 2000. Antimicrob Agents Chemother 45:1402–1406. doi: 10.1128/AAC.45.5.1402-1406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 20.Johnson JR, Nicolas-Chanoine MH, DebRoy C, Castanheira M, Robicsek A, Hansen G, Weissman S, Urban C, Platell J, Trott D, Zhanel G, Clabots C, Johnston BD, Kuskowski MA, MASTER Investigators. 2012. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967–2009. Emerg Infect Dis 18:598–607. doi: 10.3201/eid1804.111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sneath PHA, Sokal RR. 1973. Numerical taxonomy: the principles and practice of numerical classification, vol 2 W. H. Freeman & Co., San Francisco, CA. [Google Scholar]
- 22.Manges AR, Johnson JR, Foxman B, O'Bryan TT, Fullerton KE, Riley LW. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N Engl J Med 345:1007–1013. doi: 10.1056/NEJMoa011265. [DOI] [PubMed] [Google Scholar]
- 23.Riley LW. 2014. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 24.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States (2007). Clin Infect Dis 51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 25.Johnson JR, Porter SB, Zhanel G, Kuskowski MA, Denamur E. 2012. Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect Immun 80:1554–1562. doi: 10.1128/IAI.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peirano G, van der Bij AK, Freeman JL, Poirel L, Nordmann P, Costello M, Tchesnokova VL, Pitout JD. 2014. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum beta-lactamases: global distribution of the H30-Rx sublineage. Antimicrob Agents Chemother 58:3762–3767. doi: 10.1128/AAC.02428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowrouzian FL, Adlerberth I, Wold AE. 2006. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect 8:834–840. doi: 10.1016/j.micinf.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Vimont S, Boyd A, Bleibtreu A, Bens M, Goujon JM, Garry L, Clermont O, Denamur E, Arlet G, Vandewalle A. 2012. The CTX-M-15-producing Escherichia coli clone O25b: H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One 7:e46547. doi: 10.1371/journal.pone.0046547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boetius Hertz F, Lobner-Olesen A, Frimodt-Moller N. 2014. Antibiotic selection of Escherichia coli sequence type 131 in a mouse intestinal colonization model. Antimicrob Agents Chemother 58:6139–6144. doi: 10.1128/AAC.03021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau SH, Reddy S, Cheesbrough J, Bolton FJ, Willshaw G, Cheasty T, Fox AJ, Upton M. 2008. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J Clin Microbiol 46:1076–1080. doi: 10.1128/JCM.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abraham SJD, Wong HS, Johnson JR, Mark A Toleman MA, Wakeham DL, Gordon David M, Turnidge JD, Mollinger JL, Gibson JS, Trott DJ. 2015. First detection of extended-spectrum cephalosporin- and fluoroquinolone-resistant Escherichia coli in Australian food-producing animals. J Glob Antimicrob Resist 3:273–277. doi: 10.1016/j.jgar.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Ewers CBA, Stamm I, Grobbel M, Kopp PA, Guerra B, Stubbe M, Doi Y, Zong Z, Kola A, Schaufler K, Semmler T, Fruth A, Wieler LH, Guenther S. 2014. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J Antimicrob Chemother 69:1224–1230. doi: 10.1093/jac/dkt516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.