Abstract
We report the complete nucleotide sequence of a plasmid, pA31-12, carrying blaCTX-M-55 and mcr-1 from a chicken Escherichia coli isolate. pA31-12 has an IncI2 replicon that displays extensive sequence similarity with pHN1122-1-borne blaCTX-M-55 and pHNSHP45-borne mcr-1. Insertion sequences ISEcp1 and ISApl1 are responsible for the mobilization of blaCTX-M-55 and mcr-1, respectively. The colocalization of mcr-1 with an extended-spectrum β-lactamase gene on a conjugative plasmid may accelerate the dissemination of both genes by coselection.
TEXT
Infections caused by multidrug-resistant Gram-negative bacteria, particularly extended-spectrum-β-lactamase (ESBL)-producing bacteria or carbapenem-resistant Enterobacteriaceae (CRE), have become significant threats to public health (1). The rapid increase in the prevalence of ESBL-producing or CRE isolates has prompted the reconsideration of colistin as a valid therapeutic option (2). However, an increase in the global presence of ESBL/CRE strains has resulted in an increased colistin use, leading to the inevitable risk of emerging resistance. Moreover, the massive use of colistin in animal production facilities has aggravated the problem. Recently, a plasmid encoding colistin resistance (mcr-1) was identified, and its importance was quickly recognized (3). This plasmid encodes a phosphoethanolamine transferase responsible for lipid A modification and, concomitantly, decreased colistin susceptibility.
The mcr-1 gene was initially identified in Escherichia coli and Klebsiella pneumoniae strains isolated from animals, food products, and clinical patients in China. The presence of this gene was subsequently reported from various isolates with diverse origins worldwide (3–9). The immediate clinical concern was the inevitable appearance of a plasmid cointegrating the mcr-1 gene with other resistance genes, creating a multidrug-resistant isolate that would reduce therapeutic options. Here, we report a cephalosporin- and colistin-resistant E. coli strain that harbors a transferable IncI2 plasmid mediating cotransfer of blaCTX-M-55 and mcr-1.
E. coli isolate A31-12 was recovered from the rectal swab of a chicken with diarrhea from a veterinary clinic in Guangzhou, China, in August 2012. Antimicrobial susceptibility was determined by the agar dilution method by following CLSI guidelines. The breakpoints for each antimicrobial were as recommended by the Clinical and Laboratory Standards Institute (CLSI) (10), veterinary CLSI (11), and DANMAP 98 (olaquindox) (12). The results showed that this isolate was resistant to most antimicrobial drugs tested, including colistin, cefotaxime, ceftazidime, ceftiofur, ciprofloxacin, nalidixic acid, chloramphenicol, florfenicol, amikacin, gentamicin, and trimethoprim-sulfamethoxazole, and it showed intermediate susceptibility to fosfomycin. This isolate retained sensitivity to meropenem, cefoxitin, olaquindox, doxycycline, and streptomycin (see Table S1 in the supplemental material). Multilocus sequence typing (MLST) (13) showed that E. coli A31-12 belonged to sequence type 1011 (ST1011).
PCR analysis and DNA sequencing confirmed that this strain carried blaCTX-M-55 and mcr-1, which accounted for the corresponding drug sensitivity phenotype. Filter mating assays (14) revealed that the blaCTX-M-55 and mcr-1 genes were able to cotransfer to E. coli C600 (streptomycin resistant). Compared with the MICs for the recipient strain, those for the transconjugant increased 256-fold for cefotaxime (from 0.125 to 32 μg/ml) and 64-fold for colistin (from 0.0625 to 4 μg/ml). S1 pulsed-field gel electrophoresis and Southern blot analysis (15) showed that both genes in donor clinical isolate A31-12 and recipient strain C600 were located on a single plasmid of approximately 65 kb (see Fig. S1 in the supplemental material), and this plasmid was named pA31-12 in this study.
To determine the complete nucleotide sequences of the plasmid, the total genomic DNA from the transconjugant C600/pA31-12 was extracted and sequenced using an Illumina MiSeq system. After filtering C600 chromosomal DNA data and assembling the remaining reads, we obtained a single contig for pA31-12 that was manually closed into a circle. The complete plasmid structure was further confirmed and verified by long-range PCR. The plasmid pA31-12 is 67,134 bp in length and has an average G+C content of 46.06%. Gene prediction and annotation were performed using the RAST tools. Sequence comparison and map generation were performed using BLAST (http://blast.ncbi.nlm.nih.gov) and Easyfig version 2.1.
The plasmid pA31-12 (GenBank accession number KX034083) is an IncI2-type plasmid and encodes 91 open reading frames (ORFs). A plasmid comparison based on a full-plasmid BLAST query revealed that pA31-12 is closely related to the other IncI2 plasmids in GenBank (Fig. 1), including pDMC1097 (accession number CP011978), pHN1122-1 (accession number JN797501.1), pCTXM64_C0967 (accession number KP091735.1), pCTXM132_P0421(accession number KP198615.1), pHNY2 (accession number KF601686.2), and pHNSHP45 (accession number KP347127.1) from E. coli; pSTH21(accession number LN623683.2) and pSH146_65 (accession number JN983044.1) from Salmonella enterica; p1081-CTXM (accession number KJ460501.1) from Shigella sonnei; and pBK15692 (accession number KC845573.1) from K. pneumoniae. Like the other IncI2 plasmids, pA31-12 has a typical plasmid backbone set of sequences that are responsible for plasmid replication, maintenance, and transfer (16) (Fig. 1A).
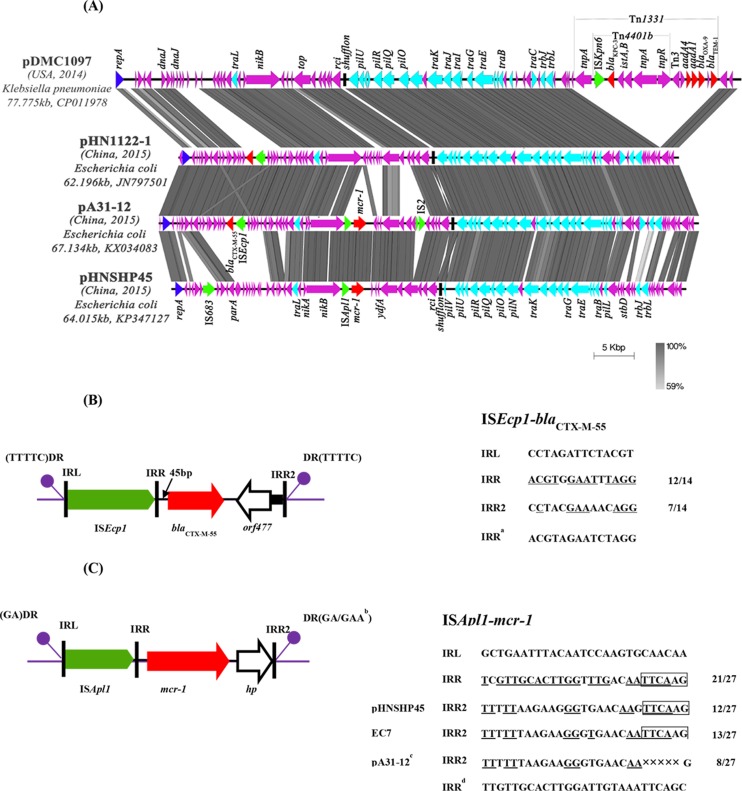
FIG 1.
(A) Linear comparison of complete plasmid sequences of pA31-12 (this study, GenBank accession number KX034083), pDMC1097 (accession number CP011978), pHN1122-1 (accession number JN797501.1), and pHNSHP45 (accession number KP347127.1); (B) schematic representations of the genetic organization surrounding blaCTX-M-55 on pA31-12 and pHN1122-1; (C) schematic representations of the genetic organization surrounding mcr-1on pA31-12, pHNSHP45, and a fragment from E. coli EC7 (GenBank accession number NZ_JWKG01000081.1), which have identical IncI2 plasmid backbones. The arrows represent the positions and transcriptional directions of the ORFs. Regions of >99% homology are marked by gray shading. Genes associated with the tra and pil loci are indicated by light-blue arrows, while replication-associated genes are indicated by dark-blue arrows. Resistance genes are indicated by red arrows, while accessory genes are indicated by purple arrows. Insertion sequences are indicated by green arrows. DR, direct repeats; IRL, terminal inverted repeats at the left; IRR, terminal inverted repeats at the right. Footnotes: a, perfect IRR of ISEcp1; b, the DR of the ISApl1-mcr-1 transposition unit are varied in IncI2 plasmids (GA in pHNSHP45 but GAA in EC7 or pA31-12); c, TTCAA was lost in pA31-12 compared to EC7; d, perfect IRR of ISApl1. Underlined nucleotides in the alternate IRR elements are identical to those of the perfect IRR. Boxed nucleotides indicated the six conserved nucleotides (TTCAAG) located at the 3′ end of alternate IRR.
The RepA replicon protein of pA31-12 shows 100% amino acid identity with RepA of pHN1122-1 (17). The genes encoding plasmid stability functions, including mok, hok, yafA, and yafB, are also present in pA31-12. pA31-12 contains two pilus-encoding genes (pil and tra) which are conserved in all IncI2 plasmids and are responsible for plasmid transfer. In addition, pA31-12 contains a plasmid-borne site-specific recombinase (Rci) gene and a shufflon region (18). Four different PilV proteins with the same N-terminal sequence but varied C-terminal sequences were most likely created by four imperfect 19-bp repeat sequences (19). In comparison with plasmid R721, segments A and B′/D′ are conserved but reversed, while segment C is missing. This suggests that pA31-12 undergoes complex rearrangements in the shufflon region mediated by the recombinase (Fig. 1A).
Two acquired resistance gene regions were identified in pA31-12. As with other pHN1122-1-like IncI2 plasmids, blaCTX-M-55 is located in a 3.084-kb ISEcp1 transposition unit (16, 17). It is a typical ISEcp1-blaCTX-M-55 transposition unit containing ISEcp1, blaCTX-M-55, orf477, and a 112-bp fragment of the IncA/C backbone. ISEcp1 is able to mobilize several blaCTX-M genes and their adjacent sequences by recognizing its own left inverted repeat (IRL) and the closest downstream sequence that resembles its right inverted repeat (IRR) (20, 21). This whole structure is inserted near yajA, generating a 5-bp duplication of the target sequence (GAAAA) (Fig. 1B). The identical configuration, which was observed not only in plasmids harboring blaCTX-M-55 (pHN1122-1 and pSTH21) but also in other IncI2 plasmids (pHNY2 carrying blaCTX-M-15, pCTXM64_C0967 carrying blaCTX-M-64, and pCTXM132_P0421 carrying blaCTX-M-132), infers that such IncI2 plasmids have a common plasmid ancestor (16).
A plasmid-mediated colistin resistance gene, mcr-1, was also identified in pA31-12. Like the original mcr-1-positive plasmid pHNSHP45, pA31-12 has an ISApl1 mobile element that flanks the 5′ mcr-1 gene (3). ISApl1 is a member of the IS30 family, which was initially identified in Actinobacillus pleuropneumoniae (22). A potential linked transposition element was found in pHNSHP45-like plasmids that initiated at the 27-bp IRL sequence of ISApl1 (5′-GCTGAATTTACAATCCAAGTGCAACAA-3′) but ended at a fixed position 0.8 kb downstream from the mcr-1 stop codon by recognizing a similar IRR. Intriguingly, the IRR-like sequences seemed flexible and shared only 12 or 13 bp of the 27 bp of the perfect IRR sequence. Nevertheless, the 6 nucleotides (TTCAAG) located at the 3′ end of these IRRs was always found, indicating that these were necessary for transposition. Compared with E. coli EC7 (NZ_JWKG01000081.1), TTCAA was lost in pA31-12, implying a new reorganization event (Fig. 1C). These data indicated that ISApl1 was involved in a transposition process with weakly related IRRs that was similar to the transposition process in other insertion sequences, such as ISEcp1 (21).
This transposition process resulted in an ∼3.7-kb fragment (Fig. 1C) encoding a hypothetical protein in addition to mcr-1. Though its exact function is unknown, the hypothetical protein encodes a phosphoesterase and may be involved in colistin resistance. However, further experiments are needed to assess this hypothesis. Consistently with what occurred with the other IS30 family mobile elements, the insertion of ISApl1-mcr-1 resulted in the duplication at the target insertion site of 2 or 3 base pairs (22) (Fig. 1C).
To summarize, this study reports for the first time the identification of a transferable IncI2 plasmid harboring both blaCTX-M-55 and mcr-1 in an E. coli isolate derived from a chicken. Comparative analysis of the IncI2 plasmids provided information on their evolutionary history, suggesting that ISApl1 might play a key role in transferring the mcr-1 gene to an IncI2 backbone. Given the fact that colistin is the last-resort antibiotic for treating human infections due to multidrug-resistant Enterobacteriaceae, cotransfer of mcr-1 with blaCTX-M-55 by a single mobile plasmid constitutes a potentially serious threat to clinical treatment regimens. Therefore, enhanced surveillance and efforts to limit the spread of such multidrug-resistant plasmids is urgently needed.
Nucleotide sequence accession number.
The annotated sequence of plasmid pA31-12 from strain A31-12 has been submitted to GenBank accession number KX034083.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation and the Natural Science Foundation of Guangdong Province, China (grant U1201214), the Program for Changjiang Scholars and Innovative Research Team at the University of the Ministry of Education of China (grant IRT13063), and the Natural Science Foundation of Guangdong Province (grant S2012030006590).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00774-16.
REFERENCES
- 1.Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S, Bhutta ZA, Coates A, Bergstrom R, Wright GD, Brown ED, Cars O. 2013. Antibiotic resistance—the need for global solutions. Lancet Infect Dis 13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 2.Doi Y, Paterson DL. 2015. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med 36:74–84. doi: 10.1055/s-0035-1544208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Shen Z, Wang Y, Shen Y, Shen J, Wu C. 2016. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis 16:293. doi: 10.1016/S1473-3099(16)00061-X. [DOI] [PubMed] [Google Scholar]
- 5.Petrillo M, Angers-Loustau A, Kreysa J. 2016. Possible genetic events producing colistin resistance gene mcr-1. Lancet Infect Dis 16:280. doi: 10.1016/S1473-3099(16)00005-0. [DOI] [PubMed] [Google Scholar]
- 6.Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Butaye P, Goossens H. 2016. Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet Infect Dis 16:283–284. doi: 10.1016/S1473-3099(16)00012-8. [DOI] [PubMed] [Google Scholar]
- 7.Haenni M, Poirel L, Kieffer N, Chatre P, Saras E, Metayer V, Dumoulin R, Nordmann P, Madec JY. 2016. Co-occurrence of extended spectrum beta lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis 16:281–282. doi: 10.1016/S1473-3099(16)00007-4. [DOI] [PubMed] [Google Scholar]
- 8.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Kasbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T, RESET Consortium. 2016. Colistin resistance gene mcr-1 in extended-spectrum beta-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 9.Du H, Chen L, Tang YW, Kreiswirth BN. 2016. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis 16:287–288. doi: 10.1016/S1473-3099(16)00056-6. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI document M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard—fourth edition and supplement, VET01A4E and VET01S3E. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 12.Danish Integrated Antimicrobial Resistance Monitoring and Research Programme (DANMAP) 98. 1999. Consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and human in Denmark. Danish Veterinary and Food Administration, Statens Serum Institut, Copenhagen, Denmark. [Google Scholar]
- 13.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith MD, Guild WR. 1980. Improved method for conjugative transfer by filter mating of Streptococcus pneumoniae. J Bacteriol 144:457–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, He D, Lv L, Liu W, Chen X, Zeng Z, Partridge SR, Liu JH. 2015. blaCTX-M-1/9/1 hybrid genes may have been generated from blaCTX-M-15 on an IncI2 plasmid. Antimicrob Agents Chemother 59:4464–4470. doi: 10.1128/AAC.00501-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv L, Partridge SR, He L, Zeng Z, He D, Ye J, Liu JH. 2013. Genetic characterization of IncI2 plasmids carrying blaCTX-M-55 spreading in both pets and food animals in China. Antimicrob Agents Chemother 57:2824–2827. doi: 10.1128/AAC.02155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SR, Komano T. 1992. Nucleotide sequence of the R721 shufflon. J Bacteriol 174:7053–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Chavda KD, Al Laham N, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob Agents Chemother 57:5019–5025. doi: 10.1128/AAC.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhanji H, Doumith M, Hope R, Livermore DM, Woodford N. 2011. ISEcp1-mediated transposition of linked blaCTX-M-3 and blaTEM-1b from the IncI1 plasmid pEK204 found in clinical isolates of Escherichia coli from Belfast, United Kingdom. J Antimicrob Chemother 66:2263–2265. doi: 10.1093/jac/dkr310. [DOI] [PubMed] [Google Scholar]
- 21.Poirel L, Lartigue MF, Decousser JW, Nordmann P. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob Agents Chemother 49:447–450. doi: 10.1128/AAC.49.1.447-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tegetmeyer HE, Jones SC, Langford PR, Baltes N. 2008. ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet Microbiol 128:342–353. doi: 10.1016/j.vetmic.2007.10.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.