Abstract
Among 15,588 Enterobacteriaceae isolates collected in 63 U.S. hospitals from 2012 to 2014, 2,129 (13.7%) displayed an extended-spectrum β-lactamase (ESBL) phenotype. These rates were similar over time (13.2 to 13.9%); however, differences among Escherichia coli (12.7 and 15.1% in 2012 and 2014; P = 0.007) and Klebsiella pneumoniae (18.9 and 15.5% in 2012 and 2014; P = 0.006) were noted when comparing 2014 and 2012. Carbapenem-resistant Enterobacteriaceae (CRE) (2.3 and 1.8%) and carbapenem-resistant K. pneumoniae (6.8 and 5.1%; P = 0.003) rates were lower in 2014 than in 2012. Isolates carrying blaCTX-M-15-like genes were stable (42.1 to 42.4%), but a decrease among E. coli isolates (59.1 and 49.7%; P = 0.008) and an increase among K. pneumoniae isolates (32.7 and 41.2%; P = 0.022) in 2014 were observed. Isolates carrying blaKPC (304) decreased over the years (16.5 and 10.9%; P = 0.008), mainly due to the decrease in K. pneumoniae isolates harboring blaKPC (n = 285; 35.6 and 28.4%; P = 0.041) in hospitals in the Mid-Atlantic and South Atlantic regions, where these isolates were highly prevalent during 2012 and 2013. Isolates carrying blaCMY-2-like and blaCTX-M-14-like genes increased (8.2 and 11.9% and 9.1 and 12.9%, respectively; P = 0.04 for both), and those producing blaSHV ESBL decreased (24.9 and 12.7%; P < 0.001) over the studied years, due to a decreased occurrence of the enzymes among K. pneumoniae isolates. Other enzymes were detected in smaller numbers of isolates, including four K. pneumoniae isolates carrying blaNDM-1 metallo-β-lactamase (two in 2012 and two in 2014). Ceftazidime-avibactam, a recently approved β-lactamase inhibitor combination, was very active against the ESBL phenotype isolates (MIC50/90, 0.12 and 1 μg/ml; 99.7% susceptible) and CRE strains (MIC50/90, 0.5 and 2 μg/ml; 98.5% susceptible) that displayed elevated MIC values for many comparator agents. In conclusion, significant changes were noted in the frequencies of isolates harboring various β-lactamases among U.S. hospitals between 2012 and 2014 that will require continued monitoring.
INTRODUCTION
Enterobacteriaceae isolates producing extended-spectrum β-lactamases (ESBLs) or carbapenemases (carbapenem-resistant Enterobacteriaceae [CRE]) are usually resistant to most or all β-lactam agents. These isolates often coharbor resistance mechanisms to other antimicrobial classes that are carried on mobile genetic elements, which promote the dissemination of these resistance genes (1, 2). These isolates pose a challenge for clinical microbiology laboratories, infection control practitioners, and clinicians due to the difficulties in detecting, containing, and treating infections caused by these emerging organisms. Monitoring the occurrence and prevalence of the isolates is prudent; however, very few studies in U.S. hospitals report on the prevalence and characteristics of broad collections of Enterobacteriaceae isolates producing the enzymes.
We recently reported on the prevalence of common β-lactamase genes among isolates collected in over 70 U.S. hospitals during 2012 (3) and 2013 (4) that displayed a positive ESBL phenotype according to the Clinical and Laboratory Standards Institute (CLSI) MIC-based epidemiological criteria. During these and other published studies, it has been demonstrated that the β-lactamase prevalence scenario in the United States differs from that in other countries regarding the occurrence and distribution of β-lactamase-producing isolates and enzyme types (3). A decade ago, isolates carrying blaCTX-M were considered endemic in nosocomial and community settings in countries in Europe and Asia (5), but the first studies showing the dissemination of isolates harboring blaCTX-M in the United States date from 2007 (6, 7). After the initial reports, a rapid spread of isolates harboring blaCTX-M-15-like and blaCTX-M-14-like genes was observed in U.S. hospitals, and currently, the rates are becoming more similar to those observed in other nations (8).
The emerging scenario in U.S. hospitals is also different regarding the presence of carbapenemases. Isolates carrying blaKPC that were first described in North Carolina and later in New York City (9), became endemic in the latter geographic region and surrounding areas. More recently, isolates producing these serine-carbapenemases have been detected throughout the country (3, 4). Furthermore, metallo-β-lactamase (MBL)-producing isolates have been uncommon in U.S. hospitals; however, this could rapidly change with the dissemination of Enterobacteriaceae isolates harboring blaNDM-1, which have been reported in 22 states (10).
In this study, we assess the occurrence of β-lactamase-producing isolates collected in 69 U.S. hospitals during 2014 and evaluate trends of these enzymes in 63 institutions participating in all three consecutive years of surveillance, including 2012 and 2013, which have been previously reported as parts of larger collections (3, 4). Additionally, we evaluated the activities of ceftazidime-avibactam and comparator antimicrobial agents against the isolates producing β-lactamases.
MATERIALS AND METHODS
Bacterial isolates.
A total of 5,771 clinical isolates, including Escherichia coli (n = 2,813), Klebsiella pneumoniae (n = 1,832), Klebsiella oxytoca (n = 502), and Proteus mirabilis (n = 624) clinical isolates, were collected as part of the International Network for Optimal Resistance Monitoring (INFORM) program in 69 U.S. medical centers during 2014. Only one isolate per patient infection episode was included in the study. Species identification was confirmed when necessary by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) using the Bruker Daltonics MALDI Biotyper (Billerica, MA) following the manufacturer's instructions.
For the multiple-year comparison, 15,588 clinical isolates of E. coli (n = 7,688), K. pneumoniae (n = 4,845), K. oxytoca (n = 1,250), and P. mirabilis (n = 1,805) from 63 U.S. hospitals participating in a surveillance study from 2012 to 2014 were analyzed. ESBL phenotype isolates collected from 2012 to 2014 were recovered from bloodstream infections (n = 340), intra-abdominal infections (n = 154), pneumonia in hospitalized patients (n = 556), skin/soft tissue infections (n = 468), urinary tract infections (n = 504), and other or unknown sites (n = 107).
Antimicrobial susceptibility testing.
Isolates were susceptibility tested using the reference broth microdilution method as described by the CLSI (11). The categorical interpretations for all antimicrobials were those found in CLSI M100-S25 (2015), and quality control (QC) was performed using E. coli ATCC 25922 and 35218, K. pneumoniae ATCC 700603, and Pseudomonas aeruginosa ATCC 27853 (12). All QC results were within the ranges published in the CLSI document (12).
Screening for β-lactamases.
Isolates displaying an ESBL phenotype (MIC, >1 μg/ml for aztreonam and/or ceftazidime and/or ceftriaxone [12]) were tested for β-lactamase-encoding genes using the microarray-based Check-MDR CT101 assay kit (Check-Points, Wageningen, Netherlands). The assay was performed according to the manufacturer's instructions. The kit has the capability to detect blaCTX-M group 1, 2, 8 plus 25, and 9; blaTEM wild type (WT) and ESBL; blaSHV WT and ESBL; blaACC; blaACT/MIR; blaCMY-2-like; blaDHA; blaFOX; blaKPC; and blaNDM-1-like genes The most common amino acid alterations that expand the spectrum of TEM and SHV enzymes are detected by the assay and include E104K, R164S/H, and G238S for TEM and G238A/S and E240K for SHV. Validation of the assay against U.S. isolates was performed previously (8).
All isolates displaying a ceftazidime-avibactam MIC value of >4 μg/ml were screened for the presence of metallo-β-lactamase- and serine-carbapenemase-encoding gene families (blaIMP, blaVIM, blaNDM, blaKPC, blaOXA-48, blaGES, blaIMI, blaNMC-A, and blaSME) by PCR as previously described (13). Amplicons were sequenced on both strands, and the results were analyzed using the Lasergene software package (DNASTAR, Madison, WI). The amino acid sequences were compared with those available in the NCBI database using BLAST.
Statistical analysis and definitions.
Statistical analysis was performed by Fisher's exact test comparing 2012 and 2014 rates using EpiInfo 7 (Centers for Disease Control and Prevention, Atlanta, GA). CRE were those isolates displaying imipenem and/or meropenem MIC values of >2 μg/ml (14).
RESULTS
Occurrence of β-lactamases in U.S. hospitals during 2014.
A total of 782 Enterobacteriaceae isolates collected during 2014 displayed an ESBL phenotype: 409 E. coli (14.5% of the overall samples for the species), 284 K. pneumoniae (15.6%), 58 K. oxytoca (11.5%), and 31 P. mirabilis (5.0%) isolates (Table 1).
TABLE 1.
Results of screening for β-lactamase genes/families among 782 ESBL phenotype isolates collected during 2014 in 69 U.S. hospitals
| β-Lactamase gene/family | No. of positive resultsa |
||||
|---|---|---|---|---|---|
| Overall (782)b | E. coli (409) | K. oxytoca (58) | K. pneumoniae (284) | P. mirabilis (31) | |
| CRE (including MBL) | |||||
| blaKPC | 90 | 4 | 3 | 83 | |
| blaNDM-1 | 2 | 1 | 1 | ||
| ESBL | |||||
| blaCTX-M-15-like | 326 | 204 | 1 | 113 | 8 |
| blaSHV ESBL | 106 | 9 | 10 | 87 | |
| blaCTX-M-14-like | 100 | 84 | 13 | 3 | |
| blaTEM ESBL | 15 | 11 | 1 | 3 | |
| blaCTX-M-8-like | 2 | 1 | 1 | ||
| blaCTX-M-2-like | 1 | 1 | |||
| Transferrable AmpC | |||||
| blaCMY-2-like | 92 | 74 | 7 | 11 | |
| blaFOX-like | 7 | 2 | 2 | 3 | |
| blaDHA-like | 3 | 2 | 1 | ||
| blaACT/MIR | 1 | 1 | |||
| Non-ESBL | |||||
| blaTEM WT | 360 | 171 | 8 | 166 | 15 |
| blaSHV WT | 271 | 8 | 263 | ||
| Negative result | 65 | 17 | 43 | 2 | 3 |
Isolates can be positive for more than one test/gene.
Number tested.
Isolates carrying blaCTX-M-15-like genes (including blaCTX-M-1, blaCTX-M-15, and blaCTX-M-3) were the most prevalent. A total of 326 isolates yielded positive results for blaCTX-M-15-like genes among all four species tested, often in combination with other β-lactamase-encoding genes/families. Additionally, 100 isolates carried blaCTX-M-14-like family genes—blaCTX-M-9 and blaCTX-M-27, among others. The majority of these isolates were E. coli (n = 84), but K. pneumoniae (n = 13) and P. mirabilis (n = 3) were also detected.
Carbapenemase-encoding genes were observed among 92 isolates. blaKPC was detected among 90 isolates, the majority being K. pneumoniae (83 isolates), but also in E. coli (n = 4) and K. oxytoca (n = 3) isolates. blaNDM-1 was detected in two isolates, a K. pneumoniae isolate from New York and an E. coli isolate from California, with results confirmed by DNA sequencing.
blaSHV genes encoding enzymes with an extended spectrum of activity (blaSHV ESBL) were detected among 106 (13.6%) isolates, and the vast majority of the isolates were K. pneumoniae (87/106; 82%). blaTEM genes encoding an ESBL variant were observed in 15 isolates, including three bacterial genera/species (Table 1). Other ESBL genes detected were blaCTX-M-8-like (two isolates and two genera/species) and blaCTX-M-2-like (one isolate) genes.
Additionally, transferable cephalosporinases (plasmidic AmpCs) were detected among 103 (13.2%) isolates, and 92 (89.3%) strains displayed a positive result for blaCMY-2-like genes (74 E. coli, 7 K. pneumoniae, and 11 P. mirabilis) (Table 1). Other AmpC genes detected were blaFOX (seven isolates and three bacterial genera/species), blaDHA (three isolates and two bacterial species), and blaACT/MIR (one isolate each). A total of 54 isolates did not carry the screened genes encoding ESBL (blaCTX-M, blaSHV ESBL, or blaTEM ESBL), transferable AmpC, or carbapenemases, and these isolates yielded only positive results for narrow-spectrum blaSHV and/or blaTEM. Additionally, 65 isolates displayed negative results for all β-lactamase genes/families screened, which were mostly K. oxytoca (43 isolates) (data not shown).
Comparison of β-lactamase occurrences from 2012 to 2014.
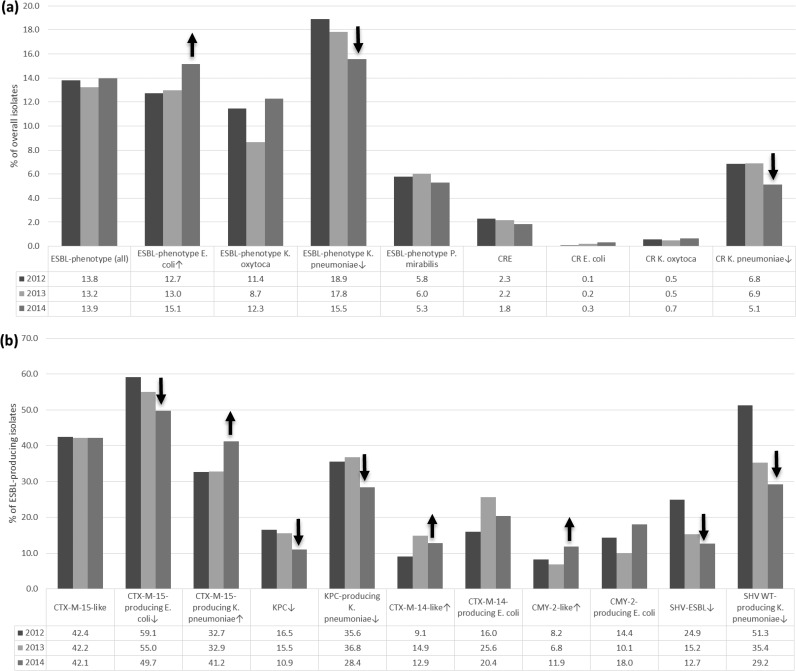
A total of 2,129 (13.7% of targeted species) isolates met the MIC-based ESBL criteria during the 2012-2014 period in hospitals participating in all years of surveillance. This total included 1,048 E. coli (13.6% for this species), 843 K. pneumoniae (17.4%), 135 K. oxytoca (10.8%), and 103 P. mirabilis (5.7%) isolates. The overall ESBL rates were similar over time and were 13.8, 13.2, and 13.9% in 2012, 2013, and 2014, respectively (comparing 2012 to 2014: P = 0.841; odds ratio [OR], 0.988; 95% confidence interval [CI], 0.883 to 1.105) (Fig. 1); however, an increase in the ESBL phenotype among E. coli isolates from 12.7 to 15.1% (P = 0.007; OR, 0.817; 95% CI, 0.695 to 0.959) and a decrease in the ESBL phenotype among K. pneumoniae isolates from 18.9 to 15.5% (P = 0.006; OR, 1.264; 95% CI, 1.054 to 1.517) was documented (Fig. 1).
FIG 1.
(a) Percent occurrences of ESBL phenotype and CRE isolates collected in 63 U.S. hospitals from 2012 to 2014 among all isolates collected during the period. (b) Percent occurrences of the most common β-lactamase genes among ESBL phenotype isolates. Statistically significant shifts upward (↑) or downward (↓) are indicated.
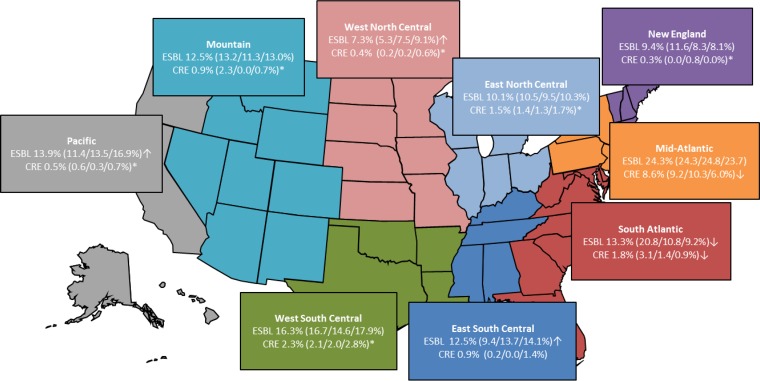
Differences in the ESBL phenotype rates among census regions were noted, and a significant decrease from 20.8 to 9.2% (P < 0.001; OR, 2.582; 95% CI, 1.792 to 3.722) in the ESBL phenotype rates between 2012 and 2014 was documented in the South Atlantic region (Fig. 2). Additionally, a minor decrease in ESBL phenotype isolates was noted in the New England region (P = 0.086; OR, 1.484; 95% CI, 0.881 to 2.502) (Fig. 2). In both census regions, the overall decrease was caused by the reduced occurrence of E. coli isolates displaying an ESBL phenotype in the last year of the study (16.7 to 10.3% in the South Atlantic and 11.5 to 7.5% in the New England regions). Conversely, a significant increase in ESBL phenotype rates from 2012 to 2014 was observed in the West North Central (5.3 to 9.1%) (P = 0.016; OR, 0.563; 95% CI, 0.338 to 0.936), East South Central (9.4 to 14.1%) (P = 0.014; OR, 0.632; 95% CI, 0.425 to 0.940), and Pacific (11.4 to 16.9%) (P = 0.003; OR, 0.628; 95% CI, 0.452 to 0.869) regions (Fig. 2). An increased frequency during 2014 was noted for ESBL phenotype E. coli in the same three regions (5.8 to 14.2%, 8.9 to 16.7%, and 12.9 to 19.1% for the West North Central, East South Central, and Pacific regions, respectively), in K. pneumoniae for the East South Central (11.0 to 14.4%) and Pacific (11.6 to 14.0%) regions, and in K. oxytoca (9.0 to 17.9%) and P. mirabilis (5.4 to 14.8%) in the Pacific region (data not shown).
FIG 2.
Occurrence and changes in ESBL phenotype and CRE isolates (overall percentages) among 15,588 isolates collected from 63 U.S. hospitals during 2012 to 2014. ↑, statistically significant increase; ↓, statistically significant decrease; *, number too small to calculate.
CRE represented 2.1% of the total isolates during the study period, and a slight but not statistically significant decline was observed from 2012 (2.3%) to 2014 (1.8%) (P = 0.110; OR, 1.253; 95% CI, 0.953 to 1.647) (Fig. 1). Among the 326 CRE organisms, 304 (93.3%) were K. pneumoniae, and the remaining isolates were E. coli (n = 15) and K. oxytoca (n = 7). The frequency of the carbapenem resistance phenotype within K. pneumoniae was significantly higher in 2012 (6.8%) than in 2014 (5.1%). Other carbapenem-resistant (CR) organisms occurred in limited numbers each year; however, carbapenem-resistant E. coli isolates displayed a steady increase over the study interval (two, five, and eight isolates in 2012, 2013, and 2014, respectively).
Among U.S. census regions, a statistically significant decline in CRE rates was noted in the Mid-Atlantic (9.2 to 6.0%) (P = 0.018; OR, 1.593; 95% CI, 1.050 to 2.419) and South Atlantic (3.1 to 0.9%) (P = 0.009; OR, 3.494; 95% CI, 1.260 to 9.685) (Fig. 2) regions when comparing 2012 to 2014 rates, although a small number of isolates were present in the South Atlantic region in 2012, 2013, and 2014. In both census regions, a decrease among carbapenem-resistant K. pneumoniae isolates was observed; however, in the Mid-Atlantic region (24.7 to 19.9%; not statistically significant), a reduction in carbapenem-resistant K. oxytoca (5.7 to 2.9%) also contributed to the regional decline in CRE rates.
Among the most common β-lactamase-encoding genes, blaCTX-M-15-like genes occurred among 295, 297, and 326 isolates in 2012, 2013, and 2014, respectively. Although the overall prevalence of these isolates was stable (42.4, 42.2, and 42.1% in 2012, 2013, and 2014, respectively) (P = 0.474; OR, 1.013; 95% CI, 0.821 to 1.250), a decrease in the frequency of E. coli isolates carrying blaCTX-M-15-like genes (59.1 to 49.7%) (P = 0.008; OR, 1.460; 95% CI, 1.081 to 1.972) and an increase in the frequency of the enzyme family among K. pneumoniae isolates (32.7 to 41.2%) (P = 0.022; OR, 0.692; 95% CI, 0.490 to 0.976) were documented in 2014. The 2014 decrease in the occurrence of E. coli isolates carrying blaCTX-M-15-like genes was noted in different U.S. census regions, including the New England (56.5 and 38.5% in 2012 and 2014, respectively), Mid-Atlantic (62.3 and 49.4%), East North Central (66.0 and 46.8%), South Atlantic (70.3 and 37.9%), and Mountain (55.6 and 42.9%) regions (data not shown); however, these differences were either not statistically significant or the number of isolates was not large enough for meaningful interpretation.
A decrease in isolates harboring blaKPC was observed in the study years: 16.5% of ESBL phenotype isolates carried blaKPC in 2012, 15.5% in 2013, and only 10.9% in 2014 (P = 0.008; OR, 3.868; 95% CI, 1.188 to 2.192). This trend was mainly due to the decrease of K. pneumoniae isolates carrying blaKPC from 35.6% in 2012 and 36.8% in 2013 to 28.4% in 2014 (P = 0.041; OR, 1.3; 95% CI, 0.975 to 1.995). The largest decline in blaKPC-carrying isolates was in the Mid-Atlantic region (68.4 to 52.5%) (P = 0.043; OR, 1.1; 95% CI, 0.972 to 3.917) (data not shown). E. coli and K. oxytoca isolates harboring blaKPC were detected in small numbers in all 3 years (two to four isolates each per year) and did not have a significant impact on the differences observed.
A significant increase in the percentage of isolates carrying blaCTX-M-14-like genes did occur from 2012 (9.1%) to 2014 (12.9%) (P = 0.042; OR, 0.700; 95% CI, 0.499 to 0.982) (Fig. 1), although higher rates of this family of genes was noted in 2013 (14.9%). blaCTX-M-14-like genes were most prevalent among E. coli isolates, and an increase in more recent years (25.6 and 20.4% in 2013 and 2014) was noted compared to 2012 (16.0%); although the difference between 2012 and 2014 was not statistically significant (P = 0.081; OR, 0.744; 95% CI, 0.503 to 1.099), higher rates were observed in 2013 (P = 0.001; OR, 0.551; 95% CI, 0.374 to 0.811) than in 2012. E. coli isolates carrying blaCTX-M-14-like genes were also increasingly observed in four of the nine census regions—New England (13.0 to 23.1%), Mid-Atlantic (16.4 to 31.0%), East North Central (10.0 to 14.9%), and East South Central (15.0 to 27.0%)—but lower rates were noted in the West North Central region (26.7 to 13.5% in 2012 and 2014, respectively).
Isolates harboring blaCMY-2-like genes increased from 2012 (8.2%) to 2014 (11.9%) (P = 0.042; OR, 0.685; 95% CI, 0.482 to 0.974), but only 6.8% of all 2013 ESBL phenotype isolates carried the gene encoding the enzyme. Among the organisms carrying blaCMY-2-like genes, E. coli isolates displayed a modestly increased rate compared to the overall ESBL phenotype population: 14.4, 10.1, and 18.0% in 2012, 2013, and 2014, respectively (P = 0.115; OR, 0.763; 95% CI, 0.507 to 1.147) (Fig. 1), and these increased rates were observed in the New England (13.0 to 30.8%), East North Central (6.0 to 12.8%), West North Central (26.7 to 35.1%), South Atlantic (8.1 to 31.0%), and East South Central (5.0 to 10.8%) regions.
Interestingly, an important decrease in the percentage of isolates carrying blaSHV ESBL-producing organisms from 2012 (24.9%) to 2014 (12.7%) (P < 0.001; OR, 2.274; 95% CI, 1.723 to 3.000) was discovered, and it was caused by a decline in blaSHV ESBL occurrence among K. pneumoniae isolates from 51.3% in 2012 to 29.2% in 2014 (P < 0.001; OR, 2.557; 95% CI, 1.801 to 3.629). It is noteworthy that a decrease in the percentage of blaSHV ESBL enzymes was noted in seven of the census regions—New England (53.8 and 20.0% in 2012 and 2014, respectively), Mid-Atlantic (50.6 and 33.9%), East North Central (46.7 and 22.5%), East South Central (50.0 and 23.1%), West South Central (42.4 and 37.5%), Mountain (55.6 and 11.1%), and Pacific (69.6 and 25.9%)—while rates in the remaining regions were stable.
Differences in the relative frequencies of other β-lactamase-encoding genes were also observed in the study period. However, the number of positive isolates was small, and statistical analysis could not be performed.
Antimicrobial activities of ceftazidime-avibactam and comparator agents.
Ceftazidime-avibactam was very active when tested against the 2,129 ESBL phenotype isolates (MIC50/90, 0.12/1 μg/ml) (Table 2) collected in U.S. hospitals during 2012 to 2014, and the compound inhibited nearly all (99.7%) of the isolates, applying the U.S. FDA breakpoints (15). Ceftazidime alone, ceftriaxone, and aztreonam inhibited only 30.2, 9.7, and 22.4% of the isolates, respectively, under the current susceptibility breakpoint criteria (12). The activity of piperacillin-tazobactam against the isolates was also limited, and the combination was active against 59.6% of the isolates using the CLSI breakpoints (12) and 49.2% applying the EUCAST interpretative criteria (16). Meropenem inhibited 84.2 and 85.3% of the isolates under the current susceptibility breakpoint criteria for CLSI and EUCAST, respectively (12, 16). Among other antimicrobial classes, tigecycline and colistin were active against 98.8 and 89.9% of the isolates using the U.S. FDA (12) and EUCAST (16) breakpoints, respectively (Table 2).
TABLE 2.
Activities of ceftazidime-avibactam and comparator antimicrobial agents tested against 2,129 ESBL phenotype-positive Enterobacteriaceae isolates and subgroups collected in 63 U.S. hospitals (2012 to 2014)
| Organism group (no. tested) and antimicrobial agent | MIC (μg/ml) |
CLSIa (%) |
EUCASTa (%) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 50% | 90% | Range | S | I | R | S | I | R | |
| ESBL phenotype (2,129)b | |||||||||
| Ceftazidime-avibactam | 0.12 | 1 | ≤0.015 to >32 | 99.7 | 0.3c | ||||
| Ceftazidime | 16 | >32 | 0.03 to >32 | 30.2 | 10.7 | 59.1 | 13.3 | 16.9 | 69.8 |
| Ceftriaxone | >8 | >8 | ≤0.06 to >8 | 9.7 | 2.4 | 87.8 | 9.7 | 2.4 | 87.8 |
| Aztreonam | >16 | >16 | ≤0.12 to >16 | 22.4 | 7.9 | 69.8 | 11.5 | 10.9 | 77.6 |
| Meropenem | ≤0.06 | >8 | ≤0.06 to >8 | 84.2 | 1.1 | 14.7 | 85.3 | 4.6 | 10.1 |
| Piperacillin-tazobactam | 16 | >64 | ≤0.5 to >64 | 59.6 | 9.4 | 31.1 | 49.2 | 10.4 | 40.4 |
| Gentamicin | 2 | >8 | ≤1 to >8 | 58.8 | 4.9 | 36.3 | 55.3 | 3.5 | 41.2 |
| Levofloxacin | >4 | >4 | ≤0.12 to >4 | 29.0 | 3.5 | 67.5 | 27.1 | 1.9 | 71.0 |
| Colistin | 0.5 | 4 | 0.12 to >8 | 89.9 | 10.1 | ||||
| Tigecycline | 0.25 | 1 | 0.03 to 8 | 98.8 | 1.2 | <0.1d | 94.1 | 4.7 | 1.2 |
| CRE (326)e | |||||||||
| Ceftazidime-avibactam | 0.5 | 2 | ≤0.015 to >32 | 98.5 | 1.5c | ||||
| Ceftazidime | >32 | >32 | 4 to >32 | 0.9 | 1.5 | 97.5 | 0.0 | 0.9 | 99.1 |
| Ceftriaxone | >8 | >8 | 4 to >8 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 100.0 |
| Aztreonam | >16 | >16 | 4 to >16 | 0.3 | 0.9 | 98.8 | 0.0 | 0.3 | 99.7 |
| Meropenem | >8 | >8 | 1 to >8 | 0.9 | 3.1 | 96.0 | 4.0 | 29.8 | 66.3 |
| Piperacillin-tazobactam | >64 | >64 | 16 to >64 | 0.3 | 1.2 | 98.5 | 0.0 | 0.3 | 99.7 |
| Gentamicin | 8 | >8 | ≤1 to >8 | 48.2 | 13.8 | 38.0 | 38.7 | 9.5 | 51.8 |
| Levofloxacin | >4 | >4 | ≤0.12 to >4 | 13.5 | 1.8 | 84.7 | 11.3 | 2.1 | 86.5 |
| Colistin | 0.5 | 8 | 0.25 to >8 | 83.9 | 16.1 | ||||
| Tigecycline | 0.5 | 1 | 0.06 to 8 | 98.8 | 0.9 | 0.3d | 93.8 | 4.9 | 1.2 |
| Isolates carrying blaCTX-M (1,120)f | |||||||||
| Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015 to 2 | 100.0 | 0.0c | ||||
| Ceftazidime | 16 | >32 | 0.06 to >32 | 28.1 | 13.9 | 57.9 | 10.4 | 17.8 | 71.9 |
| Ceftriaxone | >8 | >8 | 4 to >8 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 100.0 |
| Aztreonam | >16 | >16 | ≤0.12 to >16 | 14.0 | 8.8 | 77.1 | 3.7 | 10.4 | 86.0 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06 to 8 | 98.2 | 0.7 | 1.1 | 98.9 | 1.1 | 0.0 |
| Piperacillin-tazobactam | 8 | 64 | ≤0.5 to >64 | 77.9 | 12.2 | 9.8 | 62.9 | 15.0 | 22.1 |
| Gentamicin | 2 | >8 | ≤1 to >8 | 52.6 | 1.3 | 46.1 | 51.3 | 1.3 | 47.4 |
| Levofloxacin | >4 | >4 | ≤0.12 to >4 | 13.9 | 4.1 | 82.0 | 12.8 | 1.2 | 86.1 |
| Colistin | 0.5 | 1 | 0.12 to >8 | 94.8 | 5.2 | ||||
| Tigecycline | 0.12 | 0.5 | 0.03 to 4 | 98.9 | 1.1 | 0.0d | 95.8 | 3.1 | 1.1 |
| Isolates carrying blaKPC (304)g | |||||||||
| Ceftazidime-avibactam | 0.5 | 2 | ≤0.015 to >32 | 99.7 | 0.3c | ||||
| Ceftazidime | >32 | >32 | 4 to >32 | 1.0 | 1.6 | 97.4 | 0.0 | 1.0 | 99.0 |
| Ceftriaxone | >8 | >8 | 8 to >8 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 100.0 |
| Aztreonam | >16 | >16 | 8 to >16 | 0.0 | 0.3 | 99.7 | 0.0 | 0.0 | 100.0 |
| Meropenem | >64 | >64 | 64 to >64 | 0.0 | 1.0 | 99.0 | 0.0 | 0.0 | 100.0 |
| Piperacillin-tazobactam | >8 | >8 | 1 to >8 | 1.3 | 4.3 | 94.4 | 5.6 | 26.3 | 68.1 |
| Gentamicin | 8 | >8 | ≤1 to >8 | 48.0 | 14.5 | 37.5 | 38.2 | 9.9 | 52.0 |
| Levofloxacin | >4 | >4 | ≤0.12 to >4 | 12.5 | 1.6 | 85.9 | 9.9 | 2.6 | 87.5 |
| Colistin | 0.5 | 8 | 0.25 to >8 | 84.1 | 15.9 | ||||
| Tigecycline | 0.5 | 1 | 0.06 to 8 | 98.7 | 1.0 | 0.3d | 94.1 | 4.6 | 1.3 |
Criteria as published by CLSI (11, 12) and EUCAST (16). S, susceptible; I, intermediate; R, resistant.
ESBL phenotype isolates included E. coli (1,048), K. oxytoca (135), K. pneumoniae (843), and P. mirabilis (103).
Breakpoints from U.S. FDA package insert.
Breakpoints from U.S. FDA package insert (revised December 2014).
CRE isolates included E. coli (n = 15), K. oxytoca (n = 7), and K. pneumoniae (n = 304).
Isolates carrying blaCTX-M included isolates of the following species harboring blaCTX-M-14-like and/or blaCTX-M-15-like genes: E. coli (n = 780), K. oxytoca (n = 5), K. pneumoniae (n = 290), and P. mirabilis (n = 45).
Isolates carrying blaKPC included E. coli (n = 11), K. oxytoca (n = 8), and K. pneumoniae (n = 285).
CRE strains had elevated MIC values for most comparator agents, although ceftazidime-avibactam (MIC50/90, 0.5/2 μg/ml; 98.5% susceptible), tigecycline (MIC50/90, 0.5/1; 98.8% susceptible) (U.S. FDA breakpoint) and colistin (MIC50/90, 0.5/8; 83.9% susceptible) (EUCAST breakpoint) were relatively unaffected.
Isolates harboring blaCTX-M-15-like and/or blaCTX-M-14-like genes without carbapenemases displayed elevated MIC values for cephalosporins (0.0 to 28.1% susceptible using CLSI criteria) (Table 2) and aztreonam (14.0% susceptible); however, ceftazidime-avibactam (MIC50/90, 0.12/0.5 μg/ml; 100.0% susceptible) inhibited all of the isolates at the current U.S. FDA-established breakpoint. Meropenem, tigecycline, and colistin were also active against these strains (98.2, 98.9, and 94.8% susceptible, respectively).
Similar to the CRE analysis, isolates carrying blaKPC were very resistant to most agents tested, including non-β-lactam agents, such as gentamicin and levofloxacin (only 48.0 and 12.5% susceptible, respectively, using the CLSI criteria) (Table 2). Ceftazidime-avibactam (MIC50/90, 0.5/2 μg/ml) and tigecycline (MIC50/90, 0.5/1 μg/ml) were the most active in vitro agents against these isolates, inhibiting 99.7 and 98.7% of the isolates at the U.S. FDA breakpoints. Colistin (MIC50/90, 0.5/8 μg/ml) inhibited 84.1% of the isolates carrying blaKPC at the EUCAST susceptible breakpoint of ≤2 μg/ml.
All but five isolates displayed ceftazidime-avibactam MIC values of ≤4 μg/ml. The five were K. pneumoniae isolates (ceftazidime-avibactam MIC values of >32 μg/ml) and produced metallo-β-lactamase genes. Four isolates from Colorado (two isolates), New York, and California carried blaNDM-1, and one isolate harboring blaKPC-2 and blaVIM-4 was from New York.
DISCUSSION
Important and significant differences were detected across the 3 years of surveillance in U.S. hospitals, with the most notable changes among E. coli and K. pneumoniae isolates. E. coli isolates displayed an overall increase of ESBL phenotype rates over the study period but a reduced rate of isolates carrying blaCTX-M-15-like genes that was very common among these species during the two initial years surveyed (3, 4). Conversely, the ESBL phenotype rates decreased among K. pneumoniae isolates, mainly due to a reduction of isolates harboring blaSHV ESBL and a decrease in CR K. pneumoniae and isolates carrying blaKPC in 2014 compared to 2012. Furthermore, there was an increase of K. pneumoniae isolates carrying blaCTX-M-15-like genes that should be closely monitored, since these isolates could be replacing the organisms carrying blaSHV that usually display rates of susceptibility to other antimicrobial classes higher than those of isolates harboring blaCTX-M (3).
The reduced occurrences of CR K. pneumoniae and isolates harboring blaKPC were noticed in five hospitals where the prevalence of these genes had been elevated in prior monitored study years to epidemic/endemic levels (data not shown). Nationwide CRE prevention programs carried out in Israeli hospitals reported a reduction in the CRE rates from 55.5 carriers to 11.7 carriers per 100,000 patient days in 1 year (17). The measures in Israeli prevention programs involved routine CRE screening, patient and staff cohorting with isolation of carriers and dedicated nursing staff reducing contact with noncarriers, effective communication among medical and laboratory staff, and active surveillance in acute and long-term-care facilities (17), procedures that are also recommended by the CDC (18).
Despite the recent report of blaKPC and blaSHV variants genetically engineered to contain mutations in important motifs that affect avibactam inhibition (19–21) and a recent report of isolates harboring blaKPC with an elevated ceftazidime-avibactam MIC value (22), clinical isolates carrying blaKPC and other β-lactamases collected in 63 U.S. hospitals monitored over three consecutive recent years were predominantly susceptible to ceftazidime-avibactam. The compound displayed broad activity against isolates producing the commonly observed β-lactamases that are prevalent in the United States, including isolates carrying blaKPC, and is still a valuable therapy option to treat infections caused by these organisms that are usually resistant to multiple antimicrobial classes.
Avibactam is not active against metallo-β-lactamase-producing isolates, which are still very uncommon yet increasingly reported in U.S. hospitals. Aztreonam-avibactam has demonstrated activity against these isolates (23–26), since the metalloenzymes do not hydrolyze monobactams and avibactam would inhibit other β-lactamases present in the isolate; however, further in vivo and in vitro investigations of this new combination are warranted.
In summary, a robust 3-year analysis of the frequencies of ESBL- and carbapenemase-producing Enterobacteriaceae isolates demonstrated that important changes are occurring in U.S. medical centers.
ACKNOWLEDGMENTS
We thank all participants of the INFORM program for providing bacterial isolates.
This study was supported by Allergan. Allergan had no involvement in the collection, analysis, and interpretation of data.
Allergan was involved in the design and decision to present these results, and JMI Laboratories received compensation fees for services in relation to preparing the manuscript. JMI Laboratories, Inc., has also received research and educational grants in 2014 and 2015 from Achaogen, Actavis, Actelion, American Proficiency Institute (API), AmpliPhi, Anacor, Astellas, AstraZeneca, Basilea, Bayer, BD, Cardeas, Cellceutix, CEM-102 Pharmaceuticals, Cempra, Cerexa, Cidara, Cormedix, Cubist, Debiopharm, Dipexium, Dong Wha, Durata, Enteris, Exela, Forest Research Institute, Furiex, Genentech, GSK, Helperby, ICPD, Janssen, Lannett, Longitude, Medpace, Meiji Seika Kasha, Melinta, Merck, Motif, Nabriva, Novartis, Paratek, Pfizer, Pocared, PTC Therapeutics, Rempex, Roche, Salvat, Scynexis, Seachaid, Shionogi, Tetraphase, The Medicines Co., Theravance, ThermoFisher, VenatoRX, Vertex, Wockhardt, Zavante, and some other corporations. Some JMI employees are advisors/consultants for Allergan, Astellas, Cubist, Pfizer, Cempra, and Theravance. We have no speaker's bureaus or stock options to declare.
Funding Statement
This study was supported by Allergan. Allergan was involved in the design and decision to present these results and JMI Laboratories received compensation fees for services in relation to preparing the manuscript. Allergan had no involvement in the collection, analysis, and interpretation of data.
REFERENCES
- 1.Ramphal R, Ambrose PG. 2006. Extended-spectrum beta-lactamases and clinical outcomes: current data. Clin Infect Dis 42(Suppl 4):S164–S172. doi: 10.1086/500663. [DOI] [PubMed] [Google Scholar]
- 2.Bush K. 2008. Extended-spectrum beta-lactamases in North America, 1987-2006. Clin Microbiol Infect 14(Suppl 1):S134–S143. [DOI] [PubMed] [Google Scholar]
- 3.Castanheira M, Farrell SE, Krause KM, Jones RN, Sader HS. 2014. Contemporary diversity of β-lactamases among Enterobacteriaceae in the nine United States census regions and ceftazidime-avibactam activity tested against isolates producing the most prevalent β-lactamase groups. Antimicrob Agents Chemother 58:833–838. doi: 10.1128/AAC.01896-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castanheira M, Mills JC, Costello SE, Jones RN, Sader HS. 2015. Ceftazidime-avibactam activity tested against Enterobacteriaceae from United States hospitals (2011-2013) and characterization of beta-lactamase producing strains. Antimicrob Agents Chemother 59:3509–3517. doi: 10.1128/AAC.00163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canton R, Coque TM. 2006. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol 9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Castanheira M, Mendes RE, Rhomberg PR, Jones RN. 2008. Rapid emergence of blaCTX-M among Enterobacteriaceae in U.S. medical centers: molecular evaluation from the MYSTIC Program (2007). Microb Drug Resist 14:211–216. [DOI] [PubMed] [Google Scholar]
- 7.Lewis JS II, Herrera M, Wickes B, Patterson JE, Jorgensen JH. 2007. First report of the emergence of CTX-M-type extended-spectrum beta-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob Agents Chemother 51:4015–4021. doi: 10.1128/AAC.00576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castanheira M, Farrell SE, Deshpande LM, Mendes RE, Jones RN. 2013. Prevalence of β-lactamase encoding genes among Enterobacteriaceae bacteremia isolates collected in 26 US hospitals: report from the SENTRY Antimicrobial Surveillance Program (2010). Antimicrob Agents Chemother 57:3012–3020. doi: 10.1128/AAC.02252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CDC. 2015. Tracking CRE infections. CDC, Atlanta, GA: http://www.cdc.gov/hai/organisms/cre/TrackingCRE.html Accessed December 2015. [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2015. M07-A10. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2015. M100-S25. Performance standards for antimicrobial susceptibility testing: 25th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Kaiser RM, Castanheira M, Jones RN, Tenover F, Lynfield R. 2013. Trends in Klebsiella pneumoniae carbapenemase-positive K. pneumoniae in US hospitals: Report from the 2007-2009 SENTRY Antimicrobial Surveillance Program. Diagn Microbiol Infect Dis 76:356–360. doi: 10.1016/j.diagmicrobio.2013.03.032. [DOI] [PubMed] [Google Scholar]
- 14.CDC. 2015. FAQs about choosing and implementing a CRE definition. CDC, Atlanta, GA: http://www.cdc.gov/hai/organisms/cre/definition.html Accessed December 2015. [Google Scholar]
- 15.Allergan. 2015. AVYCAZ prescribing information. Allergan, Irvine, CA: http://www.avycaz.com Accessed 23 March 2015. [Google Scholar]
- 16.EUCAST. January 2015. Breakpoint tables for interpretation of MICs and zone diameters, version 5.0. http://www.eucast.org/clinical_breakpoints/ Accessed January 2015.
- 17.Schwaber MJ, Carmeli Y. 2014. An ongoing national intervention to contain the spread of carbapenem-resistant enterobacteriaceae. Clin Infect Dis 58:697–703. doi: 10.1093/cid/cit795. [DOI] [PubMed] [Google Scholar]
- 18.CDC. 2013. 2012 CRE toolkit—guidance for control of carbapenem-resistant Enterobacteriaceae (CRE). CDC, Atlanta, GA: http://www.cdc.gov/hai/organisms/cre/cre-toolkit/f-level-prevention.html#facility-measures Accessed December 2015. [Google Scholar]
- 19.Winkler ML, Papp-Wallace KM, Bonomo RA. 2015. Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV beta-lactamases with single amino acid substitutions in the Omega-loop. J Antimicrob Chemother 70:2279–2286. doi: 10.1093/jac/dkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkler ML, Papp-Wallace KM, Taracila MA, Bonomo RA. 2015. Avibactam and inhibitor-resistant SHV beta-lactamases. Antimicrob Agents Chemother 59:3700–3709. doi: 10.1128/AAC.04405-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papp-Wallace KM, Winkler ML, Taracila MA, Bonomo RA. 2015. Variants of the KPC-2 beta-lactamase which are resistant to inhibition by avibactam. Antimicrob Agents Chemother 59:3710–3717. doi: 10.1128/AAC.04406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, Gregson A. 2015. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae Isolate. Antimicrob Agents Chemother 59:6605–6607. doi: 10.1128/AAC.01165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupont H, Gaillot O, Goetgheluck AS, Plassart C, Emond JP, Lecuru M, Gaillard N, Derdouri S, Lemaire B, Girard de Courtilles M, Cattoir V, Mammeri H. 2016. Molecular characterization of carbapenem-non-susceptible Enterobacterial isolates collected during a prospective interregional survey in France and susceptibility to the novel ceftazidime-avibactam and aztreonam-avibactam combinations. Antimicrob Agents Chemother 60:215–221. doi: 10.1128/AAC.01559-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasoo S, Cunningham SA, Cole NC, Kohner PC, Menon SR, Krause KM, Harris KA, De PP, Koh TH, Patel R. 2015. In vitro activities of ceftazidime-avibactam, aztreonam-avibactam, and a panel of older and contemporary antimicrobial agents against carbapenemase-producing gram-negative bacilli. Antimicrob Agents Chemother 59:7842–7846. doi: 10.1128/AAC.02019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biedenbach DJ, Kazmierczak K, Bouchillon SK, Sahm DF, Bradford PA. 2015. In vitro activity of aztreonam-avibactam against a global collection of Gram-negative pathogens from 2012 and 2013. Antimicrob Agents Chemother 59:4239–4248. doi: 10.1128/AAC.00206-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Estabrook M, Jacoby GA, Nichols WW, Testa RT, Bush K. 2015. In vitro susceptibility of characterized beta-lactamase-producing strains tested with avibactam combinations. Antimicrob Agents Chemother 59:1789–1793. doi: 10.1128/AAC.04191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]