Abstract
The increasing frequency of bacteria showing antimicrobial resistance (AMR) raises the menace of entering into a postantibiotic era. Horizontal gene transfer (HGT) is one of the prime reasons for AMR acquisition. Acinetobacter baumannii is a nosocomial pathogen with outstanding abilities to survive in the hospital environment and to acquire resistance determinants. Its capacity to incorporate exogenous DNA is a major source of AMR genes; however, few studies have addressed this subject. The transformation machinery as well as the factors that induce natural competence in A. baumannii are unknown. In this study, we demonstrate that naturally competent strain A118 increases its natural transformation frequency upon the addition of Ca2+or albumin. We show that comEA and pilQ are involved in this process since their expression levels are increased upon the addition of these compounds. An unspecific protein, like casein, does not reproduce this effect, showing that albumin's effect is specific. Our work describes the first specific inducers of natural competence in A. baumannii. Overall, our results suggest that the main protein in blood enhances HGT in A. baumannii, contributing to the increase of AMR in this threatening human pathogen.
INTRODUCTION
Acinetobacter baumannii has emerged as a severe nosocomial pathogen over the course of the last few decades, with high levels of morbidity and mortality associated with infections by this pathogen (1, 2). A. baumannii is considered to be a paradigm of multidrug resistance since it has developed resistance to almost all available antibiotics, leaving few or no treatment options left. The ability of A. baumannii to persist in the clinical setting even under desiccation and nutrient starvation, as well as its ability to accumulate several antibiotic resistance determinants, allowed its evolution as a successful pathogen in the hospital environment (3).
The large number of available A. baumannii genomes (n = 1,289) shows that foreign DNA is acquired at high frequencies (4–7).
The transformation process has been well described for some species, such as Streptococcus pneumoniae, Vibrio cholerae, Neisseria meningitidis, and Helicobacter pylori (8–12). However, how natural competence is regulated has not been thoroughly studied, particularly for Gram-negative bacteria. Some well-characterized competence inducers are DNA damage in H. pylori (13), starvation, as was suggested for Haemophilus influenzae, and chitin metabolism in Vibrio cholerae (14, 15). In most known examples, natural competence is a transitory state that is regulated by different internal and external signals. Often, these regulation networks are not completely understood. For Acinetobacter spp., most of the studies focusing on this issue were performed by using Acinetobacter baylyi strain ADP1 (16–20), a bacterium not as threatening to human health as A. baumannii (18–23). Data regarding A. baumannii and competence inducers are scarce (24–27). Wilharm et al. showed previously that several A. baumannii clinical isolates were able to acquire exogenous DNA depending on the expression of pilT, encoding an ATPase involved in type IV pilus retraction, and comEC, encoding the DNA uptake channel (26). Another study showed that the natural competence of Acinetobacter nosocomialis strain M2 relies on the presence of pilA, pilD, and pilT (28).
We recently demonstrated that A. baumannii strain A118, which was recovered from a patient's blood sample and was shown to be susceptible to several antibiotics, has the capacity to acquire exogenous DNA (24, 25).
In the present study, we focused on identifying inducers that enhance or trigger natural competence in A. baumannii. Careful comparison of media led us to identify two specific signals, bovine serum albumin (BSA) and Ca2+, as transformation inducers. These compounds increased the natural competence frequency while inducing comEA and pilQ expression. Albumin induction was protein specific, since human serum also exhibited an effect on natural transformation, and other proteins, such as casein, did not enhance the rate of DNA acquisition.
Our results show that albumin, the main blood protein, and Ca2+ are specific inducers of natural competence in A. baumannii, a mechanism that could be implicated in the increasing frequency of emergence of antimicrobial resistance (AMR) in this threatening pathogen.
MATERIALS AND METHODS
Strains, media, and plasmids.
Acinetobacter baumannii A118, ATCC 17978, and A42 and Escherichia coli TOP10 cells harboring pDsRedAK were cultured on Luria-Bertani (LB) broth and LB agar at 37°C. When appropriate, antibiotics (12.5 μg/ml kanamycin and/or 6 μg/ml amikacin) were added to the bacterial cultures. Total genomic DNA (gDNA) from A. baumannii Ab144, Ab155, and A118 was obtained by using a Wizard genomic DNA purification kit according to the manufacturer's instructions (Promega, Madison, WI). Plasmid DNA (pDsRedAK) was extracted from E. coli TOP10 cells by using a Highway ADN Puriprep-P kit (Inbio Highway).
Natural transformation assays.
Standard natural transformation assays were performed as previously described (24). Briefly, 50 μl of late-stationary-death-phase cultures of A. baumannii A118 was transferred to 50 μl of sterile LB medium with 100 ng of pDsRed plasmid DNA and/or gDNA. These cultures were incubated for 1 h at 37°C and then plated onto LB agar with 12.5 μg/ml kanamycin. Transformation events were scored by counting Kanr colonies, while total CFU were assessed by plating serial dilutions onto LB agar (see Fig. S1 in the supplemental material). Transformation events were confirmed by PCR targeting aac(6′)-Ib, aac(3)-IIa, and aph(3′)-Ia as well as by measuring the level of resistance to aminoglycosides (MICs) and macroscopic observation of red-pigmented colonies. MICs were determined by the gradient diffusion method (Etest method) (29) with commercial strips (bioMérieux) according to procedures recommended by the supplier. An average of 15 colonies (transformants) were checked in every independent transformation experiment.
In order to test different conditions, assays were carried out as described above while altering each physical parameter one at a time. The temperature was switched to 25°C, 30°C, 35°C, 37°C, or 42°C. The pH varied from 2 to 14. The effect of osmolarity was tested by changing the concentration of sodium chloride. Different media were assayed, including brucella broth, lactose broth, complete transformation medium (CTM), minimal medium M9 (20% [wt/vol] α-d-glucose and 20% [wt/vol] sucrose), Terrific broth, and human serum. Individual effects of BSA (0.2%), human serum albumin (HSA) (0.2%), casein (0.2%), and CaCl2 (1 mM) were also tested (Sigma-Aldrich). Finally, the concentration of transforming DNA was studied by using total genomic DNA of A. baumannii strains Ab155KanR and Ab144KanR. All experiments were performed in triplicate, and statistical analysis was performed. Transformation events were scored as mentioned above.
Kinetics of natural transformation.
To determine the kinetics of natural transformation, 50 μl of the late-stationary-phase culture was transferred to 50 μl of sterile LB medium with 100 ng of pDsRed plasmid DNA. The transformation experiments were done at various time points (0 to 180 min) and scored by plating onto selective agar. Termination of transformation was done by adding 0.0017 U of DNase I (Promega, Madison, WI) to the mixture of transforming DNA and the bacterial inoculum.
Denaturation and digestion of BSA and casein.
To denature BSA and casein, the proteins were exposed to heat treatment at 100°C for 1 min. BSA and casein digestions were done with trypsin (2 μg) as previously described (30).
RNA procedures and transcriptional analysis.
Strain A118 was grown in LB broth, LB broth with 1 mM CaCl2, LB broth with 0.2% BSA, LB broth with 1 mM CaCl2 and 0.2% BSA, and CTM for 24 h at 37°C under agitation. Cell pellets were immediately lysed by using a solution containing 0.3 M sodium acetate (pH 4), 30 mM EDTA, and 3% SDS (wt/vol) and incubated for 3 min at 100°C. RNAs were extracted according to protocols described previously by Ramirez et al. (31). The integrity of the RNA samples was checked by agarose electrophoresis. RNA concentrations were determined by spectrophotometry. Reverse transcription was carried out by using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI) according to the manufacturer's instructions. Specific oligonucleotides were designed for retrotranscription coupled to quantitative PCR (RT-qPCR) with Primer3 (see Table S1 in the supplemental material). Samples containing no reverse transcriptase or template RNA were included as negative controls to ensure that RNA samples were free of DNA contamination. The 16S rRNA gene was used as an internal control for relative quantification. The RT-qPCRs were completed by using 5× Hot FIREPol EvaGreen qPCR Mix Plus (Solis Biodyne, Tartu, Estonia) with a two-step reaction protocol consisting of 40 cycles of 94°C for 15 s and 52°C for 20 s, followed by a dissociation phase for quality control. The 20-μl qPCR mixtures contained 0.2 μM specific primers and 2 μl of cDNA (10 ng/μl).
The comEA and pilQ transcript levels of each sample were normalized to the 16S rRNA transcript levels for each cDNA sample. Each cDNA sample was run in triplicate, and experiments were repeated with at least three independent sets of samples. The relative quantification of gene expression was performed by using the 2−ΔΔCT comparative threshold method. The ratios obtained after normalization were expressed as fold changes compared to values for cDNA samples isolated from bacterial cultures on LB medium.
Comparative analyses of BSA and HSA.
The amino acid sequence comparison was performed by using BLAST (V2.0) and ClustalX. Moreover, the structure comparison between BSA and HSA was performed by using the jFATCAT-rigid (java flexible structure alignment by chaining aligned fragment pairs allowing twists) method using UCSF Chimera software.
Bioinformatic analyses of presumptive competence genes.
Forty-two genome sequences of A. baumannii and 8 reference genomes of other Acinetobacter species (Acinetobacter sp. strain ADP1, A. baylyi, A. calcoaceticus, A. haemolyticus, A. johnsonii, A. junii, A. lwoffii, and A. nosocomialis) were used to generate an amino acid similarity matrix for the 16 presumptive natural competence genes (comEA, pilQ, pilD, pilF, pilP, pilA, comEC, pilW, pilH, pilO, pilN, pilY1 [comC], pilE, pilR, pilM, and pilT) previously identified in the A118 strain (25). Comparison and analyses of these genes were performed by blastp and represented by using a heat map. The heat map was done by using R project software (version 3.0.2).
Statistical methods.
Data were expressed as means ± standard deviations (SD). The Student t test was used to analyze differences between two groups, and one-way analysis of variance with Bonferroni correction was used to study differences between three or more groups. Statistical analyses were performed by using R project software (version 3.0.2).
RESULTS
Natural transformation is optimal at blood pH but is not affected by temperature or osmolarity.
Abundant evidence from the literature points out that physical environment factors, such as temperature, pH, and osmolarity, impact natural competence in different bacterial species (11, 17, 32, 33). Therefore, we inquired about the effect of different physical conditions on the transformation frequency of the A118 strain. Natural transformation assays with A118 at 26°C, 30°C, 37°C, and 42°C showed no significant differences, indicating that temperature does not influence transformation in A. baumannii A118 (see Fig. S2A in the supplemental material).
Variations of the NaCl concentration (0, 0.25, 0.5, 0.75, 1.0, and 1.25%) showed no significant differences in transformation frequencies, suggesting that osmotic stress does not affect transformation (see Fig. S2B in the supplemental material).
We next investigated the effect of pH on natural competence by measuring bacterial survival and the natural transformation frequency of A. baumannii A118 at a range of pHs from 2 to 14. The bacterium survived at a pH range of 5.5 to 7.5, while natural transformation was optimal at pH 7.5 and occurred at a lower frequency at pH 5.5 (see Fig. S2C in the supplemental material). These results show that although growth can be sustained at a pH interval ranging from 5.5 to 7.5, the pH for optimal transformation frequency corresponds to the highest pH supporting bacterial culture. Interestingly, this value coincides with the pH found in human blood (pH 7.35 to 7.45).
Transformation in A. baumannii A118 occurs readily and is independent of the amount of DNA.
For some species, such A. calcoaceticus and S. pneumoniae, it has been shown that transformation follows saturation kinetics (17, 18, 34). For E. coli, Baur et al. obtained higher transformation frequencies with increasing amounts of DNA (35).
To address this point, transformation assays were performed by adding plasmid DNA (pDsRed) at various concentrations ranging from 5 ng/μl to 100 ng/μl (Fig. 1). Meanwhile, we also tested transformation using increasing quantities of gDNA in A118. For this purpose, we carried out experiments by adding gDNAs of two different A. baumannii clinical strains (A144KanR and A155KanR) that contain aminoglycoside resistance genes [aac(6′)-Ib, aac(3)-IIa (aacC2), and aph(3′)-Ia] within their genomes (GenBank accession numbers JQSF01000000 and JXSV01000000, respectively). The acquisition of A155 and A144 gDNAs was confirmed phenotypically by measuring the level of resistance to aminoglycosides and by PCRs using specific primers that amplified the aac(3)-IIa and/or aph(3′)-Ia gene.
FIG 1.
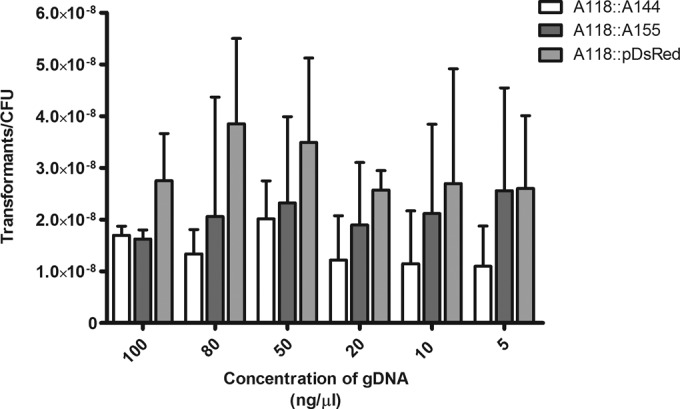
Natural transformation frequencies using different concentrations of genomic and plasmid DNAs. The transformation assay was performed with gDNAs of two A. baumannii strains (A144KanR and A155KanR) and plasmid pDsRed. The cultures were then plated onto selective media, and the number of CFU per plate was determined. Three independent experiments were performed. Data are presented as the means, and error bars represent the SD (n = 3).
No significant differences were detected in the transformation frequency independently of the concentration or the source of the DNA (Fig. 1). The same amount of DNA resulted in a similar transformation frequency when plasmid DNA or gDNA was used. However, in the latter case, the generation of resistant clones required a double-recombination event for the integration of the resistance marker into the A118 chromosome. This suggests that the entry of DNA into the bacterial cytoplasm is the limiting step, rather than further recombination steps.
We next studied natural transformation kinetics by monitoring this process over time (see Fig. S3 in the supplemental material). Surprisingly, we already obtained transformant colonies after 30 s. The frequency of transformation was then constant over further time points (see Fig. S3 in the supplemental material).
Identification of Ca2+ and BSA as competence inducers for natural transformation through comparison of media.
Data from previous studies support the notion that specific chemical compounds influence natural competence in various species (17, 35–39). A well-studied example is the case of chitin in the Gram-negative species V. cholerae (40, 41).
Information regarding the Acinetobacter genus is scarce, as A. baylyi ADP1 is the most studied strain (16–19). Most of the data obtained by using this strain support the hypothesis that an upshift of nutrients could have a role in competence induction (18, 42). Moreover, in 1993, Palmen et al. proved that transformation in A. calcoaceticus BD413 (also know as A. baylyi ADP1) was Mg2+, Mn2+, and Ca2+ dependent, not observing a dependency on the growth medium and pH (18). Previously reported conclusions and data proving natural competence for A. baylyi ADP1 have often been applied to the entire genus. However, putative key inducer chemical compounds remain to be identified.
In this study, we challenged the natural transformation of A118 with six different media: brucella broth, lactose broth, CTM, minimal medium M9 supplemented with 20% (wt/vol) α-d-glucose, minimal medium M9 supplemented with 20% (wt/vol) sucrose, and Terrific broth. Transformation assays using the above-mentioned media were performed in parallel with assays using LB medium. Among these media, CTM showed a significant increase in the transformation frequency compared to that with LB medium (Table 1). The differential components in CTM compared to LB medium are 1 mM CaCl2 and 0.2% BSA (Fig. 2). Therefore, medium composition impacts the transformation frequency.
TABLE 1.
Natural transformation frequencies of A. baumannii strain A118 challenged with different culture mediaa
| Medium | Mean transformation frequency ± SD | Mean CFU/ml ± SD |
|---|---|---|
| LB | 4.81 × 10−8 ± 4.67 × 10−8 | 2.79 × 105 ± 2.10 × 105 |
| Brucella broth | 1.17 × 10−8 ± 4.55 × 10−9 | 1.93 × 106 ± 1.40 × 106 |
| Lactose broth | 2.82 × 10−9 ± 2.87 × 10−9 | 3.02 × 106 ± 1.47 × 106 |
| Terrific broth | 2.00 × 10−8 ± 5.61 × 10−8 | 3.00 × 106 ± 1.03 × 106 |
| M9 broth supplemented with 20% (wt/vol) α-d-glucose | 9.25 × 10−8 ± 9.47 × 10−8 | 1.33 × 105 ± 1.20 × 105 |
| M9 broth supplemented with 20% (wt/vol) sucrose | 4.94 × 10−8 ± 2,84 × 10−8 | 1.58 × 105 ± 2.04 × 105 |
| CTM | 4.67 × 10−7 ± 4.46 × 10−7 | 5.26 × 105 ± 3.7 × 102 |
n = 3 biological replicates.
FIG 2.

Natural transformation frequency with LB broth, LB broth plus BSA, LB broth plus CaCl2, LB broth plus BSA and CaCl2, and CTM. Transformation assays were performed with LB broth with 0.2% (wt/vol) BSA, LB broth with 1 mM CaCl2, LB broth with 0.2% (wt/vol) BSA and 1 mM CaCl2, and CTM. The experimental control was transformed in LB broth. The cultures were then plated onto selective media, and the number of CFU per plate was determined. Data are presented as the means, and error bars represent the SD. Six independent experiments were performed. Asterisks indicate that the observed difference in transformability is statistically significant (*, P < 0.05 by the Student t test; n = 6).
To gain insight into the role of CaCl2 in this process, we performed natural transformation experiments by adding CaCl2 or EDTA, a Ca2+-chelating agent. On one hand, a significant CaCl2 addition enhanced the transformation frequency (Fig. 3 and Table 2). Meanwhile, EDTA treatment reduced transformation levels. These results demonstrate that Ca2+ ions are specifically involved in promoting competence in A. baumannii A118. Indeed, divalent cations seem to play a role in transformation in several bacterial systems (35, 36).
FIG 3.
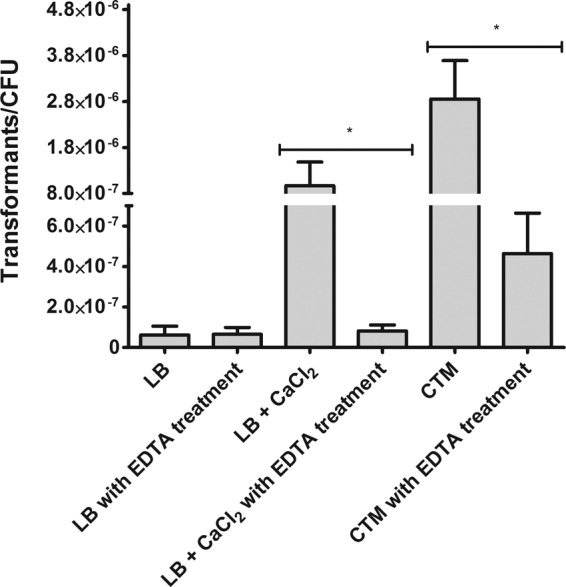
Transformability of A. baumannii cells under CaCl2 induction. Transformation assays were carried out by using different media: LB broth, LB broth plus CaCl2, and CTM. The cells were treated with EDTA to remove Ca2+. The cultures were then plated onto selective media, and the number of CFU per plate was determined. Data are presented as the means, and error bars represent the SD. Three independent experiments were performed. Asterisks indicate that the observed difference in transformability is statistically significant (*, P < 0.05 by the Mann-Whitney test; n = 3).
TABLE 2.
Natural transformation frequencies of A. baumannii A118 in the presence or absence of CaCl2 in LB medium and CTM with or without EDTA treatmenta
| Treatment | Mean transformation frequency ± SD | Mean CFU/ml ± SD |
|---|---|---|
| LB | 1.522 × 10−7 ± 4.952 × 10−8 | 1.025 × 108 ± 9.874 × 107 |
| LB + EDTA | 6.560 × 10−8 ± 3.348 × 10−8 | 4.11 × 105 ± 2.08 × 105 |
| LB + CaCl2b | 5.620 × 10−7 ± 4.140 × 10−7 | 4.762 × 106 ± 4.471 × 106 |
| LB + CaCl2 + EDTAb | 2.146 × 10−7 ± 1.482 × 10−7 | 1.502 × 107 ± 1.354 × 107 |
| CTM | 1.433 × 10−6 ± 7.375 × 10−7 | 1.010 × 108 ± 5.050 × 107 |
| CTM + EDTA | 4.640 × 10−7 ± 1.999 × 10−7 | 1.861 × 108 ± 1.641 × 108 |
n = 5 biological replicates.
A total of 0.001 M CaCl2 was used.
Since BSA is the other differential component in CTM, we first carried out transformation experiments using LB medium supplemented with 0.2% BSA, at the same concentration as that in CTM, to determine if this protein directly induces competence (Fig. 2).
We observed an ∼2.5-fold increase in the frequency of transformation upon the addition of BSA. However, these levels are still below the ones observed with CTM (Fig. 4). Assays with the addition of CaCl2 and BSA to the same levels as those in CTM showed a frequency of transformation equal to that observed with CTM (Fig. 2). These two inducers showed added effects rather than synergistic or antagonistic modes of action, suggesting that they may act through different mechanisms or at different stages of the transformation process.
FIG 4.
Transformability of A. baumannii A118 cells under different conditions. (A) Transformation assays with the addition of 0.2% (wt/vol) casein, heat-denatured 0.2% (wt/vol) casein, trypsin-digested 0.2% (wt/vol) casein, and 0.2% (wt/vol) BSA to LB medium (*, P < 0.05 by the Mann-Whitney test; n = 3). (B) Comparison of natural transformation frequencies with LB broth, LB broth and 0.2% (wt/vol) BSA, LB broth and HSA, and human serum (*, P < 0.05 by the Mann-Whitney test; n = 3). (C) Transformation assays with denatured and digested 0.2% (wt/vol) BSA added to LB broth. Simultaneously transformation assays using LB broth were performed to allow comparison. We compared transformation with BSA treatment to transformation without treatment in LB broth (*, P < 0.05 by the Mann-Whitney test; n = 3). For all assays, cultures were plated onto selective media, and the number of CFU per plate was determined. Data are presented as the means, and error bars represent the SD. Three independent experiments were performed in each case. Asterisks indicate that the observed difference in transformability is statistically significant.
To determine if the increase in the frequency of transformation observed with BSA is specific to this protein or rather due to the mere consequence of a higher peptide concentration, we carried out transformation assays by adding 0.2% (wt/vol) casein to LB medium. Notably, casein did not induce natural competence, showing that the effect of BSA is specific (Fig. 4A and Table 3).
TABLE 3.
Natural transformation frequencies of A. baumannii A118 in the presence of casein, denatured and digested casein, and BSA in LB mediuma
| Treatment | Mean transformation frequency ± SD | Mean CFU/ml ± SD |
|---|---|---|
| LB | 1.146 × 10−7 ± 6.362 × 10−8 | 4.784 × 106 ± 2.168 × 106 |
| LB + BSAb | 3.849 × 10−7 ± 2.492 × 10−7 | 5.534 × 107 ± 5.433 × 107 |
| LB + caseinc | 1.030 × 10−7 ± 4.854 × 10−8 | 1.737 × 107 ± 4.105 × 106 |
| LB + denatured casein | 1.818 × 10−8 ± 2.553 × 10−8 | 2.944 × 107 ± 2.315 × 107 |
| LB + digested casein | 3.800 × 10−8 ± 1.384 × 10−8 | 1.087 × 108 ± 2.935 × 107 |
n = 3 biological replicates.
A total of 0.2% (wt/vol) BSA was used.
A total of 0.2% (wt/vol) casein was used.
To determine dosage dependence, we performed the experiment with a wide range of BSA concentrations (0.002 to 3.2%, wt/vol). The lowest concentration of BSA tested (0.002%) was already able to induce a significant increase in the transformation frequency, and a concentration of 0.2% was enough to reach maximal competence induction (see Fig. S4 in the supplemental material). Moreover, a posttest for linear trends showed a significant linear trend between the amount of BSA added and the transformation frequency, indicating dose dependence for the observed biological effect (P < 0.0001; slope, 0.1169; R2 = 0.4).
Since albumin is a main component of serum, we performed transformation assays by supplementing LB medium with human serum and compared the effect of this mixture with the effect of BSA. Human serum triggered an increase in natural transformation (Fig. 4B). Moreover, the addition of HSA independently to LB medium also showed a significant 12.65-fold increase in the transformation frequency. This result, as well as results with BSA and serum, strongly supports our above-described hypothesis, showing the role of albumin as an inducer of competence.
We next inquired how BSA could induce this physiological process. To test if its structure was necessary, BSA was heat denatured. To determine if the whole molecule is necessary for competence induction, we digested BSA with trypsin. The samples were then used for transformation assays. Assays in which cultures are treated with BSA or trypsin-digested BSA showed similar increases in their frequencies of natural transformation. Interestingly, competence levels were even higher when denatured BSA was employed than when BSA, LB plus BSA, or LB plus trypsin-digested BSA were employed (Fig. 4C and Table 4). Our results indicate that neither BSA's native full-length structure nor its tertiary structure is needed to enhance competence. We hypothesized that some of the peptides obtained after trypsin digestion maintain the capacity to prompt the effect and that a masked protein motif may serve as a specific enhancer of natural competence. Importantly, no variations in natural transformation were observed when denatured casein was used, showing that BSA possesses a specific motif capable of inducing natural competence (Fig. 4A). In the case of Streptococcus thermophilus, it was observed that competence is induced by casein-derived octapeptides (43). This was not observed with A. baumannii A118, suggesting a different regulatory mechanism and reinforcing the idea of the specificity exerted by albumins.
TABLE 4.
Natural transformation frequencies of A. baumannii A118 in the presence of native, denatured, and digested BSAa
| Treatment | Mean transformation frequency ± SD | Mean CFU/ml ± SD |
|---|---|---|
| LB | 2.607 × 10−8 ± 7.441 × 10−9 | 1.250 × 106 ± 7.05 × 105 |
| LB + BSA | 7.049 × 10−8 ± 3.807 × 10−8 | 2.881 × 106 ± 2.560 × 106 |
| LB + denatured BSA | 2.620 × 10−7 ± 1.540 × 10−7 | 1.544 × 106 ± 1.168 × 106 |
| LB + digested BSA | 6.332 × 10−8 ± 5.684 × 10−8 | 5.708 × 106 ± 4.100 × 106 |
n = 3 biological replicates.
BSA and Ca2+ induce comEA and pilQ transcription in A. baumannii strain A118.
The recent sequencing of the A118 genome shows the conservation of genes that could be related to competence, such as comEA, which encodes a small DNA-binding periplasmic protein, and pilQ, which encodes the outer secretin protein found in type IV pili (11, 44–46). We investigated whether Ca2+ could induce the expression of such genes. Retrotranscription coupled to quantitative PCR (RT-qPCR) was performed in the presence or absence of 1 mM CaCl2. We observed 2-fold increases in the expression levels of both comEA and pilQ in the presence of CaCl2 (Fig. 5), suggesting that divalent cations, specifically Ca2+, imposed an effect on the frequency of natural transformation of the A118 strain by inducing competence-related genes. We also observed that upon the addition of BSA, the levels of comEA and pilQ transcripts were increased by 1.4- and 2.5-fold, respectively (Fig. 5). Most prominently, the addition of BSA and CaCl2 to the media increased the expression levels of the comEA and pilQ transcripts by 2.5- and 3.1-fold, respectively (Fig. 5).
FIG 5.
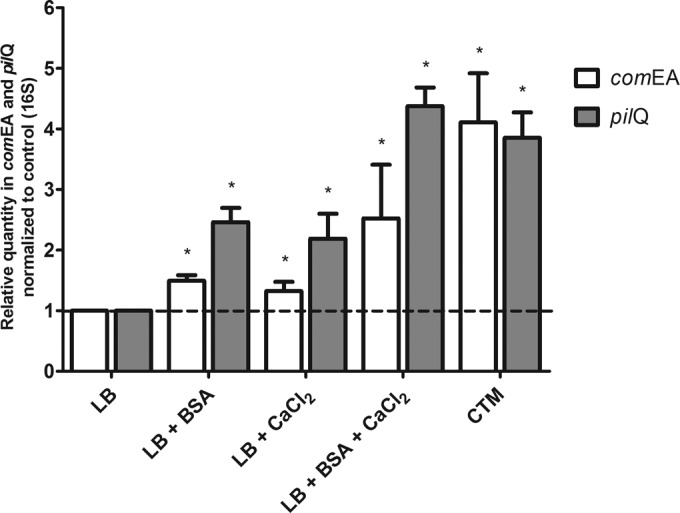
BSA and Ca2+ induction of comEA and pilQ transcription in A. baumannii strain A118. Bacterial cells grown in LB, LB plus CaCl2, LB plus BSA, LB plus CaCl2 and BSA, and CTM were used. Determination of relative expression levels of comEA and pilQ was performed by using RT-qPCR. Fold changes were calculated from threshold cycle (CT) values after normalization to 16S rRNA. Data are presented as the means and SD. (*, P < 0.05 compared with the absence of 1 mM CaCl2 and 0.2% [wt/vol] BSA in medium, as determined by the Kruskal-Wallis test; n = 4).
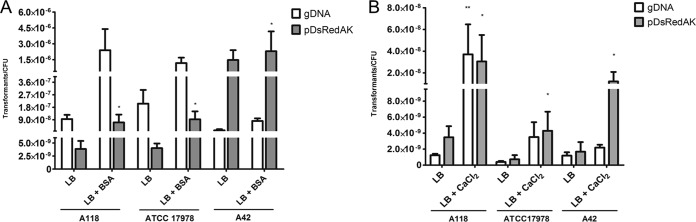
BSA and Ca2+ induce competence among several A. baumannii strains.
To rule out the possibility that BSA-dependent competence enhancement is limited to A118, two other A. baumannii strains (ATCC 17978 and A42) were tested. ATCC 17978 is one of the most studied A. baumannii strains (5, 47–52), and A42 is a multidrug-resistant clinical isolate exhibiting susceptibility to kanamycin (53). We performed transformation assays with and without BSA, and we observed an increase in the transformation frequency for both strains (Fig. 6A). Upon the addition of BSA and using plasmid DNA as the DNA source, we observed 23- and 1.5-fold increases for ATCC 17978 and A42, respectively (see Table S2 in the supplemental material).
FIG 6.
Transformability of A. baumannii A118, ATCC 17978, and A42. Transformation using gDNA (A144KanR) and plasmid DNA (pDsRed) was performed under two different conditions. (A) LB medium with 0% BSA and LB medium with 0.2% (wt/vol) BSA. Cultures were plated onto selective media, and the number of CFU per plate was determined. Data are presented as the means, and error bars represent the SD. Four independent experiments were performed. Asterisks indicate significant differences in transformability (*, P < 0.05 by the Mann-Whitney test; n = 4). (B) LB medium and LB medium with 0.001 M CaCl2. Cultures were plated onto selective media, and the number of CFU per plate was determined. Data are presented as the means, and error bars represent the SD. Five independent experiments were performed. Asterisks indicate significant differences in transformability (*, P < 0.05 by the Mann-Whitney test; **, P < 0.01 by the Mann-Whitney test; n = 5).
We carried out the transformation assay by adding Ca2+, and we also observed a significant increase in the frequency of transformation for both strains (Fig. 6B). Increases of 5.96- and 7.17-fold were observed for strains ATCC 17978 and A42, respectively (see Table S3 in the supplemental material).
These results strongly suggest that induction by BSA and Ca2+ is not limited to the A118 strain, thereby extending the relevance of our findings to other A. baumannii strains. Additionally, these findings suggest that transformation is a common trait of A. baumannii.
Moreover, for clinical strain A42, we observed a significantly higher transformation frequency when plasmid DNA was used as the DNA source. As is known for A. baumannii, transformation rates can vary depending on the isolates. These results suggest that this strain can acquire plasmid DNA more efficiently. Other plasmids and gDNA sources can be tested to see if the same results occur with other types of DNA.
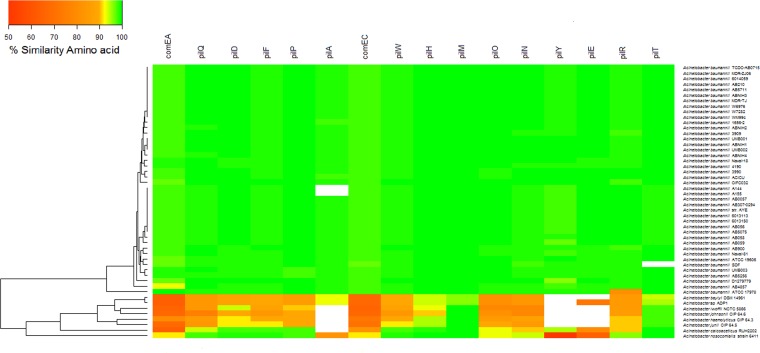
Bioinformatic analysis of putative competence genes in A. baumannii genomes and other non-A. baumannii species.
We recently identified the presence of 17 presumptive competence genes (comM, comEA, pilQ, pilD, pilF, pilP, pilA, comEC, pilW, pilH, pilO, pilN, pilY1 [comC], pilE, pilR, pilM, and pilT) in strain A118 (25). To confirm whether this set of genes is conserved within A. baumannii species and to determine the extent to which natural competence is preserved among members of this genus, we decided to perform a comparative sequence analysis using 42 genomes of A. baumannii and 8 genomes of species within the Acinetobacter genus available in GenBank. Since comM was interrupted by an insertion of a resistance island (AbaR-type genomic island) in most of the A. baumannii genomes, this gene was excluded from the analysis. We found that 15 of the 16 presumptive competence gene-encoded proteins are highly conserved (>80% similarity) among A. baumannii isolates, whereas 12 of the 16 genes were present in all members of the Acinetobacter genus included in the present study (Fig. 7). Of the 16 genes, pilA, pilY, pilE, and pilT were not present in all Acinetobacter spp. A. nosocomialis, a species closely related to A. baumannii and an important pathogen in hospital-acquired infections, possessed all 16 genes, with a considerable degree of average similarity (86.47%). Our results, as well as those reported in the literature, support the notion that most Acinetobacter species strains are likely to be transformable under certain conditions and that they share common mechanisms of DNA uptake described for other transformable bacteria (7, 25).
FIG 7.
Comparative sequence analysis of known competence genes. The heat map representation shows similarity between 15 competence genes of 42 genomes of Acinetobacter baumannii and 8 other species of the genus against the Acinetobacter baumannii A118 genome. The values are represented from 50% (red) to 100% (green) amino acid similarities, as indicated by the color scale. Clustering was carried out by using the complete-linkage method together with Euclidean distance.
DISCUSSION
Antibiotic-resistant bacteria, such as A. baumannii, pose a serious threat to human health, and the increase in the number of infections caused by such pathogens is a growing concern worldwide. The CDC reports 12,000 infections with multidrug-resistant A. baumannii annually (54). This pathogen is responsible for a wide variety of nosocomial infections, including pneumonia, blood infections, meningitis, and skin infections (1, 55). In some cases, the patient outcomes associated with extensively drug-resistant A. baumannii infections can be deadly (2).
Recent analysis of the A. baumannii genome suggests that transformation plays a key role in the development of antibiotic resistance in A. baumannii, but the mechanisms by which it occurs and the factors or signals that play a role in transformation are poorly understood.
Here we identified environmental conditions and factors that could trigger and enhance transformation in this dangerous pathogen. Transformation could help A. baumannii acquire DNA and increase its potential to be a deadly nosocomial pathogen. We employed A. baumannii strain A118, which was previously shown to be naturally competent (24), for this purpose.
We observed that temperature and osmolarity did not exert any effect on the level of natural transformation of A118. In contrast, we observed that different alkaline or acidic conditions impacted transformation levels. A higher level of transformation was obtained at pH 7.5, a pH that corresponds to the pH found in human blood (pH 7.35 to 7.45). It is known that one of the most severe infections caused by this pathogen is septicemia (1, 56); therefore, this result shows that blood pH could be a signal that could have an impact on transformation during the course of A. baumannii infection.
Our results also showed that transformation is not dependent on the amount of DNA or on its source. As has been observed for other species, it is a prompt process that occurs rapidly during the first seconds of DNA-cell exposition, proceeding continuously thereafter over time.
The capacity to undergo natural competence has been studied for many bacterial species. However, how natural competence is triggered, enhanced, or blocked is less understood, particularly for Gram-negative bacteria. In some species, such as H. influenzae, starvation of required carbon sources acts as an inducer of competence (14, 15); in Moraxella catarrhalis, iron limitation affects natural competence (57); and in V. cholerae, chitin induces natural competence (11, 40). In other cases, replication arrest generated by stress can induce competence when competence genes are located near the replication origin (58). For Acinetobacter spp., using A. baylyi strain ADP1, it was observed that an upshift of nutrients has a role in competence induction (18, 42). However, the effect of individual compounds was not clearly defined in the literature. Our results obtained by comparing different media revealed that two distinctive components, Ca2+ and BSA, produced an increase in the level of transformation observed for strain A118 as well as the other two A. baumannii strains tested. Each component induced transformation independently and presented added effects when included in LB medium. Importantly, the addition of BSA did not cause transformation induction simply by increasing the nutrient concentration or protein availability, since the addition of casein at the same concentration did not enhance the rate of transformation. Interestingly, such specificity is not provided by the tertiary structure of the protein but rather by a motif that is exposed when the protein is denatured. We are currently searching this motif and investigating if it is borne by other human proteins within blood or the extracellular matrix. On the other hand, a specific receptor might exist on the bacterium surface.
Another important element supporting the notion that BSA induces natural transformation is that it induces the transcription of comEA and pilQ, two competence genes. Meanwhile, these two genes are also overexpressed upon the addition of Ca2+. Taken together, these results and the observed decrease in the transformation frequency upon EDTA treatment provide evidence that demonstrates that Ca2+ is actively involved in the induction of competence rather than just aiding in this process by mitigating electrostatic repulsion between the negative charges present in the DNA phosphate backbone and on the cell surface. In accordance with our results, Palmen et al. also showed that divalent ions, such as Mg2+, Ca2+, or Mn2+, are required during the transformation process in A. baylyi strain BD413 (18).
Serum albumin is the main protein of mammalian blood plasma, and it is involved in the regulation of the colloidal osmotic pressure of blood as well as serving as a carrier for molecules that have low solubility in water. Calcium is among the elements carried by this protein (59). In particular, BSA (also known as “fraction V”) is a serum albumin protein derived from cows, and its crystal structure showed three calcium ions bound to it (60). As shown by our results, the association of calcium with BSA could play a role in the observed effect on natural transformation, since added effects were observed when both elements were present in the medium. These results possessed relevant significance, since it is known that serum albumins interact with Ca2+ (acid dissociation constant [Ka] = 1.5 × 103 M−1 for human albumin) and that almost 45% of the 2.4 mM flowing Ca2+ in serum is albumin bound (60, 61).
A. baumannii A118 showed an increase in the transformation frequency with BSA. Similar results were obtained for two other A. baumannii clinical strains tested under the same conditions. Therefore, it is highly likely that induction of natural transformation by BSA could be present in the whole species or even along the genus, greatly increasing the significance of our finding.
Jacobs et al. reported increased expression levels of various genes when A. baumannii ATCC 17978 was grown in human serum by using microarrays (51). Among some of the genes that showed upregulation were DNA uptake genes (51). Our experiments are in line with those results, since we also observed an increase of natural transformation when we used human serum as well as HSA in the transformation experiments. Comparison of BSA and HSA proteins showed a high level of similarity between the BSA and HSA structures (root mean square deviation [RMSD], 1.372; Q score, 0.811). Taken together, the results that we provide are evidence that serum albumins could serve as a competence inducer.
Our work provides a description of the first specific inducers, albumins and Ca2+, of natural competence and also shows some aspects of this process in A. baumannii. We observed that natural transformation is a prompt and continuous process that is affected by the pH of the environment. The competence genes comEA and pilQ showed high levels of expression in medium containing both Ca2+ and BSA, conditions under which higher levels of natural transformation were obtained.
Overall, our results suggest that the main blood proteins enhance a primary mechanism of horizontal gene transfer (HGT) in A. baumannii and therefore contribute to the emergence of AMR in this threatening human pathogen. Remarkably, the A118 strain, as well as the other two strains tested here (ATCC 17978 and A42), easily acquired resistance determinants from related bacteria, showing the high flux of AMR across different isolates from this species.
Therefore, albumins and Ca2+ can directly contribute to the acquisition and dispersion of antimicrobial resistance determinants among clinical A. baumannii strains.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Secretaría de Ciencia y Técnica de la Universidad de Buenos Aires (UBACyT) and PICT 0120, Buenos Aires, Argentina, to M.S.R. A.S.-B. was supported by the Institut Pasteur (EMBO-ALTF-1473-2010) and Marie Skłodowska-Curie Actions (FP7-PEOPLE-2011-IIF-BMC). M.S.R. is a member of the CONICET research career, and G.M.T. has a doctoral fellowship from CONICET.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00529-16.
REFERENCES
- 1.Roca I, Espinal P, Vila-Farres X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front Microbiol 3:148. doi: 10.3389/fmicb.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falagas ME, Kopterides P, Siempos II. 2006. Attributable mortality of Acinetobacter baumannii infection among critically ill patients. Clin Infect Dis 43:389. doi: 10.1086/505599 (Reply, 43:389-390, doi:.) [DOI] [PubMed] [Google Scholar]
- 3.Mussi MA, Gaddy JA, Cabruja M, Arivett BA, Viale AM, Rasia R, Actis LA. 2010. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J Bacteriol 192:6336–6345. doi: 10.1128/JB.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Liu F, Zhang Y, Wang X, Zhao C, Chen H, Zhang F, Zhu B, Hu Y, Wang H. 2015. Evolution of carbapenem-resistant Acinetobacter baumannii revealed through whole-genome sequencing and comparative genomic analysis. Antimicrob Agents Chemother 59:1168–1176. doi: 10.1128/AAC.04609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Touchon M, Cury J, Yoon EJ, Krizova L, Cerqueira GC, Murphy C, Feldgarden M, Wortman J, Clermont D, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P, Rocha EP. 2014. The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol Evol 6:2866–2882. doi: 10.1093/gbe/evu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 9.Engelmoer DJ, Donaldson I, Rozen DE. 2013. Conservative sex and the benefits of transformation in Streptococcus pneumoniae. PLoS Pathog 9:e1003758. doi: 10.1371/journal.ppat.1003758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humbert O, Dorer MS, Salama NR. 2011. Characterization of Helicobacter pylori factors that control transformation frequency and integration length during inter-strain DNA recombination. Mol Microbiol 79:387–401. doi: 10.1111/j.1365-2958.2010.07456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seitz P, Blokesch M. 2013. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev 37:336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 12.Woodhams KL, Benet ZL, Blonsky SE, Hackett KT, Dillard JP. 2012. Prevalence and detailed mapping of the gonococcal genetic island in Neisseria meningitidis. J Bacteriol 194:2275–2285. doi: 10.1128/JB.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorer MS, Fero J, Salama NR. 2010. DNA damage triggers genetic exchange in Helicobacter pylori. PLoS Pathog 6:e1001026. doi: 10.1371/journal.ppat.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herriott RM, Meyer EM, Vogt M. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol 101:517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Redfield RJ. 1991. sxy-1, a Haemophilus influenzae mutation causing greatly enhanced spontaneous competence. J Bacteriol 173:5612–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carr EL, Kampfer P, Patel BK, Gurtler V, Seviour RJ. 2003. Seven novel species of Acinetobacter isolated from activated sludge. Int J Syst Evol Microbiol 53:953–963. doi: 10.1099/ijs.0.02486-0. [DOI] [PubMed] [Google Scholar]
- 17.Lorenz MG, Wackernagel W. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev 58:563–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmen R, Vosman B, Buijsman P, Breek CK, Hellingwerf KJ. 1993. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J Gen Microbiol 139:295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- 19.Young DM, Parke D, Ornston LN. 2005. Opportunities for genetic investigation afforded by Acinetobacter baylyi, a nutritionally versatile bacterial species that is highly competent for natural transformation. Annu Rev Microbiol 59:519–551. doi: 10.1146/annurev.micro.59.051905.105823. [DOI] [PubMed] [Google Scholar]
- 20.Gerischer U, Ornston LN. 2001. Dependence of linkage of alleles on their physical distance in natural transformation of Acinetobacter sp. strain ADP1. Arch Microbiol 176:465–469. doi: 10.1007/s00203-001-0353-7. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen KM, Bones AM, Van Elsas JD. 1997. Induced natural transformation of Acinetobacter calcoaceticus in soil microcosms. Appl Environ Microbiol 63:3972–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen KM, van Weerelt MD, Berg TN, Bones AM, Hagler AN, van Elsas JD. 1997. Natural transformation and availability of transforming DNA to Acinetobacter calcoaceticus in soil microcosms. Appl Environ Microbiol 63:1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Utnes AL, Sorum V, Hulter N, Primicerio R, Hegstad J, Kloos J, Nielsen KM, Johnsen PJ. 2015. Growth phase-specific evolutionary benefits of natural transformation in Acinetobacter baylyi. ISME J 9:2221–2231. doi: 10.1038/ismej.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez MS, Don M, Merkier AK, Bistue AJ, Zorreguieta A, Centron D, Tolmasky ME. 2010. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J Clin Microbiol 48:1488–1490. doi: 10.1128/JCM.01264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traglia GM, Chua K, Centron D, Tolmasky ME, Ramirez MS. 2014. Whole-genome sequence analysis of the naturally competent Acinetobacter baumannii clinical isolate A118. Genome Biol Evol 6:2235–2239. doi: 10.1093/gbe/evu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilharm G, Piesker J, Laue M, Skiebe E. 2013. DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J Bacteriol 195:4146–4153. doi: 10.1128/JB.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez MS, Merkier AK, Quiroga MP, Centron D. 2012. Acinetobacter baumannii is able to gain and maintain a plasmid harbouring In35 found in Enterobacteriaceae isolates from Argentina. Curr Microbiol 64:211–213. doi: 10.1007/s00284-011-0052-9. [DOI] [PubMed] [Google Scholar]
- 28.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS Jr. 2013. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4:e00360-13. doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen JH, Ferraro MJ. 2009. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49:1749–1755. doi: 10.1086/647952. [DOI] [PubMed] [Google Scholar]
- 30.Turapov OA, Mukamolova GV, Bottrill AR, Pangburn MK. 2008. Digestion of native proteins for proteomics using a thermocycler. Anal Chem 80:6093–6099. doi: 10.1021/ac702527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramirez MS, Traglia GM, Perez JF, Muller GL, Martinez F, Golic AE, Mussi MA. 2015. White and blue light induce reduction in susceptibility to minocycline and tigecycline in Acinetobacter sp. and other bacteria of clinical importance. J Med Microbiol 64:525–537. doi: 10.1099/jmm.0.000048. [DOI] [PubMed] [Google Scholar]
- 32.Chen I, Dubnau D. 2004. DNA uptake during bacterial transformation. Nat Rev Microbiol 2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 33.Vegge CS, Brondsted L, Ligowska-Marzeta M, Ingmer H. 2012. Natural transformation of Campylobacter jejuni occurs beyond limits of growth. PLoS One 7:e45467. doi: 10.1371/journal.pone.0045467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saunders CW, Guild WR. 1981. Monomer plasmid DNA transforms Streptococcus pneumoniae. Mol Gen Genet 181:57–62. doi: 10.1007/BF00339005. [DOI] [PubMed] [Google Scholar]
- 35.Baur B, Hanselmann K, Schlimme W, Jenni B. 1996. Genetic transformation in freshwater: Escherichia coli is able to develop natural competence. Appl Environ Microbiol 62:3673–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hisano K, Fujise O, Miura M, Hamachi T, Matsuzaki E, Nishimura F. 2014. The pga gene cluster in Aggregatibacter actinomycetemcomitans is necessary for the development of natural competence in Ca(2+)-promoted biofilms. Mol Oral Microbiol 29:79–89. doi: 10.1111/omi.12046. [DOI] [PubMed] [Google Scholar]
- 37.Huddleston JR, Brokaw JM, Zak JC, Jeter RM. 2013. Natural transformation as a mechanism of horizontal gene transfer among environmental Aeromonas species. Syst Appl Microbiol 36:224–234. doi: 10.1016/j.syapm.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Kang F, Wang H, Gao Y, Long J, Wang Q. 2013. Ca2+ promoted the low transformation efficiency of plasmid DNA exposed to PAH contaminants. PLoS One 8:e58238. doi: 10.1371/journal.pone.0058238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Liu L, Wang Y, Wang X, Ma Y, Li Y. 2014. The study on the factors affecting transformation efficiency of E. coli competent cells. Pak J Pharm Sci 27:679–684. [PubMed] [Google Scholar]
- 40.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 41.Marvig RL, Blokesch M. 2010. Natural transformation of Vibrio cholerae as a tool—optimizing the procedure. BMC Microbiol 10:155. doi: 10.1186/1471-2180-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porstendorfer D, Gohl O, Mayer F, Averhoff B. 2000. ComP, a pilin-like protein essential for natural competence in Acinetobacter sp. strain BD413: regulation, modification, and cellular localization. J Bacteriol 182:3673–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fontaine L, Goffin P, Dubout H, Delplace B, Baulard A, Lecat-Guillet N, Chambellon E, Gardan R, Hols P. 2013. Mechanism of competence activation by the ComRS signalling system in streptococci. Mol Microbiol 87:1113–1132. doi: 10.1111/mmi.12157. [DOI] [PubMed] [Google Scholar]
- 44.Burkhardt NY, Baldridge GD, Williamson PC, Billingsley PM, Heu CC, Felsheim RF, Kurtti TJ, Munderloh UG. 2011. Development of shuttle vectors for transformation of diverse Rickettsia species. PLoS One 6:e29511. doi: 10.1371/journal.pone.0029511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charpentier X, Kay E, Schneider D, Shuman HA. 2011. Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J Bacteriol 193:1114–1121. doi: 10.1128/JB.01146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salzer R, Joos F, Averhoff B. 2015. Different effects of MglA and MglB on pilus-mediated functions and natural competence in Thermus thermophilus. Extremophiles 19:261–267. doi: 10.1007/s00792-014-0711-4. [DOI] [PubMed] [Google Scholar]
- 47.Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, Parreira JR, Rodriguez-Velo P, Bou G. 2010. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol 10:279. doi: 10.1186/1471-2180-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giles SK, Stroeher UH, Eijkelkamp BA, Brown MH. 2015. Identification of genes essential for pellicle formation in Acinetobacter baumannii. BMC Microbiol 15:116. doi: 10.1186/s12866-015-0440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hare JM, Ferrell JC, Witkowski TA, Grice AN. 2014. Prophage induction and differential RecA and UmuDAb transcriptome regulation in the DNA damage responses of Acinetobacter baumannii and Acinetobacter baylyi. PLoS One 9:e93861. doi: 10.1371/journal.pone.0093861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heindorf M, Kadari M, Heider C, Skiebe E, Wilharm G. 2014. Impact of Acinetobacter baumannii superoxide dismutase on motility, virulence, oxidative stress resistance and susceptibility to antibiotics. PLoS One 9:e101033. doi: 10.1371/journal.pone.0101033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacobs AC, Sayood K, Olmsted SB, Blanchard CE, Hinrichs S, Russell D, Dunman PM. 2012. Characterization of the Acinetobacter baumannii growth phase-dependent and serum responsive transcriptomes. FEMS Immunol Med Microbiol 64:403–412. doi: 10.1111/j.1574-695X.2011.00926.x. [DOI] [PubMed] [Google Scholar]
- 52.Johnson TL, Waack U, Smith S, Mobley H, Sandkvist M. 2016. Acinetobacter baumannii is dependent on the type II secretion system and its substrate LipA for lipid utilization and in vivo fitness. J Bacteriol 198:711–719. doi: 10.1128/JB.00622-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vilacoba E, Almuzara M, Gulone L, Traglia GM, Figueroa SA, Sly G, Fernandez A, Centron D, Ramirez MS. 2013. Emergence and spread of plasmid-borne tet(B)::ISCR2 in minocycline-resistant Acinetobacter baumannii isolates. Antimicrob Agents Chemother 57:651–654. doi: 10.1128/AAC.01751-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.CDC. 2013. Antibiotic resistance threats in the United States, 2013. CDC, Atlanta, GA. [Google Scholar]
- 55.Doughari HJ, Ndakidemi PA, Human IS, Benade S. 2011. The ecology, biology and pathogenesis of Acinetobacter spp.: an overview. Microbes Environ 26:101–112. doi: 10.1264/jsme2.ME10179. [DOI] [PubMed] [Google Scholar]
- 56.Seifert H, Strate A, Pulverer G. 1995. Nosocomial bacteremia due to Acinetobacter baumannii. Clinical features, epidemiology, and predictors of mortality. Medicine (Baltimore) 74:340–349. [DOI] [PubMed] [Google Scholar]
- 57.Luke NR, Howlett AJ, Shao J, Campagnari AA. 2004. Expression of type IV pili by Moraxella catarrhalis is essential for natural competence and is affected by iron limitation. Infect Immun 72:6262–6270. doi: 10.1128/IAI.72.11.6262-6270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slager J, Kjos M, Attaiech L, Veening JW. 2014. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell 157:395–406. doi: 10.1016/j.cell.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 59.He XM, Carter DC. 1992. Atomic structure and chemistry of human serum albumin. Nature 358:209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- 60.Majorek KA, Porebski PJ, Dayal A, Zimmerman MD, Jablonska K, Stewart AJ, Chruszcz M, Minor W. 2012. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol Immunol 52:174–182. doi: 10.1016/j.molimm.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peters T. 1995. All about albumin: biochemistry, genetics, and medical applications. Academic Press, San Diego, CA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.