ABSTRACT
Mobile genetic elements play a pivotal role in the adaptation of bacterial populations, allowing them to rapidly cope with hostile conditions, including the presence of antimicrobial compounds. IncA/C conjugative plasmids (ACPs) are efficient vehicles for dissemination of multidrug resistance genes in a broad range of pathogenic species of Enterobacteriaceae. ACPs have sporadically been reported in Vibrio cholerae, the infectious agent of the diarrheal disease cholera. The regulatory network that controls ACP mobility ultimately depends on the transcriptional activation of multiple ACP-borne operons by the master activator AcaCD. Beyond ACP conjugation, AcaCD has also recently been shown to activate the expression of genes located in the Salmonella genomic island 1 (SGI1). Here, we describe MGIVchHai6, a novel and unrelated mobilizable genomic island (MGI) integrated into the 3′ end of trmE in chromosome I of V. cholerae HC-36A1, a non-O1/non-O139 multidrug-resistant clinical isolate recovered from Haiti in 2010. MGIVchHai6 contains a mercury resistance transposon and an integron In104-like multidrug resistance element similar to the one of SGI1. We show that MGIVchHai6 excises from the chromosome in an AcaCD-dependent manner and is mobilized by ACPs. Acquisition of MGIVchHai6 confers resistance to β-lactams, sulfamethoxazole, tetracycline, chloramphenicol, trimethoprim, and streptomycin/spectinomycin. In silico analyses revealed that MGIVchHai6-like elements are carried by several environmental and clinical V. cholerae strains recovered from the Indian subcontinent, as well as from North and South America, including all non-O1/non-O139 clinical isolates from Haiti.
IMPORTANCE
Vibrio cholerae, the causative agent of cholera, remains a global public health threat. Seventh-pandemic V. cholerae acquired multidrug resistance genes primarily through circulation of SXT/R391 integrative and conjugative elements. IncA/C conjugative plasmids have sporadically been reported to mediate antimicrobial resistance in environmental and clinical V. cholerae isolates. Our results showed that while IncA/C plasmids are rare in V. cholerae populations, they play an important yet insidious role by specifically propagating a new family of genomic islands conferring resistance to multiple antibiotics. These results suggest that nonepidemic V. cholerae non-O1/non-O139 strains bearing these genomic islands constitute a reservoir of transmissible resistance genes that can be propagated by IncA/C plasmids to V. cholerae populations in epidemic geographical areas as well to pathogenic species of Enterobacteriaceae. We recommend future epidemiological surveys take into account the circulation of these genomic islands.
INTRODUCTION
The diarrheal disease cholera remains a serious public health threat worldwide (1). The O1 and O139 toxigenic strains of Vibrio cholerae, the etiologic agent of cholera, produce a toxin that causes profuse diarrhea, vomiting, and subsequent severe dehydration (2). While proper hydration is usually sufficient to treat cholera patients, antibiotic therapy is necessary to treat severe cases and to reduce the release of this infectious agent into the environment (1). Toxigenic V. cholerae is very efficient at rapidly infecting and spreading among and between human populations (1, 3). Stepwise evolution of pathogenic lineages of V. cholerae has involved acquisition of adaptive traits, such as antibiotic resistance genes and virulence factors encoded by diverse mobile genetic elements, including those coding for cholera toxin and carried on the CTXφ phage, pathogenicity islands, and integrative and conjugative elements (ICEs) (2, 4–7).
ICEs of the SXT/R391 family have been recognized as major drivers of the dissemination of antibiotic resistance genes among several species of Enterobacteriaceae and Vibrionaceae, including environmental and clinical V. cholerae isolates (8–10). ICEVchInd5, an ICE conferring resistance to sulfamethoxazole-trimethoprim (co-trimoxazole), streptomycin, and chloramphenicol has been shown to be the most prevalent variant of SXT/R391 ICE in the seventh-pandemic multidrug-resistant V. cholerae lineage (5–7, 11). While the role of SXT/R391 ICEs has been extensively studied over the last two decades, recent studies have also highlighted the sporadic involvement of conjugative plasmids of the IncA/C group (ACPs) in genome plasticity and multidrug resistance (MDR) acquisition in V. cholerae (12–18). ACPs are large plasmids (>110 kb) that transfer efficiently by conjugation to and maintenance in a broad range of Gammaproteobacteria (19, 20). ACPs are a threat to human and animal health due to their worldwide prevalence in clinical isolates of bacterial enteric pathogens and their carriage of a large variety of antibiotic resistance genes (18, 19, 21–24). Several reports have associated ACPs with resistance to penicillins, cephalosporins, and carbapenems, conferred by allelic variants of the blaNDM gene (25–28). These plasmids are frequently detected in food products, food-producing animals, and environmental samples that likely constitute a large reservoir for their subsequent dissemination to human pathogens (19).
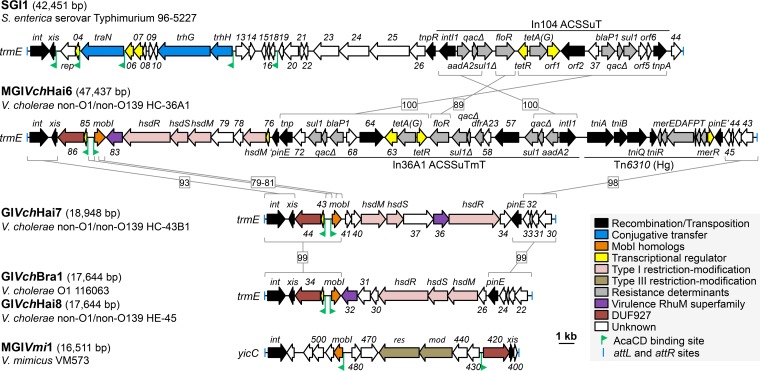
Our group has previously described pVCR94, a type 2 IncA/C2 plasmid conferring MDR to the V. cholerae O1 El Tor strain responsible for the 1994 explosive cholera outbreak in Goma refugee camps (Democratic Republic of the Congo) (12). pVCR94 was used to characterize the main regulatory pathway that controls ACP conjugative transfer (12, 20, 21, 29). We have shown that acaCD encodes the master activator of ACP transfer that activates transcription of 18 ACP-borne genes and operons, including those coding for the conjugative machinery (21). In addition, AcaCD was shown to trans-activate excision and dissemination of MDR-conferring Salmonella genomic island 1 (SGI1) and unrelated MGIVmi1 of Vibrio mimicus (Fig. 1) (20, 21, 29).
FIG 1 .
AcaCD-activated GIs. Schematic representation of the genetic map of GIs bearing predicted AcaCD binding sites. GIs are drawn to scale. The left and right junctions (attL and attR) within the host chromosome are indicated by blue bars at the extremities. ORFs with similar function are color coded as indicated in the figure. Green chevrons indicate the position and orientation of predicted AcaCD-binding sites. Homologous regions are bracketed and linked by a gray line with the corresponding percentage of nucleotide identity. Gene numbers correspond to the last digits of respective locus tags in the GenBank accession numbers for SGI1 (AF261825), MGIVchHai6 (AXDR01000001), GIVchHai7 (ALDP01000008), GIVchBra1 (APFK01000082), and GIVchHai8 (ALED01000018). ACSSuT, resistance to ampicillin, chloramphenicol, spectinomycin/streptomycin, sulfamethoxazole, and tetracycline; Tm, trimethoprim resistance; Hg, mercury resistance.
The presence of an MDR-conferring ACP in a 2012 V. cholerae isolate from Haiti has been reported only once to date (17). We report here the insidious role played by ACPs in the spread of MDR through mobilization of a new family of MDR-conferring genomic islands (GIs) in clinical non-O1/non-O139 V. cholerae isolates from Haiti. The presence of an ACP specifically triggers the excision and conjugative transfer of MGIVchHai6, the prototypical member of this new family of mobilizable GIs (MGIs). Further in silico analyses revealed the presence of MGIVchHai6-like elements in several environmental and clinical strains of V. cholerae isolated in North and South America and the Indian subcontinent. Our results demonstrate that ACPs have influenced the evolution of Haitian V. cholerae strains by propagating genomic islands, thereby allowing circulation of a vast reservoir of mobilizable MDR genes.
RESULTS
MDR determinants of V. cholerae HC-36A1 are neither carried on nor mobilized by SXT/R391 ICEs.
V. cholerae HC-36A1 is a non-O1/non-O139 clinical isolate recovered in 2010 in the province of Port-au-Prince, Haiti at the beginning of the ongoing cholera outbreak in that country (4). Antibiotic susceptibility tests showed HC-36A1 is resistant to multiple antibiotics, including ampicillin, sulfamethoxazole, trimethoprim, chloramphenicol, kanamycin, streptomycin, and spectinomycin, and exhibits an inducible resistance to tetracycline when the strain is preexposed to subinhibitory concentrations of tetracycline (Table 1). The vast majority of Haitian outbreak strains carry ICEVchInd5 (also known as ICEVchHai1), a member of the SXT/R391 family of ICEs that mediates resistance to sulfamethoxazole, trimethoprim, chloramphenicol, and streptomycin (4, 30). Although most SXT/R391 ICEs are easily transferable between V. cholerae and Escherichia coli, our attempts to transfer resistance markers by conjugation, from V. cholerae HC-36A1 to either a rifampin-resistant derivative of E. coli MG1655 (MG1655 Rf) or the tetracycline-resistant E. coli strain CAG18439, did not generate any transconjugants (Fig. 2A). Analysis of the genome of HC-36A1 (GenBank accession no. AXDR01000001) confirmed the presence of an SXT/R391 ICE virtually identical to ICEVchHai2, an 83-kb ICE first identified in non-O1/non-O139 V. cholerae isolate HC-1A2 but lacking the typical antibiotic resistance gene cluster found in SXT/R391 ICEs (30). This result indicates that the resistance determinants carried by HC-36A1 are not carried by an SXT/R391 ICE or a genomic island that they can mobilize in trans or in cis (20, 31–33).
TABLE 1 .
MICs of 12 antibiotics against V. cholerae and E. coli with or without MGIVchHai6
| Antibiotic |
V. cholerae HC-36A1 |
E. coli CAG18439 |
E. coli CAG18439 MGIVchHai6 |
|||
|---|---|---|---|---|---|---|
| MIC (µg/ml)a | Phenotypeb | MIC (µg/ml)a | Phenotypeb | MIC (µg/ml)a | Phenotypeb | |
| Ampicillin | >120 | R | <25 | S | >800 | R |
| Chloramphenicol | >32 | R | 10 | S | >160 | R |
| Ciprofloxacin | 0.5 | S | <0.13 | S | <0.13 | S |
| Erythromycin | <200 | S | 100 | S | 100 | S |
| Gentamicin | 2.5 | S | <1.25 | S | <1.25 | S |
| Kanamycin | 120 | R | 25 | S | 25 | S |
| Nalidixic acid | <10 | S | 20 | S | 20 | S |
| Rifampin | 100 | S | 25 | S | 25 | S |
| Streptomycin | >40 | R | <50 | S | >1,600 | R |
| Spectinomycin | >80 | R | 100 | S | >400 | R |
| Sulfamethoxazole | ND | R | ND | S | ND | R |
| Tetracyclinec | 3–40 | I | − | R | − | R |
| Trimethoprim | >40 | R | <8 | S | >256 | R |
ND, not determined. Tests were done using solid agar plates.
R, resistant; S, susceptible; I, inducible resistance.
Tetracycline resistance of strain HC-36A1 ranges from 3 µg/ml to 40 µg/ml, without and with induction using 3 µg/ml of tetracycline as the inducible treatment, respectively. Tetracycline resistance tests were not carried out for the tetracycline-resistant strain CAG18439 (−).
FIG 2 .
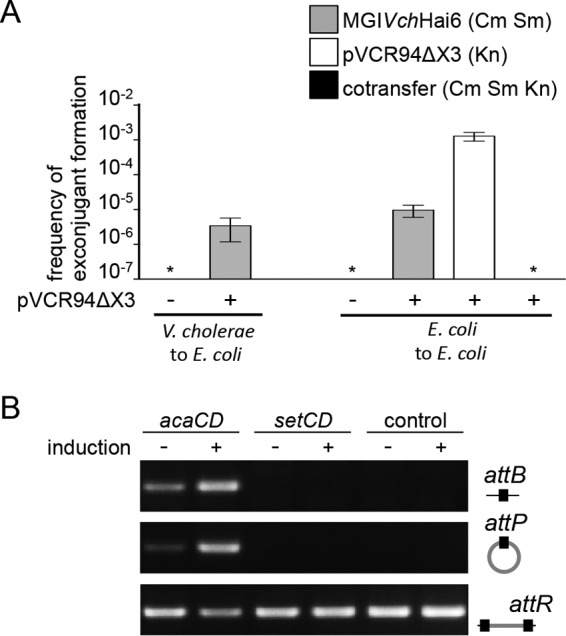
IncA/C-dependent excision and transfer of GIs. (A) MGIVchHai6 is mobilizable by ACPs. Inter- and intraspecific mobilization of MGIVchHai6 was assayed from V. cholerae to E. coli and from E. coli to E. coli, respectively. Interspecific transfer was done using V. cholerae HC-36A1 bearing pVCR94ΔX3 as the donor and E. coli CAG18439 as the recipient. Intraspecific transfer was performed using E. coli CAG18439 bearing pVCR94ΔX3 and MGIVchHai6 as the donor and E. coli MG1655 Rf as the recipient. Donor cells lacking pVCR94ΔX3 were used as a control. Exconjugants were selected for the acquisition of either MGIVchHai6, pVCR94ΔX3, or both. Transfer frequencies are expressed as the number of exconjugants per recipient CFU. Bars represent the mean and standard deviation values obtained from three independent experiments. Asterisks indicate the frequency of exconjugant formation below the detection limit (<10−7). Cm, chloramphenicol; Sm, streptomycin; Kn, kanamycin. (B) AcaCD specifically induces MGIVchHai6 excision. Excision was detected by PCR on genomic DNA to amplify chromosomal site attB and the attP site resulting from the excision of MGIVchHai6 in E. coli. Integrated MGIVchHai6 was detected by amplification of the attR site. Assays were done employing E. coli MG1655 Rf strains bearing the single-copy integrated IPTG-inducible pacaDC3×FLAG vector (acaCD), the single-copy integrated IPTG-inducible psetDC3×FLAG vector (setCD), or devoid of plasmid (control), without (−) or with (+) induction using IPTG.
V. cholerae HC-36A1 holds a complex MDR-associated integron reminiscent of In104 from Salmonella genomic island 1.
Sequence analysis of the HC-36A1 genome aimed at localizing MDR determinants uncovered a new resistance gene cluster in chromosome I (Fig. 1). This locus contains a complex integron, In36A1, which exhibits structure and gene content relatively similar to those of the In104 resistance complex integron of SGI1 (Fig. 1) (34). Like In104, In36A1 contains the virtually identical resistance determinants blaP (β-lactams), sul1 (sulfamethoxazole), tetA(G) (tetracycline), and aadA2 (streptomycin/spectinomycin). In36A1 also contains a floR (florfenicol/chloramphenicol) variant sharing only 89% identity with floR of SGI1 and a trimethoprim resistance determinant sharing 93% identity with dfrA23, as a larger insertion between two sul1 copies (Fig. 1).
In36A1 is surrounded by additional putative resistance loci: (i) a mercury resistance Tn5053-like transposon, Tn6310, and (ii) a gene cluster coding for an hsd-like type I restriction-modification (RM) system that can confer resistance to bacteriophage infections (35–37). Such accumulation of adaptive determinants within a 38-kb fragment hinted at the presence of a larger genomic island.
In36A1 is part of a new family of genomic islands.
Analysis of the nearby sequence revealed proximity of a tRNA modification GTPase-encoding gene trmE, the 3′ end of which is the target site for SGI1 integration in Salmonella enterica (38–40). Located next to the trmE integration site is a gene, int, encoding a predicted tyrosine recombinase/integrase (VCHC36A1_0088) distantly related to the integrase of SGI1 (67% identity over 386 amino acid residues). Moreover, the gene adjacent to int, xis, codes for a predicted recombination directionality factor (RDF) (VCHC36A1_0087) sharing 37% identity over 106 amino acid residues with Xis of SGI1. Similar to SGI1, two imperfect direct repeats (differences underlined) were identified as part of the attL (TTCTGTATTGGGAAGTAA) and attR (TTCTGTATTGGCAAGTAA) attachment sites flanking the 47,437-bp genomic island MGIVchHai6. While MGIVchHai6 and SGI1 share distantly related recombination modules (int/xis) and closely related complex resistance integrons (In36A1 and In104, respectively), the remainder of the gene content is strikingly different (Fig. 1). Therefore, MGIVchHai6 is concluded not to be a SGI1-like element but rather a representative of a novel and large family of mobilizable genomic islands.
ACPs trigger excision of MGIVchHai6 in V. cholerae HC-36A1.
Since we were unable to observe MDR transfer from ICEVchHai2-containing HC-36A1 to E. coli, we conclude that MGIVchHai6 is not mobilizable by SXT/R391 ICEs. On the other hand, the recent discovery of key regulators of ACP conjugation and the ability of the ACP-encoded positive regulator AcaCD to trigger excision of genomic islands SGI1 and MGIVmi1 led us to consider ACPs are possible drivers of MGIVchHai6 mobility (20, 21, 29). In silico analysis of MGIVchHai6 revealed the presence of two divergent AcaCD binding motifs located in a 441-bp intergenic region. The first predicted AcaCD binding motif, GAAGTTCCCAAAAAGGGCAGTTCCAGCG (FIMO P value, 7.69 × 10−10), is located upstream of an operon-like cluster of three genes that likely code for a putative transcriptional regulator (VCHC36A1_0085), a DUF927 domain-containing protein of unknown function (VCHC36A1_0086), and Xis (Fig. 1). The second predicted AcaCD binding motif, CGTGTGCCCCAAAAGGGCACGAAGGCAG (FIMO P value, 3.26 × 10−8), is located upstream of a gene coding for a distant homolog of the mobilization protein MobI (27% identity over two fragments of 109 and 53 amino acid residues), a conserved key factor for conjugative transfer of ACPs (12, 20, 29). We also searched for binding motifs of the master activator of conjugation of SXT/R391 ICEs (SetCD) (41, 42), yet a SetCD binding motif with a FIMO P value below 10−5 could not be detected.
The presence of an AcaCD binding motif upstream of a putative xis gene strongly suggested that excision of MGIVchHai6 from the chromosome could be triggered by ACPs. To test this hypothesis, we introduced pVCR94ΔX3, a kanamycin-resistant derivative of ACP pVCR94, into V. cholerae HC-36A1 (21). To circumvent the presence of redundant kanamycin resistance in the two strains, as well as the possible retrotransfer of MGIVchHai6 from HC-36A1 to the donor cell, pVCR94ΔX3 was transferred to V. cholerae using as donor E. coli β2163, a dapA mutant auxotrophic for diaminopimelic acid (DAP) synthesis (43). The presence of pVCR94ΔX3, together with MGIVchHai6 in HC-36A1, was confirmed by growth on selective media and PCR amplification of specific DNA fragments—i.e., the promoter region of traI in pVCR94ΔX3 and a fragment in open reading frame 86 (ORF86) of MGIVchHai6. Employing PCR, we determined whether the presence of an ACP could trigger excision of MGIVchHai6, a mandatory step for transfer of a complete and functional genomic island. A fragment specific for an attP site resulting from excision of MGIVchHai6 from the chromosome was detected and sequenced, thereby confirming that excision of MGIVchHai6 occurred by recombination between the two imperfect direct repeats previously identified in the attL and attR attachment sites.
MGIVchHai6 is trans-mobilizable by ACPs.
The excision of MGIVchHai6 in the presence of ACPs in the same cell prompted us to test whether pVCR94ΔX3 could mediate MGIVchHai6 conjugative transfer into E. coli. Therefore, V. cholerae HC-36A1 carrying pVCR94ΔX3 was used as a donor in mating assays using E. coli K-12 as a recipient. The presence of pVCR94ΔX3 specifically allowed the transfer of MGIVchHai6 and the associated MDR phenotype at a frequency of 3.5 × 10−6 exconjugant per recipient cell (Fig. 2A; Table 1). To exclude Hfr-like transfer of the MDR locus and subsequent integration by homologous or nonhomologous recombination into the chromosome of the recipient cell, the functionality of transferred MGIVchHai6 was verified in intraspecific transfer assays between E. coli strains (31, 44). While MGIVchHai6 was again unable to transfer autonomously, the presence of pVCR94ΔX3 triggered its transfer at a frequency of 1.0 × 10−5 exconjugant per recipient cell (Fig. 2A). In contrast, while pVCR94ΔX3 transferred at a rate of 1.3 × 10−3 exconjugant per recipient cell, the rate of cotransfer of both MGIVchHai6 and pVCR94ΔX3 was below the limit of detection, suggesting it might be a rare event (Fig. 2A).
Master activator AcaCD specifically triggers excision of MGIVchHai6.
We further investigated whether excision of MGIVchHai6 was specifically dependent upon AcaCD. E. coli bearing MGIVchHai6 with or without a single chromosomally integrated copy of pacaDC3×FLAG, which expresses AcaCD under control of Ptac, was used in PCR experiments to detect attB and attP sites resulting from MGIVchHai6 excision.
Excision assays revealed that the presence of AcaCD specifically triggers excision of MGIVchHai6, as shown by detection of specific attB and attP sites after IPTG (isopropyl-β-d-1-thiogalactopyranoside) induction (Fig. 2B). Excision of MGIVchHai6 was also observed without induction of acaCD expression, likely due to leaky transcription from Ptac allowing sufficient production of AcaCD to induce the AcaCD-dependent promoters of MGIVchHai6 (21, 41). In contrast, attB and attP could not be detected in the strain expressing SetCD, the master activator of SXT/R391 ICE conjugation, thereby confirming that MGIVchHai6 excision is specifically triggered by ACP-encoded AcaCD (Fig. 2B). No spontaneous excision of MGIVchHai6 was detected using the control strain devoid of acaCD- or setCD-bearing plasmids.
MGIVchHai6-like elements are widespread among V. cholerae epidemic and environmental strains.
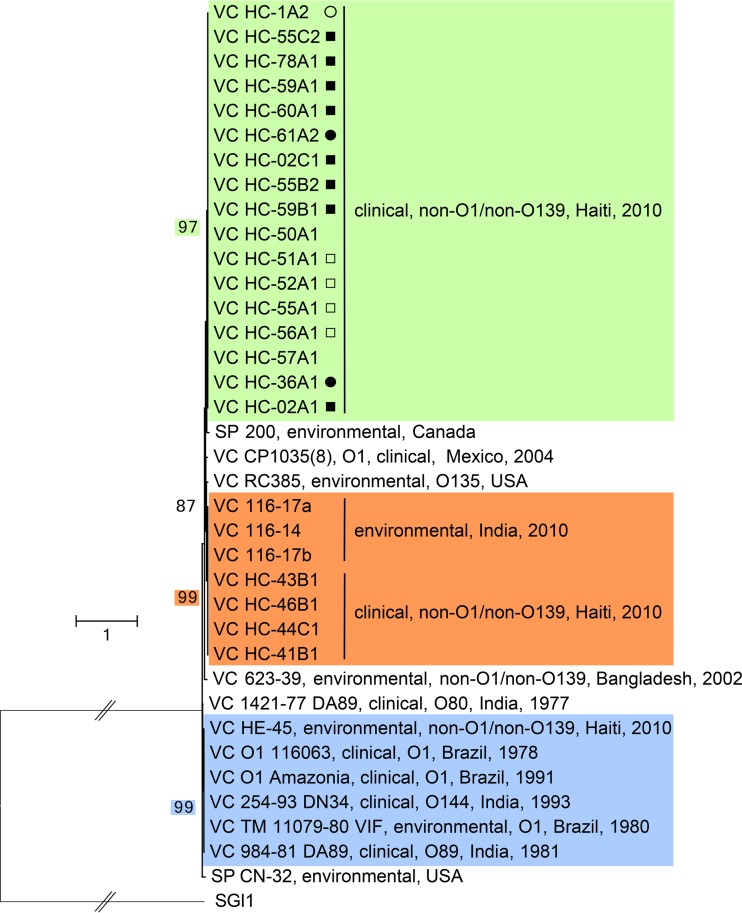
To assess the diversity and abundance of MGIVchHai6-like elements among available genome sequences, in silico analysis was conducted using the attL-int DNA portion as a query to retrieve related genomic islands integrated into the 3′ end of trmE. Using this 1,352-nucleotide (nt) sequence in a search of the GenBank nucleotide collection database (nr/nt) targeting Gammaproteobacteria yielded only two significant matches, namely, against Shewanella putrefaciens genomes, one of which was previously detected in S. putrefaciens 200 as GISpu1 (Fig. 3) (29). Further analyses targeting the whole-genome shotgun sequences and narrowing the analysis to Vibrionaceae revealed the presence of MGIVchHai6-related elements in the genome of various environmental and clinical V. cholerae strains (Fig. 3). Closer examination of these strains revealed that MGIVchHai6-related elements are not associated with a specific serotype or geographic location, because different strains were recovered from the Indian subcontinent, as well as from North and South America, including all non-O1/non-O139 clinical isolates from the 2010 outbreak in Haiti (Fig. 3). Interestingly, MGIVchHai6-related GIs were also detected in strains isolated from various locations in the 1980s, demonstrating that they had existed in V. cholerae populations prior to the 2010 Haitian cholera outbreak.
FIG 3 .
Molecular phylogenetic analysis of the attL-int locus of MGIVchHai6-related GIs by the maximum likelihood method. Based on the Hasegawa-Kishino-Yano model, the tree with the highest log likelihood (−5,618.1383) is shown (64). The percentage of trees in which the associated taxa clustered together is shown only for bootstrap values above 80. Subgroups were defined as clades supported by at least 95 bootstraps and are color coded as follows: strains containing MGIVchHai6-like elements are in green, GIVchHai7-like elements are in orange, and GIVchBra1 to GIVchHai8-like elements are in blue. Because analyses were done on nonassembled draft genome sequences, symbols indicate the confidence that the strain contains a complete element (solid circles, complete MGIVchHai6; open circles, partial detection of In36A1; solid squares and open squares, complete or partial detection, respectively, of Tn6310 upstream of attR). The tree is drawn to scale, with branch lengths measured by the number of substitutions per site. For clarity, the length of the branch linking the tree to the outgroup (SGI1) was artificially divided by 4. Vibrio cholerae and Shewanella putrefaciens are abbreviated VC and SP, respectively.
The evolutionary history of the attL-int locus of MGIVchHai6-related GIs was inferred using sequences recovered from BLAST analyses and using SGI1 as an outgroup (Fig. 1 and 3). Three strongly supported clades of elements were delineated. One representative GI of each cluster was chosen for schematic representation: MGIVchHai6 from strain HC-36A1 (green cluster), the 19-kb element GIVchHai7 from strain HC-43B1 (orange cluster), and GIVchBra1 and GIVchHai8 (17.6 kb) found in their respective strains 116063 and HE-45 (blue cluster) (Fig. 1 and 3).
All V. cholerae isolates possessing MGIVchHai6-like elements were clinical non-O1/non-O139 strains from the 2010 Haitian outbreak (Fig. 3, green cluster). Because analyses had been done on nonassembled draft genome sequences, we were not able to determine the exact structure of each member, but detection ranged from the complete MGIVchHai6 to partial detection of Tn6310 upstream of attR.
While they are very different in size, GIVchHai7 shares similar structure with the green group prototype, MGIVchHai6 (Fig. 1). In particular, the two GIs carry closely related recombination modules (attL to int/xis), as well as nearly identical right ends (pinE to attR) (Fig. 1). Nevertheless, these two elements are concluded to be distantly related as they share limited homology in the region containing the two AcaCD-dependent promoters and only in the 3′ end of the mobI gene. Surprisingly, in spite of the fact that GIVchHai7 and MGIVchHai6 carry a gene predicted to encode a DUF927 protein upstream of xis, their respective ORFs share no significant nucleotide similarity (Fig. 1). GIVchHai7 encodes a putative hsd type I RM system but lacks the resistance gene cluster insertion in pinE in MGIVchHai6, suggesting the entire MDR-conferring gene cluster of MGIVchHai6 and SGI1 is mobile and highly plastic.
The two representative members of the third group, GIVchBra1 and GIVchHai8, share identical nucleotide sequence, with the exception of three sites of single nucleotide polymorphisms (SNPs) in int and one SNP in the gene coding for the DUF927 protein (Fig. 1). These elements are closely related to GIVchHai7 as they share almost identical left and right ends. While the regions between mobI and pinE code for a type I hsd RM system in both GIVchBra1 and GIVchHai8 and GIVchHai7, their organizations differ significantly.
DISCUSSION
The availability of a massive amount of sequencing data for bacterial genomes, combined with functional data on various mobile genetic elements that has accumulated over the last four decades, allowed an extensive epidemiological survey of MDR genetic determinants circulating among pathogenic and environmental bacteria. Previous studies pointed out the major role of mobile genetic elements in propagation of MDR, notably conjugative elements such as conjugative plasmids and ICEs (5, 45–47).
Recent progress in deciphering the biology of SXT/R391 ICEs and ACPs has refined our understanding of the biology of these major drivers of the distribution of MDR among the Gammaproteobacteria (7, 11, 21, 41, 48–50). In particular, both SXT/R391 ICEs and ACPs were shown to trans-mobilize many MGIs conferring adaptive traits to their bacterial host, including MDR (21, 29, 31, 32, 51). These MGIs were found to be activated by the master activator of SXT/R391 ICEs (SetCD) or ACPs (AcaCD), thus being part of the extended regulatory network of these autonomous conjugative elements (20). Functional studies of the biology of SXT/R391 ICE-dependent MGIs has revealed key features, but the precise mechanisms allowing dissemination of ACPs-dependent MGIs remain largely unknown (31, 32). Based on recent findings, it is hypothesized that the superfamily of MGIs bearing AcaCD-activated homologs of mobI and xis, including the MGIs described here, as well as other elements previously discovered (MGIVmi1, GIVmi2, GIVpa1, and GISpu1), share a mechanism of mobilization (21, 29). Transcriptional data obtained on MGIVmi1 showed that AcaCD specifically triggers transcription of a recombination directionality factor (RDF)-encoding gene (xis), allowing excision of the MGI (21). The presence of an AcaCD-activated mobI homolog strongly suggests that, as shown for SXT/R391 ICEs and ACPs, the upstream intergenic region likely constitutes the origin of transfer (oriT) of the MGI (12, 20, 52). The current working model surmises that, upon AcaCD activation, MobI of the MGI likely recognizes its cognate oriT and recruits the ACP MobI-less relaxosome that processes the DNA to transfer one strand of the MGI from the donor cell to the recipient (20). Once in the recipient cell, the complementary strand is synthesized, and constitutive expression of the int gene allows site-specific integration of the MGI, regardless of the presence of helper ACP. Functional characterization of each gene and precise determination of the oriT locus are ongoing using different representative of the above-mentioned MGIs.
Current sequence analyses indicate that the size of the core sequence of the MGI family is ca. 8 kb and that it corresponds to DNA regions from attL to mobI and from pinE to attR (Fig. 1). pinE, which codes for a putative recombinase/invertase, is unlikely to be involved in excision, integration, or mobilization of the MGIs as it serves as an insertion site for the In36A1 resistance gene cluster, which moves by transposition (34, 53). While MGIVchHai6 carries antibiotic resistance genes, as well as a mercury resistance transposon, the majority of adaptive traits encoded by related GIs appear to be limited to RM systems (Fig. 1) (20). The presence of RM systems is considered to be ubiquitous in such GIs, likely conferring strong selective advantage to their host against bacteriophages, which thrive in aquatic environments. Nevertheless, a single event of transposition could lead to acquisition of an In104-like element and its associated MDR phenotype. Moreover, the presence of an integron likely allows further antibiotic resistance gene acquisition from, and exchange with, other integrons carried by other mobile genetic elements (46).
Although MGIs have been recently discovered, these elements are not specific to recent multidrug-resistant isolates of bacteria. For instance, GIVchBra1 was found in the genome of a V. cholerae strain isolated in 1978 in Brazil and appears to have circulated since then, with isolation of its sibling GIVchHai8 in 2010 in Haiti (Fig. 1 and 3). Such sequence conservation implies a very recent transfer event or a highly active element that does not accumulate mutations while quiescent in the chromosome of its bacterial host (54). Further large-scale analyses of sequence databases facilitated by very rapid addition of many additional bacterial genomes provided by massive sequencing, together with ongoing functional dissection of their biology, will most likely clarify the dynamics of these MGIs.
SXT/R391 ICEs were defined as major drivers of MDR among V. cholerae strains, generating a strong bias toward their detection in epidemiological reports (9, 10, 55). More recent studies highlight the presence of ACPs as MDR determinants in clinical and environmental V. cholerae strains (12–18). While ACPs are widely distributed in many species of pathogenic bacteria, their occurrence in V. cholerae is less well characterized. It is plausible that ACPs are not stably maintained in V. cholerae or that the role of ACPs in MDR acquisition by V. cholerae was overlooked. In agreement with the first hypothesis, sequence analyses of V. cholerae strains analyzed in this study using the IncA/C2 repA gene revealed that only the Indian environmental strains 116-14, 116-17a, and 116-17b contain an ACP (18). Nevertheless, ACPs likely have a more insidious role in the dissemination of MDR by not remaining in Vibrio strains but efficiently promoting circulation of MDR-associated MGIs and the adaptive traits that they may confer. In the future, circulation of MGIVchHai6 and related elements driven by ACPs should be monitored as they could enhance and accelerate dissemination of MDR in V. cholerae and related pathogens.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are described in Table 2. The strains were routinely grown in lysogeny broth (LB-Miller; EMD) at 37°C in an orbital shaker/incubator and preserved at −80°C in LB broth containing 15% (vol/vol) glycerol. For E. coli, antibiotics were used at the following concentrations: ampicillin, 100 µg/ml; chloramphenicol, 20 µg/ml; erythromycin, 200 µg/ml; gentamicin, 10 µg/ml; kanamycin, 50 µg/ml; nalidixic acid, 40 µg/ml; rifampin, 50 µg/ml; spectinomycin, 50 µg/ml; streptomycin, 200 µg/ml; sulfamethoxazole, 160 µg/ml; tetracycline, 12 µg/ml; and trimethoprim, 32 µg/ml. For V. cholerae, antibiotics were used at the following concentrations: chloramphenicol, 2 µg/ml; kanamycin, 30 µg/ml; streptomycin, 10 µg/ml; and tetracycline, 10 µg/ml. When required, bacterial cultures were supplemented with 0.3 mM dl-2,6-diaminopimelic acid (DAP) or 0.02 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG). Antibiotic susceptibility profiling and MIC determination were performed using broth dilution tests (56).
TABLE 2 .
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Reference(s) |
|---|---|---|
| Strains | ||
| V. cholerae HC-36A1 | Clinical, non-O1/non-O139, Haiti (Tabarre) 2010 (Ap Cm Kn Sp Sm Su Tc Tm) | 4 |
| E. coli | ||
| MG1655 Rf | Rfr derivative of MG1655 (Rf) | 52 |
| CAG18439 | MG1655 lacZU118 lacI42::Tn10 (Tc) | 63 |
| β2163 | (F−) RP4-2-Tc::Mu dapA::(erm-pir) (Kn Em) | 43 |
| Plasmids | ||
| pVCR94ΔX3 | Knr derivative of IncA/C plasmid pVCR94 (Kn Su) | 21 |
| pacaDC3×FLAG | pAH56::acaDC3×FLAG (Kn) | 21 |
| psetDC3×FLAG | pAH56::setDC3×FLAG (Kn) | 21, 41 |
Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Kn, kanamycin; Rf, rifampin; Sm, streptomycin; Sp, spectinomycin; Su, sulfamethoxazole; Tc, tetracycline; Tm, trimethoprim.
Bacterial conjugation assays.
Conjugation assays were performed as described by Carraro et al. (12). Donors, recipients, and exconjugants were selected on LB agar plates containing the appropriate antibiotics. If needed, additional controls were done on isolated clones using selective media, including thiosulfate-citrate-bile salts-sucrose (TCBS) agar (Difco) for identification of V. cholerae and PCR amplification of the promoting region of pVCR94ΔX3 and/or an internal fragment of ORF86 of MGIVchHai6 using primer pairs promvcrx060traIpstI.for/promvcrx060traIpstI.rev and GIVch1verif.for/GIVch1verif.rev, respectively (Table 3).
TABLE 3 .
Primers used in this study
| Name | Nucleotide sequence (5′ to 3′) | Reference |
|---|---|---|
| thdF_attBF | TATAGCCCAGCAACACCTTA | This study |
| int1_attPR | GGATCTCGTTTATGTATGCTGA | This study |
| 43_attPF | GCAATTAATGATAAAGACGGGTA | This study |
| int2_attBR | GGTATAACCGTGGTCATAAATGA | This study |
| EcU7-L12.for | ACATCTACAACAGGGCAAAG | 38 |
| Ec104D.rev | AACCATTTTGAGGTCACACA | 38 |
| promvcrx060traIpstI.for | NNNNNNCTGCAGCATCAAAAATTGTCGATGA | 21 |
| promvcrx060traIpstI.rev | NNNNNNCTGCAGCTATCGTATTTCTCGTCGCTA | 21 |
| GIVch1verif.for | TGCCATGGTCCGAAGAAGAGTC | This study |
| GIVch1verif.rev | AATCCGCGTTTTATAGGTTCCC | This study |
Molecular biology methods.
All enzymes used in this study were purchased from New England Biolabs. PCR assays were performed employing the primers listed in Table 3 under the following conditions: (i) 3 min at 94°C; (ii) 30 cycles of 30 s at 94°C, 30 s at the appropriate annealing temperature, and 1 min/kb at 68°C; and (iii) 5 min at 68°C. When necessary, PCR products were purified using an EZ-10 spin column PCR product purification kit (Biobasic) according to the manufacturer’s instructions. Sequencing reactions were performed by the Plateforme de Séquençage et de Génotypage du Centre de Recherche du CHUL (Québec, QC, Canada).
Detection of MGIVchHai6 excision.
Excision of MGIVchHai6 was detected by PCR on genomic DNA of the appropriate strains, using the primers listed in Table 3. The attR site was amplified using primer pair 43_attPF/int2_attBR in V. cholerae and pair 43_attPF/Ec104D.rev in E. coli. The attB chromosomal site was detected using thdF_attBF/int2_attBR in V. cholerae and EcU7-L12.for/Ec104D.rev in E. coli (38). The attP site carried by the extrachromosomal circular form of MGIVchHai6 was amplified using the primer pair 43_attPF/int1_attPR.
Sequence annotations.
Detection of the attL and attR attachment sites flanking MGIVchHai6 and related genomic islands was carried out using the software YASS to identify the direct repeats (57). Genes were predicted using the RAST pipeline (58), and spurious annotations were manually curated. Antibiotic resistance determinants were detected using ResFinder 2.1 (https://cge.cbs.dtu.dk/services/ResFinder/) and The Comprehensive Antibiotic Resistance Database (http://arpcard.mcmaster.ca/?q=CARD/tools/RGI). Precise locations of AcaCD binding motifs were searched using FIMO (59) against the sequence of MGIVchHai6, GIVchHai7, and GIVchBra1 with the AcaCD and SetCD MEME logos, as described elsewhere (21, 41).
Phylogenetic analyses.
Molecular phylogenetic analysis of the attL-int locus was performed in MEGA6 (60). The 1,351-bp nucleotide sequence of MGIVchHai6 encompassing 18 nt at the end of trmE that correspond to the attL site direct repeat and ending at the top codon of int was used to search for homologous sequences in the GenBank nucleotide collection database (nr/nt targeting Gammaproteobacteria and WGS targeting Vibrionaceae) using nucleotide BLAST (61). The corresponding sequence in Salmonella genomic island 1 (SGI1) was manually added to the data set. Phylogenetic analyses were computed using a nucleotide alignment generated by MUSCLE (62). The evolutionary history was inferred by using the maximum likelihood method. Initial trees for the heuristic search were obtained automatically by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and selecting the topology with superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 0.7615]). The analysis involved 37 nucleotide sequences. All positions with less than 95% site coverage were eliminated, providing a total of 1,350 positions in the final data set.
ACKNOWLEDGMENTS
We are grateful to A. Lavigueur for insightful comments on the manuscript.
This work was supported by Discovery Grants from the Natural Sciences and Engineering Council of Canada (326810-2011 and 2016-04365 to V.B.). V.B. holds a Canada Research Chair in Molecular Bacterial Genetics. This work was also supported in part by a grant from the National Institutes of Health (2RO1A1039129-11A2 to R.R.C.).
Footnotes
Citation Carraro N, Rivard N, Ceccarelli D, Colwell RR, Burrus V. 2016. IncA/C conjugative plasmids mobilize a new family of multidrug resistance islands in clinical Vibrio cholerae non-O1/non-O139 isolates from Haiti. mBio 7(4):e00509-16. doi:10.1128/mBio.00509-16.
REFERENCES
- 1.Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. 2012. Cholera. Lancet 379:2466–2476. doi: 10.1016/S0140-6736(12)60436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 3.Weil AA, Ivers LC, Harris JB. 2012. Cholera: lessons from Haiti and beyond. Curr Infect Dis Rep 14:1–8. doi: 10.1007/s11908-011-0221-9. [DOI] [PubMed] [Google Scholar]
- 4.Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. 2012. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci U S A 109:E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spagnoletti M, Ceccarelli D, Rieux A, Fondi M, Taviani E, Fani R, Colombo MM, Colwell RR, Balloux F. 2014. Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. mBio 5:e01356-14. doi: 10.1128/mBio.01356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceccarelli D, Spagnoletti M, Bacciu D, Danin-Poleg Y, Mendiratta DK, Kashi Y, Cappuccinelli P, Burrus V, Colombo MM. 2011. ICEVchInd5 is prevalent in epidemic Vibrio cholerae O1 El Tor strains isolated in India. Int J Med Microbiol 301:318–324. doi: 10.1016/j.ijmm.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh A, Ramamurthy T. 2011. Antimicrobials and cholera: are we stranded? Indian J Med Res 133:225–231. [PMC free article] [PubMed] [Google Scholar]
- 8.Carraro N, Burrus V. 2014. Biology of three ICE families: SXT/R391, ICEBs1, and ICESt1/ICESt3. Microbiol Spectr doi: 10.1128/microbiolspec.MDNA3-0008-2014. [DOI] [PubMed] [Google Scholar]
- 9.Burrus V, Quezada-Calvillo R, Marrero J, Waldor MK. 2006. SXT-related integrating conjugative element in New World Vibrio cholerae. Appl Environ Microbiol 72:3054–3057. doi: 10.1128/AEM.72.4.3054-3057.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burrus V, Marrero J, Waldor MK. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55:173–183. doi: 10.1016/j.plasmid.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Déry C, Burrus V, Waldor MK. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carraro N, Sauvé M, Matteau D, Lauzon G, Rodrigue S, Burrus V. 2014. Development of pVCR94ΔX from Vibrio cholerae, a prototype for studying multidrug resistant IncA/C conjugative plasmids. Front Microbiol 5:44. doi: 10.3389/fmicb.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan J-C, Ye R, Wang H-Q, Xiang H-Q, Zhang W, Yu X-F, Meng D-M, He Z-S. 2008. Vibrio cholerae O139 multiple-drug resistance mediated by Yersinia pestis pIP1202-like conjugative plasmids. Antimicrob Agents Chemother 52:3829–3836. doi: 10.1128/AAC.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang R, Yu D, Zhu L, Li J, Yue J, Kan B. 2015. IncA/C plasmids harboured in serious multidrug-resistant Vibrio cholerae serogroup O139 strains in China. Int J Antimicrob Agents 45:249–254. doi: 10.1016/j.ijantimicag.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Xie L, Zhang F, Ni Y, Sun J. 2015. Molecular characterization of ISCR1-mediated blaPER-1 in a non-O1, non-O139 Vibrio cholerae strain from China. Antimicrob Agents Chemother 59:4293–4295. doi: 10.1128/AAC.00166-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aberkane S, Compain F, Barraud O, Ouédraogo A-S, Bouzinbi N, Vittecoq M, Jean-Pierre H, Decré D, Godreuil S. 2015. Non-O1/non-O139 Vibrio cholerae avian isolate from France cocarrying the bla(VIM-1) and bla(VIM-4) genes. Antimicrob Agents Chemother 59:6594–6596. doi: 10.1128/AAC.00400-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folster JP, Katz L, McCullough A, Parsons MB, Knipe K, Sammons SA, Boncy J, Tarr CL, Whichard JM. 2014. Multidrug-resistant IncA/C plasmid in Vibrio cholerae from Haiti. Emerg Infect Dis 20:1951–1953. doi: 10.3201/eid2011.140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 19.Harmer CJ, Hall RM. 2015. The A to Z of A/C plasmids. Plasmid 80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Poulin-Laprade D, Carraro N, Burrus V. 2015. The extended regulatory networks of SXT/R391 integrative and conjugative elements and IncA/C conjugative plasmids. Front Microbiol 6:837. doi: 10.3389/fmicb.2015.00837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carraro N, Matteau D, Luo P, Rodrigue S, Burrus V. 2014. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet 10:e1004714. doi: 10.1371/journal.pgen.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso M-L, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, LeClerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. doi: 10.1371/journal.pone.0000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fricke WF, Welch TJ, McDermott PF, Mammel MK, LeClerc JE, White DG, Cebula TA, Ravel J. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J Bacteriol 191:4750–4757. doi: 10.1128/JB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkar A, Pazhani GP, Chowdhury G, Ghosh A, Ramamurthy T. 2015. Attributes of carbapenemase encoding conjugative plasmid pNDM-SAL from an extensively drug-resistant Salmonella enterica serovar Senftenberg. Front Microbiol 6:969. doi: 10.3389/fmicb.2015.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wailan AM, Sartor AL, Zowawi HM, Perry JD, Paterson DL, Sidjabat HE. 2015. Genetic contexts of blaNDM-1 in patients carrying multiple NDM-producing strains. Antimicrob Agents Chemother 59:7405–7410. doi: 10.1128/AAC.01319-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C, Qin S, Xu H, Xu L, Zhao D, Liu X, Lang S, Feng X, Liu H-M. 2015. New Delhi metallo-β-lactamase 1(NDM-1), the dominant carbapenemase detected in carbapenem-resistant Enterobacter cloacae from Henan Province, China. PloS One 10:e0135044. doi: 10.1371/journal.pone.0135044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahman M, Shukla SK, Prasad KN, Ovejero CM, Pati BK, Tripathi A, Singh A, Srivastava AK, Gonzalez-Zorn B. 2014. Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents 44:30–37. doi: 10.1016/j.ijantimicag.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Carraro N, Matteau D, Burrus V, Rodrigue S. 2015. Unraveling the regulatory network of IncA/C plasmid mobilization: when genomic islands hijack conjugative elements. Mob Genet Elem 5:34–38. doi: 10.1080/2159256X.2015.1045116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceccarelli D, Spagnoletti M, Hasan NA, Lansing S, Huq A, Colwell RR. 2013. A new integrative conjugative element detected in Haitian isolates of Vibrio cholerae non-O1/non-O139. Res Microbiol 164:891–893. doi: 10.1016/j.resmic.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daccord A, Ceccarelli D, Burrus V. 2010. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol Microbiol 78:576–588. doi: 10.1111/j.1365-2958.2010.07364.x. [DOI] [PubMed] [Google Scholar]
- 32.Daccord A, Mursell M, Poulin-Laprade D, Burrus V. 2012. Dynamics of the SetCD-regulated integration and excision of genomic islands mobilized by integrating conjugative elements of the SXT/R391 family. J Bacteriol 194:5794–5802. doi: 10.1128/JB.01093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellanger X, Payot S, Leblond-Bourget N, Guédon G. 2014. Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev 38:720–760. doi: 10.1111/1574-6976.12058. [DOI] [PubMed] [Google Scholar]
- 34.Levings RS, Lightfoot D, Partridge SR, Hall RM, Djordjevic SP. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J Bacteriol 187:4401–4409. doi: 10.1128/JB.187.13.4401-4409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kholodii GY, Yurieva OV, Lomovskaya OL, Gorlenko Z, Mindlin SZ, Nikiforov VG. 1993. Tn5053, a mercury resistance transposon with integron’s ends. J Mol Biol 230:1103–1107. doi: 10.1006/jmbi.1993.1228. [DOI] [PubMed] [Google Scholar]
- 36.Loenen WA, Dryden DT, Raleigh EA, Wilson GG. 2014. Type I restriction enzymes and their relatives. Nucleic Acids Res 42:20–44. doi: 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray NE. 2000. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol Mol Biol Rev 64:412–434. doi: 10.1128/MMBR.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doublet B, Boyd D, Mulvey MR, Cloeckaert A. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol 55:1911–1924. doi: 10.1111/j.1365-2958.2005.04520.x. [DOI] [PubMed] [Google Scholar]
- 39.Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A. 2006. The genetics of Salmonella genomic island 1. Microbes Infect 8:1915–1922. doi: 10.1016/j.micinf.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 40.Boyd DA, Shi X, Hu QH, Ng LK, Doublet B, Cloeckaert A, Mulvey MR. 2008. Salmonella genomic island 1 (SGI1), variant SGI1-I, and new variant SGI1-O in Proteus mirabilis clinical and food isolates from China. Antimicrob Agents Chemother 52:340–344. doi: 10.1128/AAC.00902-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poulin-Laprade D, Matteau D, Jacques P-É, Rodrigue S, Burrus V. 2015. Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon. Nucleic Acids Res 43:2045–2056. doi: 10.1093/nar/gkv071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beaber JW, Hochhut B, Waldor MK. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol 184:4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Demarre G, Guérout A-M, Matsumoto-Mashimo C, Rowe-Magnus DA, Marlière P, Mazel D. 2005. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol 156:245–255. doi: 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Hochhut B, Marrero J, Waldor MK. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J Bacteriol 182:2043–2047. doi: 10.1128/JB.182.7.2043-2047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Juhas M. 2015. Horizontal gene transfer in human pathogens. Crit Rev Microbiol 41:101–108. doi: 10.3109/1040841X.2013.804031. [DOI] [PubMed] [Google Scholar]
- 46.Escudero JA, Loot C, Nivina A, Mazel D. 2015. The integron: adaptation on demand. Microbiol Spectr 3:MDNA3-0019-2014. doi: 10.1128/microbiolspec.MDNA3-0019-2014. [DOI] [PubMed] [Google Scholar]
- 47.Colomer-Lluch M, Jofre J, Muniesa M. 2011. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One 6:e17549. doi: 10.1371/journal.pone.0017549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burrus V, Waldor MK. 2003. Control of SXT integration and excision. J Bacteriol 185:5045–5054. doi: 10.1128/JB.185.17.5045-5054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garriss G, Waldor MK, Burrus V. 2009. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet 5:e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carraro N, Poulin D, Burrus V. 2015. Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: the line between ICEs and conjugative plasmids is getting thinner. PLoS Genet 11:e1005298. doi: 10.1371/journal.pgen.1005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daccord A, Ceccarelli D, Rodrigue S, Burrus V. 2013. Comparative analysis of mobilizable genomic islands. J Bacteriol 195:606–614. doi: 10.1128/JB.01985-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ceccarelli D, Daccord A, René M, Burrus V. 2008. Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J Bacteriol 190:5328–5338. doi: 10.1128/JB.00150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plasterk RH, van de Putte P. 1985. The invertible P-DNA segment in the chromosome of Escherichia coli. EMBO J 4:237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Touchon M, Bobay L-M, Rocha EP. 2014. The chromosomal accommodation and domestication of mobile genetic elements. Curr Opin Microbiol 22:22–29. doi: 10.1016/j.mib.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 55.Waldor MK, Tschäpe H, Mekalanos JJ. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol 178:4157–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen JH, Ferraro MJ. 2009. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49:1749–1755. doi: 10.1086/647952. [DOI] [PubMed] [Google Scholar]
- 57.Noé L, Kucherov G. 2005. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res 33:W540–W543. doi: 10.1093/nar/gki478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 62.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev 53:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]