ABSTRACT
Staphylococcus aureus is the leading cause of skin and soft tissue infections, bacteremia, osteomyelitis, and endocarditis in the developed world. The ability of S. aureus to cause substantial disease in distinct host environments is supported by a flexible metabolism that allows this pathogen to overcome challenges unique to each host organ. One feature of staphylococcal metabolic flexibility is a branched aerobic respiratory chain composed of multiple terminal oxidases. Whereas previous biochemical and spectroscopic studies reported the presence of three different respiratory oxygen reductases (o type, bd type, and aa3 type), the genome contains genes encoding only two respiratory oxygen reductases, cydAB and qoxABCD. Previous investigation showed that cydAB and qoxABCD are required to colonize specific host organs, the murine heart and liver, respectively. This work seeks to clarify the relationship between the genetic studies showing the unique roles of the cydAB and qoxABCD in virulence and the respiratory reductases reported in the literature. We establish that QoxABCD is an aa3-type menaquinol oxidase but that this enzyme is promiscuous in that it can assemble as a bo3-type menaquinol oxidase. However, the bo3 form of QoxABCD restricts the carbon sources that can support the growth of S. aureus. In addition, QoxABCD function is supported by a previously uncharacterized protein, which we have named CtaM, that is conserved in aerobically respiring Firmicutes. In total, these studies establish the heme A biosynthesis pathway in S. aureus, determine that QoxABCD is a type aa3 menaquinol oxidase, and reveal CtaM as a new protein required for type aa3 menaquinol oxidase function in multiple bacterial genera.
IMPORTANCE
Staphylococcus aureus relies upon the function of two terminal oxidases, CydAB and QoxABCD, to aerobically respire and colonize distinct host tissues. Previous biochemical studies support the conclusion that a third terminal oxidase is also present. We establish the components of the S. aureus electron transport chain by determining the heme cofactors that interact with QoxABCD. This insight explains previous observations by revealing that QoxABCD can utilize different heme cofactors and confirms that the electron transport chain of S. aureus is comprised of two terminal menaquinol oxidases. In addition, a newly identified protein, CtaM, is found to be required for the function of QoxABCD. These results provide a more complete assessment of the molecular mechanisms that support staphylococcal respiration.
INTRODUCTION
Staphylococcus aureus is a significant cause of morbidity and mortality worldwide (1). In the United States, S. aureus is the leading cause of skin and soft tissue infections, endocarditis, septicemia, and osteomyelitis (1, 2). In order to colonize these diverse niches, S. aureus relies upon a dynamic metabolism that supports proliferation by overcoming the unique challenges presented by the host innate immune response (3–5). S. aureus utilizes three energy-generating pathways to support proliferation: aerobic respiration, fermentation, and anaerobic respiration (6–8). Previous reports demonstrate that aerobic respiration is required for colonization of the heart and liver (3, 4). The enzymes that catalyze the final reaction of aerobic respiration, the reduction of oxygen to water, are called terminal oxidases (9, 10). Most bacteria encode more than one terminal oxidase in their genome, with expression dependent on the growth conditions. This allows an organism to adapt its respiratory chain to changes in its environment, such as the oxygen concentration. The S. aureus genome encodes two terminal oxidases, cydAB and qoxABCD (3). This branched respiratory chain supports organ-specific colonization by S. aureus, as mutants with mutations that inactivate cydAB are unable to colonize the murine heart and impairment of qoxABCD dramatically decreases the capacity to infect the liver (3, 4).
In S. aureus, inactivation of cydAB together with qoxABCD arrests respiration and induces the small colony variant (SCV) phenotype (3, 11). This finding supports the conclusion that CydAB and QoxABCD are the exclusive terminal oxidases that power aerobic respiration in this pathogen. CydAB and QoxABCD are representatives of the two major superfamilies of respiratory oxygen reductases. cydAB encodes a bd-type menaquinol oxidase (9, 12), and qoxABCD encodes a menaquinol oxidase that is a member of the heme-copper oxygen reductase superfamily (13). Prior to the availability of the genome sequence of S. aureus, biochemical studies indicated that there were o-type and/or aa3-type terminal oxidases in the organism, but there is no mention of cytochrome bd (14–18). Typically, cytochrome bd can be identified spectroscopically due to the presence of heme D and by the resistance of the enzyme to cyanide (9). In fact, cytochrome bd cyanide resistance is a feature that distinguishes S. aureus from nonpathogenic staphylococci; S. aureus cytochrome bd is more sensitive to cyanide than the cytochrome bd expressed by the avirulent Staphylococcus carnosus (19). It was not until 2014 that the spectroscopic signature of S. aureus cytochrome bd was reported (20). Taken together, there is biochemical evidence for three types of respiratory oxygen reductases in S. aureus, a bd type, an o type, and/or an aa3 type, despite genomic evidence for only cydAB and qoxABCD. Here, we address the origin of the three putative terminal oxidases and delineate the cyanide sensitivity of the S. aureus cytochrome bd. It is shown that the S. aureus cytochrome bd is resistant to cyanide below 1 mM and that the QoxABCD enzyme exists in two forms. One form contains two equivalents of heme A (aa3 type), and the other incorporates hemes B and O (bo3 type). Both forms are functional and sensitive to cyanide. We also identify a new gene in the S. aureus genome that is conserved in many Firmicutes and is required for the assembly of functional QoxABCD in any form. This gene (locus NWMN_0982), which we name ctaM, is adjacent to the gene encoding the heme O synthase, ctaB. These results define the molecular requirements for terminal oxidase function in S. aureus, provide an explanation for the previous observations describing the presence of an o-type reductase in the staphylococcal respiratory chain, and establish a conserved protein involved in terminal oxidase assembly.
RESULTS
QoxABCD is an aa3-type terminal oxidase and CydAB is a canonical cyanide-resistant cytochrome bd.
The composition of the aerobic respiratory chain depends upon both the growth medium and the phase of growth at which the cells are harvested. Table 1 shows the comparison of the properties of membranes isolated from the wild type (WT) and different mutants of S. aureus, each grown in LB with 0.5% glucose and harvested after 8 h. The rate of O2 consumption (per mg of membrane protein) in the presence of NADH was measured in the presence and absence of 100 µM potassium cyanide (KCN). About 40% of the O2 consumption is inhibited by 100 µM KCN in the WT, whereas in the ΔcydA mutant, 100% inhibition is observed. In contrast, in the ΔqoxA mutant, 100% of the activity remains in the presence of 100 µM KCN. Hence, the oxygen reductase encoded by cydAB is cyanide resistant and the oxygen reductase encoded by qoxABCD is cyanide sensitive. The reduced-minus-oxidized difference spectra of the membranes from both the WT and ΔcydA strains (Fig. 1) clearly show the spectroscopic features of an aa3-type oxygen reductase (peaks at about 443 nm and 606 nm), which are absent from the spectrum of the ΔqoxABCD strain. The spectroscopic signature typical of heme D, a peak in the reduced-minus-oxidized spectrum near 630 nm, is not evident in any of the spectra in Fig. 1. This feature was previously observed with membranes from the WT strain, as shown in Fig. S4 in reference 20, so its absence is likely due to its relatively small amount and the noise in the spectra shown in Fig. 1. These results establish that qoxABCD encodes a cyanide-sensitive, a-type reductase.
TABLE 1 .
Respiratory activities of S. aureus terminal oxidase assembly mutants
| Strain | Terminal oxidase(s) present | Membrane respiratory activity |
|
|---|---|---|---|
| After addition of 100 µM KCN (%) | Relative to that of WT | ||
| WT | aa3 and bd | 40 | 1.0 |
| ΔcydA mutant | aa3 | 0 | 0.8 |
| ΔqoxA mutant | bd | 100 | 1.1 |
| ΔctaA mutant | bo3 and bd | 32 | 1.0 |
| ΔctaB mutant | bd | 95 | 1.0 |
| ΔctaM mutant | bd | 98 | 0.9 |
| ΔcydAB ΔctaA mutant | bo3 | 7 | 0.5 |
| ΔqoxB ΔctaA mutant | bd | 96 | 1.3 |
FIG 1 .
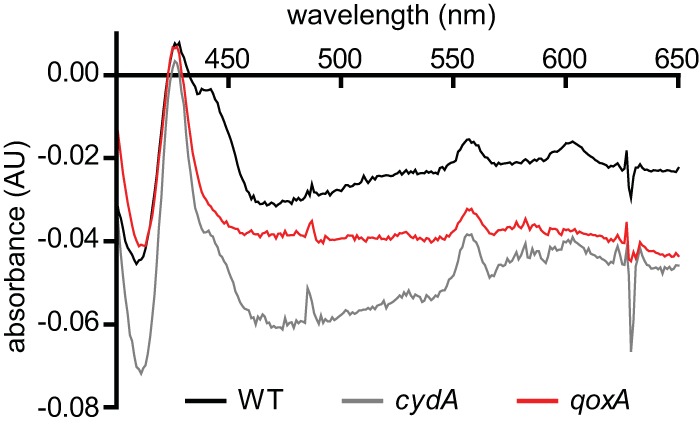
The cytochrome profiles of terminal oxidase mutants reveal the preferred heme cofactors utilized by each enzyme. The reduced-minus-oxidized spectra of Staphylococcus aureus and isogenic terminal oxidase mutants (ΔcydA and ΔqoxA) are shown. Heme A peaks are located at 430 nm and 609 nm. AU, arbitrary units.
QoxABCD assembles as a functional oxidase in the absence of heme A.
It is expected that the aa3-type menaquinol oxidase encoded by qoxABCD incorporates two equivalents of heme A. The conversion of heme B to heme A is catalyzed by CtaB and CtaA, which have been characterized from other bacteria, including Bacillus subtilis (21, 22). The reaction sequence is shown in Fig. 2A, showing heme B converted to heme O by CtaB and then to heme A via CtaA. Both the ctaB and ctaA genes are present in S. aureus and, in addition, there is a third gene adjacent to ctaB which we have named ctaM (locus NWMN_0982), whose function is not known (Fig. 2B).
FIG 2 .
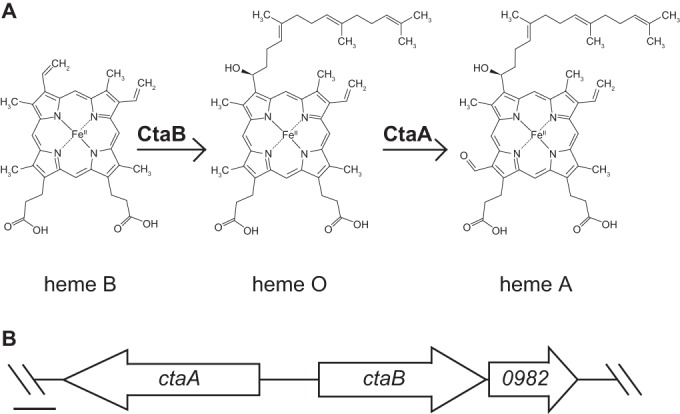
CtaB is predicted to be the heme O synthase in S. aureus. (A) Putative heme cofactor biosynthesis pathway in S. aureus. CtaA is the heme A synthase in S. aureus (21). (B) Genomic locus containing the heme cofactor biosynthesis genes in S. aureus strain Newman. 0982 denotes the locus tag NWMN_0982 (referred to herein as ctaM). Scale bar = 250 bp.
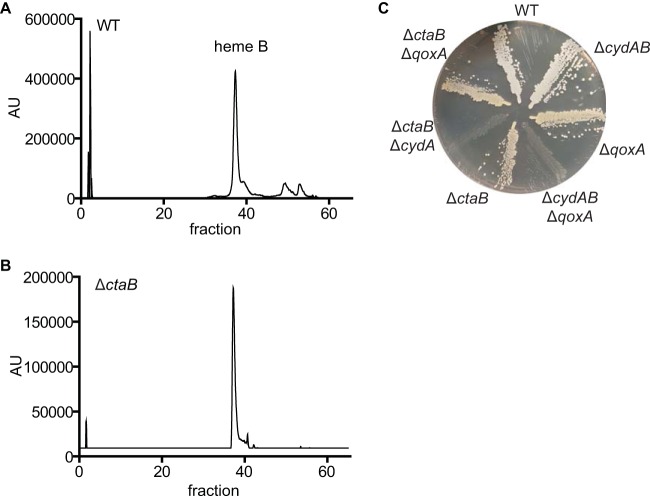
A mutant with a deletion of ctaB was constructed, and the ΔctaB strain behaves like the ΔqoxA strain insofar as the remaining respiratory activity is entirely resistant to the presence of 100 µM KCN (Table 1). Figure 3A shows a liquid chromatography-mass spectrometry (LC-MS) analysis of the hemes extracted from the membranes of the WT strain, excluding heme D, which is not observed under these conditions. The major peak is due to heme B, expected to be a component of cytochrome bd, as well as succinate dehydrogenase. Two smaller peaks elute at positions indicating that they represent heme O and heme A. This is consistent with the analysis of the hemes present in the membranes of the ΔctaB strain, in which these two small peaks are absent (Fig. 3B). In addition, the double ΔctaB ΔcydA mutant exhibits the SCV phenotype (Fig. 3C), indicating that this strain is respiration deficient. These results demonstrate that the synthesis of heme O is essential for the assembly of functional QoxABCD.
FIG 3 .
Heme O is synthesized by CtaB and is required for QoxABCD function. (A) Liquid chromatography (LC) analysis of membranes isolated from wild-type (WT) staphylococci. Tandem mass spectroscopy was used to identify heme B in fraction 37. (B) LC analysis of membranes isolated from an isogenic ΔctaB deletion mutant. (C) Overnight cultures of WT S. aureus and isogenic mutants were grown at 37°C in tryptic soy broth (TSB) and streaked for isolated colonies on tryptic soy agar (TSA). The ΔcydAB ΔqoxA and ΔctaB ΔcydA mutants are respiration-arrested small colony variants.
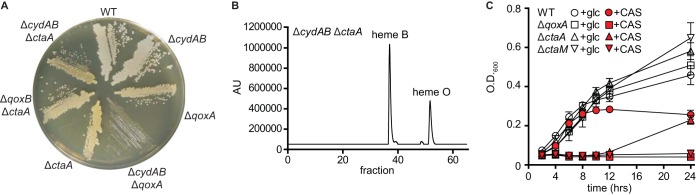
Unexpectedly, the ΔctaA strain, which also cannot synthesize heme A (Fig. 2) (21), has 32% of its respiratory activity inhibited by 100 µM KCN (Table 1), indicating that a cyanide-sensitive terminal oxidase is present, similar to what is observed in the WT strain. The ΔctaA ΔcydAB double mutant does not exhibit the SCV phenotype (Fig. 4A) and is not respiration deficient. Furthermore, the respiratory activity of the ΔctaA ΔcydAB double mutant is virtually entirely inhibited by 100 µM KCN (Table 1). Therefore, blocking the conversion of heme O to heme A (Fig. 2) does not prevent QoxABCD from assembling to an active, cyanide-sensitive oxygen reductase. This result implies that QoxABCD can assemble in the absence of heme A by incorporating heme B and heme O. LC-MS analysis of the hemes extracted from membranes from the ΔctaA strain shows a large peak identified as heme O, supporting this supposition (Fig. 4B). Previous studies on Bacillus cereus (23, 24) showed that preventing the synthesis of heme A results in the assembly of the aa3-type menaquinol oxidase into a bo3-type menaquinol oxidase. Our observations indicate that S. aureus responds in a similar manner.
FIG 4 .
In the absence of heme A synthesis, QoxABCD utilizes heme O. (A) Overnight cultures of WT S. aureus and isogenic mutants were grown at 37°C in TSB and streaked for isolated colonies on TSA. The ΔcydAB ΔqoxA mutant is a respiration-arrested small colony variant. (B) LC analysis of membranes isolated from a ΔcydAB ΔctaA mutant. Tandem mass spectroscopy was used to identify heme B in fraction 37 and heme O in fraction 52. (C) The growth of the WT and isogenic mutants of S. aureus was monitored over time by optical density at 600 nm. The final concentrations of glucose (glc) and Casamino Acids (CAS) added to the growth medium were 25 mM (open symbols) and 0.5% (red symbols), respectively. The average results from three independent experiments are shown. Error bars represent one standard deviation from the mean.
QoxABCD utilizes heme O at the expense of carbon restriction.
The ability of B. subtilis to grow in different media is impacted by mutations that eliminate one or more of the respiratory oxygen reductases (25). To determine whether the inability to synthesize heme A leads to any growth phenotype in S. aureus, WT, ΔqoxA, and ΔctaA strains were grown in a minimal medium supplemented with glucose or Casamino Acids (CAS) as the carbon source (Fig. 4C). When glucose is supplied as a carbon source, the WT, ΔctaA, and ΔqoxA strains grow equally well (Fig. 4C). However, when the cells are cultured in a minimal medium supplemented with CAS, no growth of the ΔqoxA strain is observed after 24 h and the growth of the ΔctaA strain exhibits a lag of at least 12 h before growth is observed (Fig. 4C).
CtaM is required for QoxABCD function.
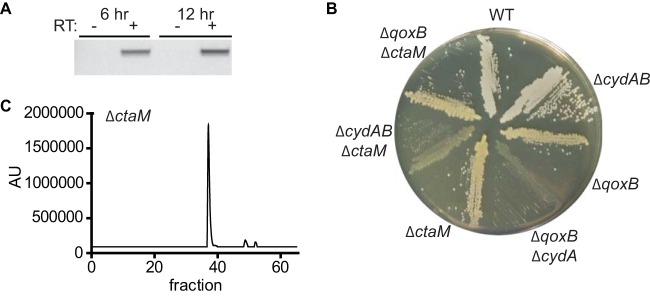
The gene adjacent to ctaB, NWMN_0982 (locus tag in S. aureus strain Newman), encodes a putative membrane protein of unknown function (Fig. 2B). The location adjacent to ctaB suggests that NWMN_0982 also facilitates QoxABCD function and that these genes are likely cotranscribed. To verify this, primers were designed to amplify between the coding regions of ctaB and NWMN_0982. Amplification between ctaB and NWMN_0982 was observed when cDNA was prepared from S. aureus grown to stationary phase and was dependent on reverse transcriptase (Fig. 5A). The finding that ctaB and NWMN_0982 are cotranscribed demonstrates that these genes reside in an operon and supports the hypothesis that these genes encode proteins that function in the same pathway. Consistent with this, we refer to NWMN_0982 as ctaM (cytochrome assembly). To test the hypothesis that ctaM supports QoxABCD function, ctaM was inactivated in the ΔcydAB strain. The ΔcydAB ΔctaM mutant displays the SCV phenotype (Fig. 5B), and a plasmid-encoded copy of ctaM complements the respiration-arrested growth of the ΔcydAB ΔctaM strain (see Fig. S2B in the supplemental material).
FIG 5 .
QoxABCD function is supported by CtaM. (A) PCR was performed on cDNA enriched from staphylococci grown at 37°C to early stationary (6 h) or late stationary (12 h) phase. RT, reverse transcriptase. (B) Overnight cultures of WT S. aureus and isogenic mutants were grown at 37°C in TSB and streaked for isolated colonies on TSA. The ΔqoxB ΔcydA and ΔcydAB ΔctaM double mutants are respiration-arrested small colony variants. (C) LC analysis of membranes isolated from the ΔctaM strain.
These results demonstrate that CtaM is required for the assembly of functional QoxABCD. Accordingly, the ΔqoxB ΔctaM double mutant does not exhibit the SCV phenotype, indicating that CtaM is not required for CydAB activity (Fig. 5B). Consistent with this, the respiratory activity of the ΔctaM strain is resistant to KCN, revealing that CydAB is the only terminal oxidase active in this mutant (Table 1). Furthermore, the ΔctaM strain behaves like the ΔqoxA strain in that neither strain exhibits growth over a 24-h period in liquid broth consisting of a minimal medium supplemented with CAS (Fig. 4C). The hemes extracted from membranes prepared from the ΔctaM strain are similar to the hemes isolated from the WT, showing a predominant LC peak due to heme B and small peaks that appear to correspond to heme O and heme A (Fig. 5C). These results suggest that CtaM is not required for the function of CtaA or CtaB but, rather, is needed either for the insertion of the hemes into the enzyme or for the activity of the final, assembled enzyme.
ctaM is conserved in respiring Firmicutes that synthesize heme O and heme A.
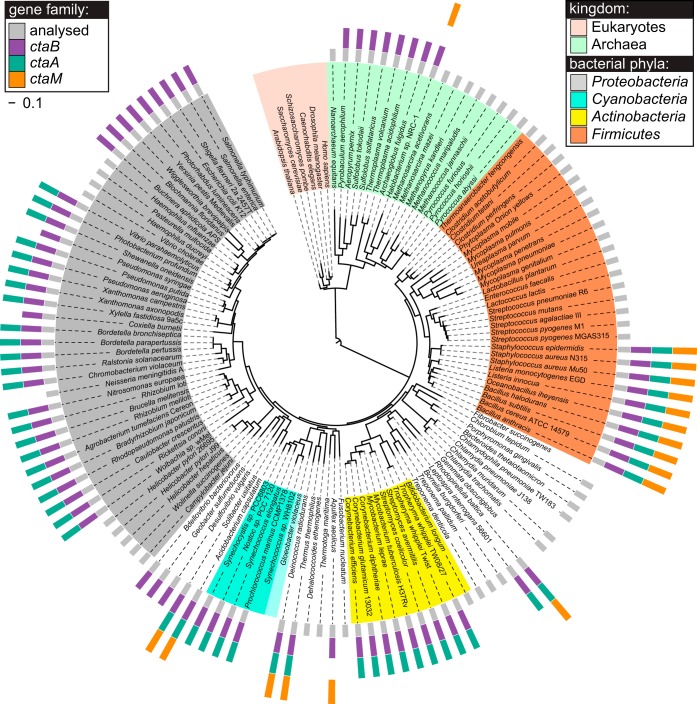
These results establish that ctaM is required for QoxABCD terminal oxidase function and imply that ctaM is conserved in other species of bacteria that utilize QoxABCD to respire. Phylogenic investigation in the SEED database (26, 27) determined that ctaM is conserved in many diverse species of bacteria, as the analysis of a set of 978 nonredundant representative prokaryotic genomes reveals a ctaM homologue in 120 organisms (12% of the genomes analyzed). The results of this analysis are illustrated in Fig. 6 and are available in full online in the SEED Subsystem “Heme O and Heme A biosynthesis (with selected terminal oxidases)” (28).
FIG 6 .
ctaB, ctaA, and ctaM homologues are conserved among respiring Firmicutes and show a strong tendency to co-occur in the same genome. The phylogenic distribution of ctaM (orange, outer ring) was mapped onto the Tree of Life (29, 45). The distributions of ctaB, the gene that encodes the heme O synthase (purple, outer ring), and of ctaA, the gene that encodes the heme A synthase (teal, outer ring), are also presented.
The presence of ctaM in the Archaea, Firmicutes, Aquificae, and Planctomycetes, some of the oldest evolutionary taxa, supports the conclusion that CtaM is an ancient protein. Consistent with this, ctaM is not present in Beta- or Gammaproteobacteria (the youngest taxa) but does occur in a few representatives of the Alpha- and Delta-/Epsilonproteobacteria (the older subgroups of Proteobacteria) (29). Notably, within each taxon, CtaM proteins are found only in genomes encoding aa3-type terminal oxidases and never in nonrespiring organisms or organisms utilizing other types of terminal oxidases (28). In particular, within the Firmicutes, ctaM is conserved among those that respire but not in obligate anaerobes like the Clostridiales or aerotolerant nonrespiring bacteria like the lactic acid bacteria (LAB). In some cases, when grown in medium supplemented with the proper cofactors, LAB respire by utilizing bd-type oxidase (CydAB) (30). This finding supports the idea that CtaM specifically interacts with QoxABCD. Additional support for this notion is provided by the fact that in all of the organisms in our representative set that encode qoxABCD (31 out of 978), ctaM is also present without exception. A similar tendency for co-occurrence in the same genome is observed between ctaM and the genes encoding cytochrome c oxidase subunits CoxI, -II, -III, and -IV and the alternative cytochrome c oxidase CoxMNOP, both aa3-type enzymes (10, 13). Furthermore, this functional association inferred by co-occurrence is strengthened by colocalization of the corresponding genes in many genomes (see Subsystem spreadsheet at reference 28). On the other hand, the ctaM homologues show no tendency to co-occur or to colocalize with genes that encode bo3-type ubiquinol oxidase (cyoABCD), bd-type ubiquinol oxidase (cydABCD), cytochrome bc1 complexes (mcb, ucb, or cbsAB-soxLN), or cbb3-type cytochrome c oxidase (ccoPONQ) (28).
The fact that only a-type oxidases, such as qoxABCD and coxMNOP, are linked with ctaM homologues by strong comparative genomics evidence implies a functional association of the CtaM protein family with heme A synthesis. Consistent with this, the co-occurrence of ctaM with ctaB (heme O synthase) has no exceptions (Fig. 6). However, 4 of the 120 genomes (3%) that contain ctaM do not have a ctaA (heme A synthase) homologue. These four exceptions are all Archaea, organisms in which heme A synthesis is not well understood (31, 32), and therefore do not contradict the detected functional association of the CtaM family with enzymes of heme O and A biosynthesis. In addition to a nearly absolute co-occurrence between ctaM and ctaA and/or ctaB, the ctaM genes show a strong tendency to colocalize with ctaB and/or ctaA genes within bacterial chromosomes: this is true in 79 of 120 genomes (66%) that contain ctaM within the set of genomes analyzed. In total, the phylogenic profile, colocalization on the chromosome, and functional analysis of the ctaM family clearly demonstrate that the CtaM protein functions to support the activity of a-type terminal oxidases in many species.
DISCUSSION
In order to survive and proliferate in the presence of the host immune response, bacterial pathogens fine-tune their metabolism for maximal energy production and growth. The utilization of multiple terminal oxidases is one mechanism that allows pathogens to optimize metabolism in the host environment (3, 33, 34). These branched aerobic respiratory chains allow for rapid adaption to the availability of oxygen and changes in available carbon sources. The availability of oxygen is likely a major environmental cue for S. aureus as it transitions from a commensal organism colonizing the skin or nasopharynx to a systemic pathogen proliferating within tissue abscesses in different host organs. Certainly, the amount of oxygen on the skin and within the nose is dramatically different than in an abscess, which has recently been demonstrated to be a hypoxic environment (35). Utilizing more than one terminal oxidase allows S. aureus to generate the maximum amount of energy to proliferate in distinct host environments, a conclusion supported by the finding that S. aureus mutants restricted to a single terminal oxidase are severely impaired for colonization of the murine heart or liver (3, 4). The findings presented herein conclusively demonstrate that S. aureus utilizes two terminal oxidases to respire aerobically, QoxABCD and CydAB.
The S. aureus CydAB is resistant to micromolar concentrations (100 µM) of cyanide, distinguishing it from the cyanide-sensitive cytochrome aa3 encoded by qoxABCD. This profile of cyanide resistance is typical of canonical representatives of the cytochrome bd and cytochrome aa3 family of oxidases. However, elegant work by Voggu et al. demonstrated that nonpathogenic S. carnosus can grow in the presence of 1.5 mM cyanide, whereas S. aureus cannot (19). This is likely due to a greater sensitivity to cyanide of the cytochrome bd from S. aureus. Given the importance of CydAB to S. aureus virulence, a particularly intriguing notion raised from these findings is that the cyanide sensitivity of S. aureus, compared to that of S. carnosus, is an expense paid to enhance fitness during colonization of the human host.
QoxABCD is promiscuous regarding its heme cofactors and can be assembled either as an aa3-type oxidase using two equivalents of heme A or as a bo3-type oxidase using one equivalent each of heme B and heme O. In the current work, QoxABCD was constrained to assemble as the bo3-type enzyme by genetically eliminating heme A biosynthesis (ΔctaA mutation), but a number of previous studies identified an o-type oxidase in the WT strain (14–18). Such promiscuity could possibly be an advantage, since the CtaA enzyme requires O2 to synthesize heme A (36). This might provide an advantage to S. aureus when shifting from an anoxic to oxic environment.
However, although QoxABCD is assembled as a functional enzyme in the ΔctaA strain, the mutation is not benign, indicating some significant differences either in the properties of the assembled bo3 and aa3 variants of QoxABCD or in the efficiency of assembly or stability of the different forms of the enzyme. It is also possible that the inability to convert heme O to heme A reflects a different function for heme A, though none are known, or toxicity of heme O. The inability of the ΔctaA strain (no heme A) to grow rapidly when CAS is substituted for glucose suggests that there is no functional respiratory oxidase present, and in the absence of a fermentable substrate (glucose), aerobic respiration is essential. Presumably, the CydAB oxygen reductase is not expressed under the conditions employed or it would be sufficient to support aerobic growth by respiration. It appears to take at least 12 h for the ΔctaA strain to assemble a sufficient amount of QoxABCD to permit growth. Understanding the drastic difference in the growth phenotype will take more study, including the control of gene expression of the various respiratory components.
The elimination of the synthesis of heme A in the ΔctaA strain also has consequences during infection, most prominently that this strain cannot colonize the murine liver (4). The reason for this is not clear but may reflect different concentrations of O2 required for the synthesis of heme A and for aerobic respiration. The growth nutrients present, as indicated by the contrast between glucose and CAS in vitro, may also play a role. This study validates further investigation of the requirements for QoxABCD and CydAB function as an approach to define the environmental conditions encountered by S. aureus during infection.
A key finding of the current work is that the assembly of a functional QoxABCD oxygen reductase requires CtaM. Our data suggest that CtaM may act as a chaperone for the insertion of heme O or heme A into QoxABCD. In support of this prediction, ctaM is highly conserved in respiring Firmicutes that also encode qoxABCD (Fig. 6). Additionally, heme O and heme A are still synthesized in the ctaM mutant (Fig. 5C), but QoxABCD is not functional (Fig. 5). This finding suggests that the hemes are not trafficked to the QoxABCD complex. However, it is currently unknown whether the hemes in the ctaM mutant reside within an inactive form of the QoxABCD complex or are restrained in CtaB or CtaA. The latter possibility would provide additional support for the hypothesis that CtaM acts as a heme chaperone and is required to traffic heme A or heme O to QoxABCD. The fact that B. subtilis ctaB and ctaA can be ectopically expressed and are able to synthesize heme O and heme A in Escherichia coli, an organism that does not express a QoxABCD oxidase, supports the possibility that the heme O and heme A are stable in the absence of an active QoxABCD complex (22). Alternatively, the presence of heme O and heme A may indicate that the function of CtaM is to assemble QoxABCD independent of heme. Further study is needed to determine the precise mechanism by which CtaM supports the assembly of QoxABCD.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are described in Table 2. All strains are derivatives of the human clinical isolate S. aureus Newman or the human clinical isolate USA300 LAC JE2 adapted for laboratory use (37, 38). The ΔcydAB and ΔctaB isogenic in-frame deletion mutants were constructed using previously described methods (39). The cydA, qoxA, ctaA, and ctaM transposon mutants were obtained from the Nebraska transposon library (NE117, NE92, NE769, and NE1084, respectively) (38). PCR confirmed that the transposon insertion mapped to the respective gene. Backcross strains of the transposon-disrupted cydA, qoxA, ctaA, and ctaM alleles into WT S. aureus Newman or USA300 LAC JE2 were produced via phage φ85-mediated transduction (38). The backcrossed strains were used in the studies described here. Phage φ85-mediated transduction of the cydA and qoxA transposon-disrupted alleles was used to create the ΔctaB ΔcydA and ΔctaB ΔqoxA double mutants. Likewise, phage φ85-mediated transduction of the ctaA and ctaM transposon-disrupted allele was also used to construct the ΔcydAB ΔctaA, ΔcydAB ΔctaM, ΔqoxB ΔctaA, and ΔqoxB ΔctaM double mutants. For complementation studies, the ctaB gene was PCR amplified and cloned into the pOS1 Plgt plasmid using primers 5′ GCGCTCGAGATGAGCAAAGAGCATACTTTGTC 3′ and 5′ GCGGGATCCCTAGATCAAAGTAAGTAATGAAAC 3′ and the restriction enzymes XhoI and BamHI. BamHI and XhoI were also used to clone the ctaM gene into pOS1 Plgt. ctaM was PCR amplified using primers 5′ GCGCTCGAGATGGGCGTTCCAATTTTACCA 3′ and 5′ GCGGGATCCTTAATGACCAAATGTTGCTTTAAT 3′. Antibiotic selection of erythromycin cassette-containing resistant recipient cells was achieved with 10 µg ml−1 erythromycin. Antibiotic selection of strains containing the pOS Plgt plasmid was achieved by using 10 µg ml−1 of chloramphenicol. Bacteria were routinely grown in tryptic soy broth (TSB) at 37°C. All strains were diluted 1:100 from overnight cultures into fresh TSB-containing 96-well round-bottom plates, and growth was measured by optical density at 600 nm. The PN minimal medium was prepared as previously described (38). The cells were grown overnight in TSB, washed thrice, and resuspended in PN minimal medium lacking a carbon source. Glucose and CAS were supplemented at the indicated concentrations.
TABLE 2 .
S. aureus strains used in this study
| Strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Newman strains | Wild type | 37 |
| qoxB mutant | ΔqoxB | 3 |
| qoxA mutant | qoxA::erm | This study |
| cydAB mutant | ΔcydAB | This study |
| cydA mutant | cydA::erm | This study |
| ctaB mutant | ΔctaB | This study |
| ctaA mutant | ctaA::erm | This study |
| ctaM mutant | ctaM::erm | This study |
| ctaB cydA mutant | ΔctaB cydA::erm | This study |
| ctaB qoxA mutant | ΔctaB qoxA::erm | This study |
| qoxB ctaM mutant | ΔqoxB ctaM::erm | This study |
| cydAB ctaM mutant | ΔcydAB ctaM::erm | This study |
| qoxB ctaA mutant | ΔqoxB ctaA::erm | This study |
| cydAB ctaA mutant | ΔcydAB ctaA::erm | This study |
| JE2 strains | Wild type | 38 |
| qoxA mutant | qoxA::erm | 38 |
| cydA mutant | cydA::erm | 38 |
| ctaA mutant | ctaA::erm | 38 |
| ctaM mutant | ctaM::erm | 38 |
Membrane preparations.
Cells were grown in LB plus 0.5% glucose for 8 h. Membranes were obtained by incubating cells with lysostaphin for 2 h at 37°C and then passing them more than five times through a microfluidizer at a pressure of 80,000 lb/in2. The cell extract was centrifuged at 14,000 × g for 10 min to remove the unbroken cells. Membranes were obtained by centrifugation at 230,000 × g for 4 h. Pellets, containing 30 to 40 mg/ml of protein, were resuspended in sodium phosphate buffer, pH 7.5, 50 mM NaCl, 10% glycerol, and the suspension was stored at −80°C. The NADH-reduced minus air-oxidized difference spectrum of S. aureus membranes was obtained at room temperature using an Agilent Technologies spectrophotometer (model 8453). Reduction was achieved by the addition of 5 mM NADH.
Heme extraction and identification.
The hemes from S. aureus membranes were extracted and analyzed using high-performance liquid chromatography (HPLC) elution profiles according to established protocols (40, 41). One milliliter of the membrane suspension was solubilized by the addition of a stock solution of 20% DDM (dodecyl-β-d-maltoside) dropwise to a final concentration of 1%. The solution was incubated at 4°C for 2 h with mild agitation. The suspension was cleared by centrifugation at 230,000 × g for 1 h. The supernatant was mixed with 9 ml of acetone-HCl (19:1, vol/vol) and shaken on a rotary shaker for 20 min at room temperature. The mixture was centrifuged at 14,000 × g for 2 min, followed by the addition of 20 ml of ice-cold water and 6 ml of ethyl acetate to the supernatant. The water-ethyl acetate mixture was vortexed and centrifuged again for 10 min at 4°C. The ethyl acetate phase was recovered, and the solvent evaporated by flushing it with nitrogen gas. The residues were dissolved in 0.05 to 0.10 ml of acetonitrile and stored at −20°C. The extracted hemes were analyzed by HPLC using a Waters 2795 separation module and Waters 2996 photodiode array (PDA) detector. For heme separation, a 1-mm-inner-diameter reverse-phase C18 column was used with an acetonitrile (0.05% trifluoroacetic acid [TFA])-water (0.05% TFA) gradient from 50 to 100% (41). The hemes were confirmed by their molecular weights using a Waters quadrupole time of flight (Q-TOF) Ultima electrospray ionization (ESI) mass spectrometer.
Oxygen consumption.
The oxygen concentration was monitored using a dual-channel respirometer system, model 782 from Strathkelvin Instruments, equipped with a temperature-controlled 0.5-ml electrode chamber at 37°C. The reaction mixture for this assay consisted of sodium phosphate buffer, pH 7.5, 50 mM NaCl, and 200 to 1,000 µg/ml membranes. The concentration of oxygen in the air-saturated buffer at this temperature was assumed to be 237.5 µM, and the reaction was initiated by injecting 500 µM NADH. One hundred micromoles of potassium cyanide (KCN) was added to distinguish between the cyanide-sensitive aa3 menaquinol oxidase and the cyanide-resistant bd menaquinol oxidase present in S. aureus membranes. The percentage of inhibition after the addition of KCN was calculated for each mutant.
RNA purification and ctaB-ctaM intergenic amplification.
Bacterial RNA was harvested as previously described (42). Briefly, overnight cultures of S. aureus were subcultured (1:100) into TSB and cultures were harvested at late exponential phase (6 h) or stationary phase (12 h). The cells were pelleted, resuspended in LETS buffer (1 M LiCl, 0.5 M EDTA, 1 M Tris HCl, pH 7.4, 10% SDS), and lysed using mechanical bead beating. RNA was extracted using TRIzol (TRI Reagent; Sigma), chloroform precipitated with isopropyl alcohol, washed with 70% ethanol, and dissolved in distilled water. Contaminating DNA was removed with DNase I (Amersham Biosciences) treatment. RNA then was purified using the RNeasy minikit according to the manufacturer’s protocol (Qiagen). The RNA concentration and purity were measured by the optical density at 260 nm and 280 nm, respectively. For cDNA synthesis, 2 µg of RNA was left untreated (without reverse transcriptase [RT]) or was treated with Moloney murine leukemia virus reverse transcriptase according to the manufacturer’s recommendations (Promega). The region between ctaB and ctaM was PCR amplified using the following standard cycling parameters: 25 cycles of amplification with the ctaB forward primer 5′ ACCATCAGGCGTACTTGGTC 3′ and the ctaM reverse primer 5′ AATTGCCATTGGTTGGAGAC 3′.
Bioinformatic analysis of ctaM.
With over 60,000 prokaryotic genomes currently available in public databases and over 15,000 more in the pipelines (www.genomesonline.org), it is not practical or possible to perform meaningful comparative analysis on all of them simultaneously. Thus, a set of diverse representative prokaryotic genomes has been developed in the SEED database as follows. The algorithm for computing molecular operational taxonomic units (OTUs) based on DNA bar code data (43, 44) was used to group ~12,600 prokaryotic genomes available in the SEED database in October 2013 into about 1,000 taxon groups. A representative genome for each OTU was selected based on the largest amount of published experimental data and the highest level of research interest within the scientific community for different microorganisms within each OTU. The resultant collection of 978 diverse eubacterial (924) and archaeal (54) genomes creates a manageable set that accurately represents the immense diversity of the over 12,000 prokaryotic organisms with sequenced genomes. Importantly, it is not skewed by an overabundance of genomes for a few microbial genera (medically or industrially important), such as Enterobacteriaceae, staphylococci, mycobacteria, etc.
The hypothetical DUF420 family (CtaM) has been exhaustively annotated for this set of 978 representative microbial genomes (54 archaeal and 928 eubacterial) in the SEED database (26). Functional associations for this gene family were predicted based on the patterns of co-occurrence and/or colocalization of its members with other protein families using the set of tools for comparative genome analysis available in SEED (27) within the functional and genomic contexts provided by the subsystem “Heme O and Heme A biosynthesis (with selected terminal oxidases)” (28).
SUPPLEMENTAL MATERIAL
Comprehensive LC analysis of heme cofactors isolated from S. aureus terminal oxidase mutants and controls. (A to C) LC analysis of heme molecules isolated from the membranes of the wild type (WT) (A) and the isogenic ΔcydA (B) and ΔqoxA (C) terminal oxidase mutants. (D to F) LC analysis of heme molecules isolated from purified aa3-type quinol oxidase from Rhodobacter sphaeroides (D), bd-type quinol oxidase from Escherichia coli (E), and membranes isolated from E. coli, expressing only the bo3-type quinol oxidase (F) (Hoeser J, Hong S, Gehmann G, Gennis RB, Friedrich T. 2014. Subunit CydX of Escherichia coli cytochrome bd ubiquinol oxidase is essential for assembly and stability of the di-heme active site. FEBS Lett 588:1537–1541.) Tandem mass spectroscopy was used to identify heme D in fraction 32, heme B in fraction 37, heme A in fraction 49, and heme O in fraction 52. Download
The SCV phenotype of the ΔctaB ΔcydA and ΔcydAB ΔctaM double mutants can be complemented by plasmid-encoded copies of ctaB or ctaM, respectively. (A) Overnight cultures of wild-type (WT) S. aureus and isogenic mutants harboring an empty-vector control plasmid (pOS) or a plasmid constitutively expressing ctaB (pctaB) were grown at 37°C in tryptic soy broth (TSB) and streaked for isolated colonies on tryptic soy agar (TSA). The ΔqoxB ΔcydA and ΔctaB ΔcydA double mutants harboring pOS are respiration-arrested small colony variants (SCVs). (B) Overnight cultures of WT S. aureus and isogenic mutants were grown at 37°C in TSB and streaked for isolated colonies on TSA. The ΔqoxB ΔcydA and ΔcydAB ΔctaM double mutants containing pOS are respiration-arrested SCVs. Download
ACKNOWLEDGMENTS
We thank members of the Skaar and Gennis laboratories for critical evaluation of the manuscript.
This work was supported by National Institutes of Health grants R01AI069233 (E.P.S.), AI073843 (E.P.S.), GM095600 (R.B.G.) and HL16101 (R.B.G.). The Q-TOF Ultima mass spectrometer was purchased in part with a grant from the National Science Foundation, Division of Biological Infrastructure (DBI-0100085). N.D.H. was supported by a Cystic Fibrosis Foundation fellowship (HAMMER13F0). Strains NE117, NE92, NE769, and NE1084 were obtained through Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources, NIAID, NIH: Nebraska Transposon Mutant Library (NTML) Screening Array, NR-48501.
Footnotes
Citation Hammer ND, Schurig-Briccio LA, Gerdes SY, Gennis RB, Skaar EP. 2016. CtaM is required for menaquinol oxidase aa3 function in Staphylococcus aureus. mBio 7(4):e00823-16. doi:10.1128/mBio.00823-16.
REFERENCES
- 1.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, Active Bacterial Core surveillance (ABCs) MRSA Investigators . 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Indriati Hood M, Skaar EP. 2013. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. mBio 4:e00241-13. doi: 10.1128/mBio.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan L, Cheng A, Dunman PM, Missiakas D, He C. 2010. Golden pigment production and virulence gene expression are affected by metabolisms in Staphylococcus aureus. J Bacteriol 192:3068–3077. doi: 10.1128/JB.00928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107. doi: 10.1016/j.chom.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramsey HH. 1962. Endogenous respiration of Staphylococcus aureus. J Bacteriol 83:507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson GM. 1936. The nutrition of Staphylococcus aureus. Necessity for uracil in anaerobic growth. Biochem J 30:2184–2190. doi: 10.1042/bj0302184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strasters KC, Winkler KC. 1963. Carbohydrate metabolism of Staphylococcus aureus. J Gen Microbiol 33:213–229. doi: 10.1099/00221287-33-2-213. [DOI] [PubMed] [Google Scholar]
- 9.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta 1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Horsman JA, Barquera B, Rumbley J, Ma J, Gennis RB. 1994. The superfamily of heme-copper respiratory oxidases. J Bacteriol 176:5587–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 12.Borisov VB, Verkhovsky MI. 23 October 2015, posting date Oxygen as acceptor. EcoSal Plus 2015 doi: 10.1128/ecosalplus.ESP-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sousa FL, Alves RJ, Ribeiro MA, Pereira-Leal JB, Teixeira M, Pereira MM. 2012. The superfamily of heme-copper oxygen reductases: types and evolutionary considerations. Biochim Biophys Acta 1817:629–637. doi: 10.1016/j.bbabio.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 14.Artzatbanov VY, Petrov VV. 1990. Branched respiratory chain in aerobically grown Staphylococcus aureus—oxidation of ethanol by cells and protoplasts. Arch Microbiol 153:580–584. doi: 10.1007/BF00245268. [DOI] [PubMed] [Google Scholar]
- 15.Frerman FE, White DC. 1967. Membrane lipid changes during formation of a functional electron transport system in Staphylococcus aureus. J Bacteriol 94:1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotelevets LM, Babenko IS, Eremin VA, Lukoianova MA. 1987. Variability of Staphylococcus aureus membranes depending on the growth phase of the culture. Mikrobiol Zh 49:14–18. (Article in Russian.) [PubMed] [Google Scholar]
- 17.Taber HW, Morrison M. 1964. Electron transport in staphylococci. Properties of a particle preparation from exponential phase Staphylococcus aureus. Arch Biochem Biophys 105:367–379. doi: 10.1016/0003-9861(64)90021-9. [DOI] [PubMed] [Google Scholar]
- 18.Tynecka Z, Szcześniak Z, Malm A, Los R. 1999. Energy conservation in aerobically grown Staphylococcus aureus. Res Microbiol 150:555–566. doi: 10.1016/S0923-2508(99)00102-3. [DOI] [PubMed] [Google Scholar]
- 19.Voggu L, Schlag S, Biswas R, Rosenstein R, Rausch C, Götz F. 2006. Microevolution of cytochrome bd oxidase in staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J Bacteriol 188:8079–8086. doi: 10.1128/JB.00858-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schurig-Briccio LA, Yano T, Rubin H, Gennis RB. 2014. Characterization of the type 2 NADH:menaquinone oxidoreductases from Staphylococcus aureus and the bactericidal action of phenothiazines. Biochim Biophys Acta 1837:954–963. doi: 10.1016/j.bbabio.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clements MO, Watson SP, Poole RK, Foster SJ. 1999. CtaA of Staphylococcus aureus is required for starvation survival, recovery, and cytochrome biosynthesis. J Bacteriol 181:501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svensson B, Lübben M, Hederstedt L. 1993. Bacillus subtilis CtaA and CtaB function in haem A biosynthesis. Mol Microbiol 10:193–201. doi: 10.1111/j.1365-2958.1993.tb00915.x. [DOI] [PubMed] [Google Scholar]
- 23.Contreras-Zentella M, Mendoza G, Membrillo-Hernández J, Escamilla JE. 2003. A novel double heme substitution produces a functional bo3 variant of the quinol oxidase aa3 of Bacillus cereus. Purification and partial characterization. J Biol Chem 278:31473–31478. doi: 10.1074/jbc.M302583200. [DOI] [PubMed] [Google Scholar]
- 24.Del Arenal IP, Contreras ML, Svlateorova BB, Rangel P, Lledías F, Dávila JR, Escamilla JE. 1997. Haem O and a putative cytochrome bo in a mutant of Bacillus cereus impaired in the synthesis of haem A. Arch Microbiol 167:24–31. doi: 10.1007/s002030050412. [DOI] [PubMed] [Google Scholar]
- 25.Winstedt L, von Wachenfeldt C. 2000. Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J Bacteriol 182:6557–6564. doi: 10.1128/JB.182.23.6557-6564.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.SEED Subsystem Editor. Heme O and Heme A biosynthesis (with selected terminal oxidases). http://pubseed.theseed.org/SubsysEditor.cgi?page=ShowSubsystem&subsystem=Heme_O_and_Heme_A_biosynthesis_%28with_selected_terminal_oxidases%29.
- 29.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 30.Pedersen MB, Gaudu P, Lechardeur D, Petit M, Gruss A. 2012. Aerobic respiration metabolism in lactic acid bacteria and uses in biotechnology. Annu Rev Food Sci Technol 3:37–58. doi: 10.1146/annurev-food-022811-101255. [DOI] [PubMed] [Google Scholar]
- 31.Hemp J, Gennis RB. 2008. Diversity of the heme-copper superfamily in archaea: insights from genomics and structural modeling. Results Probl Cell Differ 45:1–31. doi: 10.1007/400_2007_046. [DOI] [PubMed] [Google Scholar]
- 32.Lübben M, Morand K. 1994. Novel prenylated hemes as cofactors of cytochrome oxidases. Archaea have modified hemes A and O. J Biol Chem 269:21473–21479. [PubMed] [Google Scholar]
- 33.Shi L, Sohaskey CD, Kana BD, Dawes S, North RJ, Mizrahi V, Gennaro ML. 2005. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc Natl Acad Sci U S A 102:15629–15634. doi: 10.1073/pnas.0507850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Way SS, Sallustio S, Magliozzo RS, Goldberg MB. 1999. Impact of either elevated or decreased levels of cytochrome bd expression on Shigella flexneri virulence. J Bacteriol 181:1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitko NP, Spahich NA, Richardson AR. 2015. Glycolytic dependency of high-level nitric oxide resistance and virulence in Staphylococcus aureus. mBio 6:e00045-15. doi: 10.1128/mBio.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hederstedt L. 2012. Heme A biosynthesis. Biochim Biophys Acta 1817:920–927. doi: 10.1016/j.bbabio.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 37.Duthie ES, Lorenz LL. 1952. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol 6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 38.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Reinhold-Hurek B, Zhulin IB. 1997. Terminal oxidases of Azoarcus sp. BH72, a strictly respiratory diazotroph. FEBS Lett 404:143–147. doi: 10.1016/S0014-5793(97)00111-7. [DOI] [PubMed] [Google Scholar]
- 41.Sone N, Fujiwara Y. 1991. Haem O2 can replace haem A in the active site of cytochrome c oxidase from thermophilic bacterium PS3. FEBS Lett 288:154–158. doi: 10.1016/0014-5793(91)81024-3. [DOI] [PubMed] [Google Scholar]
- 42.Farrand AJ, Haley KP, Lareau NM, Heilbronner S, McLean JA, Foster T, Skaar EP. 2015. An iron-regulated autolysin remodels the cell wall to facilitate heme acquisition in Staphylococcus lugdunensis. Infect Immun 83:3578–3589. doi: 10.1128/IAI.00397-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, Abebe E. 2005. Defining operational taxonomic units using DNA bar code data. Philos Trans R Soc Lond B Biol Sci 360:1935–1943. doi: 10.1098/rstb.2005.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones M, Ghoorah A, Blaxter M. 2011. jMOTU and Taxonerator: turning DNA barcode sequences into annotated operational taxonomic units. PLoS One 6:e19259. doi: 10.1371/journal.pone.0019259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comprehensive LC analysis of heme cofactors isolated from S. aureus terminal oxidase mutants and controls. (A to C) LC analysis of heme molecules isolated from the membranes of the wild type (WT) (A) and the isogenic ΔcydA (B) and ΔqoxA (C) terminal oxidase mutants. (D to F) LC analysis of heme molecules isolated from purified aa3-type quinol oxidase from Rhodobacter sphaeroides (D), bd-type quinol oxidase from Escherichia coli (E), and membranes isolated from E. coli, expressing only the bo3-type quinol oxidase (F) (Hoeser J, Hong S, Gehmann G, Gennis RB, Friedrich T. 2014. Subunit CydX of Escherichia coli cytochrome bd ubiquinol oxidase is essential for assembly and stability of the di-heme active site. FEBS Lett 588:1537–1541.) Tandem mass spectroscopy was used to identify heme D in fraction 32, heme B in fraction 37, heme A in fraction 49, and heme O in fraction 52. Download
The SCV phenotype of the ΔctaB ΔcydA and ΔcydAB ΔctaM double mutants can be complemented by plasmid-encoded copies of ctaB or ctaM, respectively. (A) Overnight cultures of wild-type (WT) S. aureus and isogenic mutants harboring an empty-vector control plasmid (pOS) or a plasmid constitutively expressing ctaB (pctaB) were grown at 37°C in tryptic soy broth (TSB) and streaked for isolated colonies on tryptic soy agar (TSA). The ΔqoxB ΔcydA and ΔctaB ΔcydA double mutants harboring pOS are respiration-arrested small colony variants (SCVs). (B) Overnight cultures of WT S. aureus and isogenic mutants were grown at 37°C in TSB and streaked for isolated colonies on TSA. The ΔqoxB ΔcydA and ΔcydAB ΔctaM double mutants containing pOS are respiration-arrested SCVs. Download