ABSTRACT
Historically, the members of the Agrobacterium genus have been considered the only bacterial species naturally able to transfer and integrate DNA into the genomes of their eukaryotic hosts. Yet, increasing evidence suggests that this ability to genetically transform eukaryotic host cells might be more widespread in the bacterial world. Indeed, analyses of accumulating genomic data reveal cases of horizontal gene transfer from bacteria to eukaryotes and suggest that it represents a significant force in adaptive evolution of eukaryotic species. Specifically, recent reports indicate that bacteria other than Agrobacterium, such as Bartonella henselae (a zoonotic pathogen), Rhizobium etli (a plant-symbiotic bacterium related to Agrobacterium), or even Escherichia coli, have the ability to genetically transform their host cells under laboratory conditions. This DNA transfer relies on type IV secretion systems (T4SSs), the molecular machines that transport macromolecules during conjugative plasmid transfer and also during transport of proteins and/or DNA to the eukaryotic recipient cells. In this review article, we explore the extent of possible transfer of genetic information from bacteria to eukaryotic cells as well as the evolutionary implications and potential applications of this transfer.
INTRODUCTION
Vertical gene inheritance is the main pathway of transmission of genomic information from the parents to their offspring via germline or cell division. However, genetic information can be transmitted also between organisms that are not directly related; these exchanges are termed horizontal gene transfer (HGT; also known as lateral gene transfer) (1). Among prokaryotes, HGT—first observed as the spreading of drug resistance within a bacterial population (2)—is now recognized as a major evolutionary force (3–5). Indeed, several genome-wide studies have shown that HGT occurs at a high frequency between prokaryotic species, particularly if they are closely related or if they coexist in the same habitat or community, which provides many opportunities for DNA transfer (4, 5). Unlike evolution via gene duplications and mutations, a slow and incremental process, HGT permits fast acquisition of a new function important for species adaptation and survival.
Numerous cases of HGT from bacteria to eukaryotes have been demonstrated, although this process is assumed to be much less frequent than HGT between bacteria. The early evolution of eukaryotes was marked by endosymbiotic events leading to permanent acquisition of major organelles, e.g., mitochondria that originated from proteobacteria and plastids that originated from cyanobacteria, followed by organelle-to-nucleus gene transfer, usually referred to as endosymbiotic gene transfer (EGT) (6). Whereas the episodic gene transfer via EGT has a demonstrated evolutionary significance, the importance of HGT in the evolution of eukaryotes is still debated (7). A recent study analyzing a large number of protein sequences from bacterial and eukaryotic organisms indicates that gene inheritance in eukaryotes is predominantly vertical and suggests that HGT occurs only occasionally and that sequences acquired by HGT do not accumulate in eukaryotic genomes and do not contribute to long-term evolution of gene content (8). However, it is generally agreed that HGT from prokaryotes to eukaryotes does occur to a certain extent and, in some cases, plays a role in adaptive evolution (9). Whereas the identification of HGT genomic signatures indicates the existence of such events in the course of evolution, it does not inform about the pathway(s) and mechanisms(s) by which these sequences have been transferred. Instead, this information derives from numerous studies of known systems of natural and experimental gene transfer from bacteria to eukaryotic cells—such as the Agrobacterium-host plant interaction, the best-studied and best-understood system of transkingdom DNA transfer. Here, we review the major known cases of HGT from bacteria to eukaryotes that do not originate from prokaryote-derived permanent organelles, with a focus on natural and artificial prokaryote-to-eukaryote gene transfer systems that may help us understand the potential mechanisms involved in these transkingdom exchanges of genetic information.
SIGNATURES OF BACTERIUM-TO-EUKARYOTE HGT IN GENOME SEQUENCES
In most cases, the first step in identifying an HGT event is the detection of a sequence that does not follow the expected phyletic distribution. However, the presence of such a sequence may also result from differential gene loss in most species of a single clade, leading to the impression that this gene is present only in one remaining species. The classical method to validate a suspected HGT event is phylogenetic inference: the finding of a topological disagreement between a strongly supported gene tree and the known species tree is a good indicator of an HGT event (4, 7, 10). Other accessory methods may help to confirm the occurrence of HGT, such as base composition, the presence or absence of introns, codon usage, synteny analysis, and such ecological features as a shared niche or location for the species involved. Still, the exact identity of the prokaryotic species from which the sequence acquired by HGT originates is sometimes difficult to determine because of subsequent evolution of the transferred sequence or because the “donor” species is extinct.
The complete sequencing of Dictyostelium discoideum (11) revealed 18 genes resulting from potential HGT from bacteria, which sometimes conferred new functions, such as a dipeptidase enzymatic activity potentially able to degrade the bacterial cell wall. Similarly, Galdieria sulphuraria, a red alga that lives in extreme, i.e., hot, acidic, and heavy-metal-rich, environments harbors genes obtained through HGT from bacteria and archaea and that may represent, after duplication and diversification, as much as 5% of its protein-encoding genes. Most of these proteins of suspected bacterial origin, such as an arsenic membrane protein pump similar to those found in thermoacidophilic bacteria, are expressed and are believed to have facilitated ecological adaptation of G. sulphuraria to extreme environments in the course of the evolution of this species (12).
Several cases of potential HGT from bacterial sources also have been identified in Saccharomyces cerevisiae and other yeast species (13, 14). For example, the URA1 gene, which encodes the enzyme dihydroorotate dehydrogenase required for anaerobic synthesis of uracil, appears to originate from the lactic acid bacteria Lactobacillales. In plant-associated fungi, the acquisition of bacterial genes by HGT is considered widespread and likely represents a significant force in their adaptive evolution (15). These genes often encode factors involved in niche specification, pathogenicity, and adaptation to different metabolic requirements (10). Frequently, HGT occurs into plant-pathogenic fungi living in the community with many other plant-associated prokaryotic and eukaryotic microorganisms (16). In particular, within the genomes of three species of Colletotrichum, a genus of plant-pathogenic fungi that cause the crop-destructive disease anthracnose, at least 11 independent HGT events from bacterial genomes were identified (17). These transferred genes encode factors involved either in interaction with host plant and fungal virulence or in various metabolic processes, and they likely play a role for niche adaptation. Similarly, two species of the vascular wilt fungus Verticillium acquired by HGT from proteobacteria a gene that encodes a glucan glycosyltransferase involved in synthesis of extracellular glucans important for virulence (18).
Microbial eukaryotes, such as D. discoideum or G. sulphuraria, and most fungi either are unicellular or display a predominant unicellular stage in their life cycle; thus, in these species, genes can be vertically transmitted during cell division. However, for more complex, multicellular organisms, HGT can be transmitted vertically only by two general mechanisms: when the recipient cells are germline cells or when they are able to dedifferentiate and/or regenerate to a functional organism by asexual reproduction.
In multicellular organisms, bacterium-to-animal cell HGT appears to be limited to invertebrates, and it has originated either from gene transfer from endosymbiotic bacteria to their hosts or from transfer from bacteria to asexual animals (19). One of the most striking examples of HGT from bacterial endosymbionts to animal hosts is the gene transfer from Wolbachia to different arthropods and nematodes (20). Wolbachia species are good candidates for heritable HGT: they are intracellular symbionts, maternally inherited, and transmitted through egg cytoplasm. Among 11 sequenced arthropod genomes, eight contain Wolbachia sequences acquired via HGT; interestingly, the transferred sequences sometimes represented a significant portion (up to 30%) of the genome. HGT also had occurred from nonendosymbiont bacteria to freshwater asexual animals, such as Hydra magnipapillata (21) and bdelloid rotifers (22). On a few occasions, however, initial indications that animal genomes contain numerous genes originating from HGT were refuted by subsequent more-detailed analyses. For example, a large number of potential HGT events were first reported for the human genome (23), but this claim was disproved after closer examination of the data based on phylogenetic analysis that included a larger number of eukaryotic species (24).
A comparative genomic study of the early land plant Physcomitrella patens and of a flowering plant, Arabidopsis, uncovered 57 families of nuclear genes that potentially had been acquired by HGT, mainly from bacterial species (25). Several of these genes are involved in land-plant-specific activities, e.g., xylem formation, defense, and regulation of growth, suggesting that HGT played an important role in the transition from the aquatic to the terrestrial environment. Furthermore, phylogenetic evidence supports the idea that the major biosynthetic pathway of auxin, the main hormone of land plants, is derived from the bacterium-to-plant HGT (26). Another series of HGT events in plants resulted from the insertion of Agrobacterium transfer DNA (T-DNA) into the plant genome and its vertical transfer via sexual reproduction (27). It was first reported in Nicotiana glauca (28, 29) and then found in most species of Nicotiana tested to date (30, 31). In a screen of more than 100 dicotyledonous plant species, Agrobacterium T-DNA sequences were detected in two species of the genus Linaria (32). More recently, the presence of T-DNA sequences acquired by HGT was discovered in the genomes of several varieties of cultivated sweet potato, Ipomoea batatas (33). The origin of the T-DNA-derived genes was identified as a mikimopine strain of Agrobacterium rhizogenes for both Nicotiana and Linaria and likely as an ancestral form of A. rhizogenes for I. batatas. Some of these T-DNA genes are still expressed at a detectable level in modern plants, although whether they have a functional role in the plant biology remains unknown. These HGT events originating from Agrobacterium species represent a rare case for which the source of the transferred genes is clearly identified and the transfer pathway is well studied (see below).
NATURAL AND EXPERIMENTAL BACTERIUM-TO-EUKARYOTE DNA TRANSFER SYSTEMS
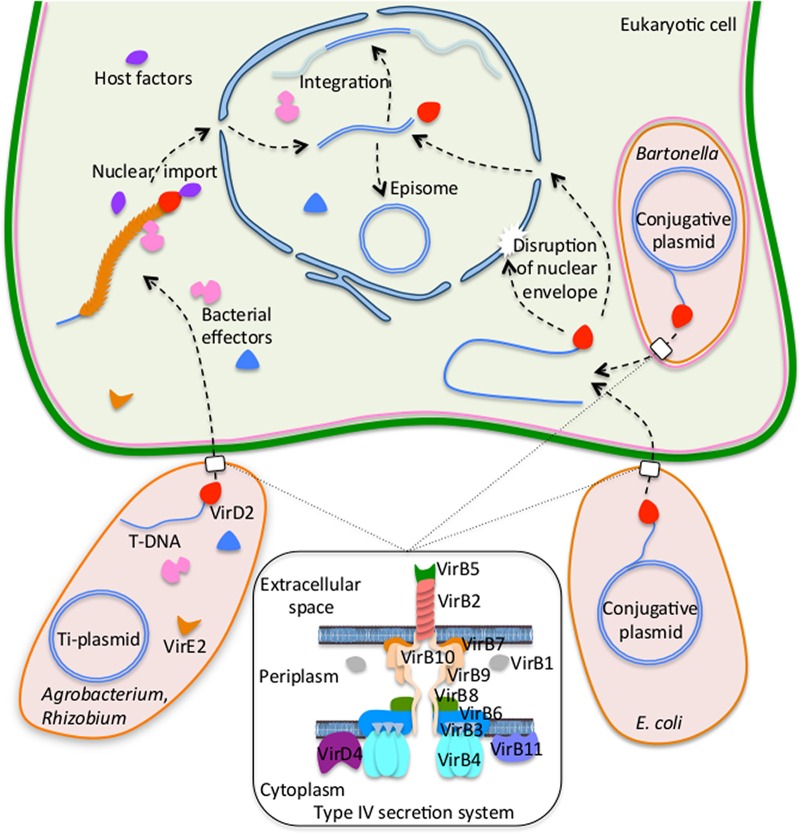
The major known natural and artificial systems for gene transfer from bacterial to eukaryotic cells include such bacteria as Agrobacterium and Rhizobium species and Escherichia coli, and they are summarized in Fig. 1. The first system is the Agrobacterium-to-plant cell DNA transfer, which represents the paradigm of eukaryotic genetic transformation by bacteria and has long been considered a unique case in living nature; thus, it is the most-studied example of transkingdom gene transfer (34, 35). Agrobacterium is a plant-pathogenic bacterium that causes neoplastic growths, i.e., uncontrolled cell divisions that form galls or root proliferations, in its host plants by transferring a segment of DNA into the host cell genome. Most of the bacterial genes essential for gene transfer are located on a large tumor-inducing plasmid, termed the Ti plasmid, which also contains the transferred DNA segment, termed the T-DNA, that is delimited and specified by two short direct repeat sequences, left and right borders. Plant-derived phenolic and sugar signal molecules trigger the expression of the virulence (vir) genes in Agrobacterium cells, and the encoded Vir proteins mediate the transfer of the T-DNA to the plant cell. The T-DNA is transferred as a single-stranded molecule, produced by the VirD2 endonuclease, which, in association with the VirD1 DNA topoisomerase (36), mediates the mobilization of the transferrable T-DNA from the Ti plasmid by a strand replacement mechanism; VirD2 then remains covalently bound to the 5′ terminus of the T-DNA molecule. Upon interaction with the coupling factor VirD4, the VirD2–T-DNA complex is directed to the type IV secretion system (T4SS) composed of 11 proteins encoded by the virB operon. The T4SS then mediates the translocation of the VirD2–T-DNA complex, as well as several other Vir protein effectors, from the bacterium to the host cell cytoplasm. The fate of the T-DNA in the host cell relies on multiple interactions with Agrobacterium and host cell proteins, taking advantage of several host cell pathways to ensure the T-DNA nuclear import and integration into the host genome. Both VirD2 and the single-stranded DNA binding protein VirE2—which packages T-DNA into a helical nucleoprotein complex, termed the transfer (T) complex—can interact, directly or indirectly, with host factors to allow nuclear import of the T complex. This process likely occurs in a polar manner such that VirD2 directs the T-DNA to the nuclear pore while VirE2 facilitates the passage of the entire T complex through the pore via the importin-α-dependent nuclear import pathway. Inside the nucleus, the T complex is proteolytically uncoated from its associated bacterial and host proteins, presumably by interacting with the host ubiquitin/proteasome system (UPS). Then, the single-stranded T-DNA most likely is converted to a double-stranded form and integrated into the plant genome by the host DNA repair machinery. Interestingly, under laboratory conditions, Agrobacterium is able to transfer DNA to many nonplant cells, from fungi to human cultured cells (37), suggesting that T-DNA nuclear import and subsequent processing and integration are mediated by factors found in all diverse eukaryotic species, rather than by factors specific for host plants. Furthermore, Agrobacterium is also able to transfer sequences from a mobilizable plasmid (RSF1010) to plant cells via the activity of Vir proteins and of the plasmid mobilization functions (38).
FIG 1 .
Schematic summary of known natural and experimental pathways for DNA transfer from bacteria to eukaryotic cells. Agrobacterium and related bacteria, E. coli, and Bartonella henselae can transfer DNA to different types of eukaryotic cells via the activity of their type IV secretion systems composed of VirD4/VirB proteins. Inside the host eukaryotic cell, the bacterial transferred DNA, usually a single-stranded molecule packaged into a nucleoprotein complex, is imported into the host nucleus. Nuclear import and further DNA processing, i.e., conversion to a double-stranded form, integration into the recipient cell genome, or formation of an episome, depend on interactions of the transferred DNA and its associated proteins with numerous host cell factors that represent different types of cellular machineries, such as nuclear import machinery, the ubiquitin/proteasome system, and DNA repair machinery. For further details, see the text.
As early as 1977, it was reported that the Ti plasmid of Agrobacterium could be transferred by conjugation to cells of the related species Rhizobium trifolii, which conferred on those bacteria the ability to trigger virulence, i.e., development of crown galls, on several plant species (39). Later, it was shown that the introduction of both a disarmed Ti plasmid, i.e., harboring the vir genes but no T-DNA, and a binary plasmid, i.e., containing T-DNA but no vir genes, into such Rhizobiaceae species as Rhizobium leguminosarum, R. trifolii, and Phyllobacterium myrsinacearum, produced virulent bacteria able to transfer T-DNA to Arabidopsis, tobacco, and rice (40), whereas potato plants were genetically transformed by Sinorhizobium meliloti, Rhizobium sp. strain NGR234, and Mesorhizobium loti supplied with a similar binary vector set (41). Another bacterial species of the Rhizobiaceae family, Ensifer adherens, when equipped with a cointegrated vector containing both the vir region and T-DNA from Agrobacterium, was used to transform potato and rice plants (42, 43). All these species are symbiotic bacteria that belong to the same Rhizobiales order as Agrobacterium and are able to mediate nitrogen assimilation for their host plants.
In all these studies, the vir region of a virulent strain of Agrobacterium had to be supplied along with the T-DNA in order to confer competence for plant transformation on the bacterial species, which naturally encoded no native plant transformation machinery. However, several species of rhizobia contain their own vir genes, with different levels of homology to the Agrobacterium vir genes. Specifically, Rhizobium etli strain CFN42 naturally contains, on its p42a plasmid, a functional T-DNA transfer machinery comprising all the necessary vir gene functions. Indeed, when a binary plasmid, containing a T-DNA but not the vir region, was introduced into R. etli, the resulting strain was able to transfer and integrate T-DNA into plant cells, albeit with a lower frequency than that with Agrobacterium (44). This T-DNA transfer was not observed with mutants of R. etli that lack one of the essential vir genes (virG or virE2) or with R. leguminosarum, a species very close to R. etli overall but with only very weak homology to the vir genes. Importantly, our analysis of the known DNA sequences of R. etli detected no homologies to known Agrobacterium T-DNA sequences, i.e., T-DNA borders or T-DNA genes; however, we cannot rule out the possibility that R. etli might contain its own, specific T-DNA sequences undetectable by in silico analyses.
The transfer of plasmid DNA can also occur from bacteria (E. coli) to yeast (S. cerevisiae) (45). This transfer was effective with both broad-host-range and F-factor plasmids, which represent the two main types of conjugative plasmids in Gram-negative bacteria. The transfer mechanism exhibited similarities with bacterial conjugation: for example, physical contact between the cells was required as well as genetic factors, i.e., mob and oriT, necessary for bacterial conjugation. DNA transfer was also observed from E. coli to other yeast species, such as Kluyveromyces lactis and Pichia angusta (46) or Saccharomyces kluyveri (47). The ability of E. coli to deliver DNA molecules via conjugation-like pathways to other types of eukaryotic cells was also reported for gene transfer to cultured human cells (48) and, more recently, to the unicellular algae diatoms (49). Although there is no definitive proof that such bacterium-to-eukaryote conjugation occurs in nature, these examples suggest that the ability to transfer genetic information to eukaryotic hosts is not restricted to Agrobacterium.
In another example of potential ability to genetically transform the host cell, Bartonella henselae, a facultative intracellular human bacterial pathogen, was shown to transfer a modified cryptic plasmid (50) or derivatives of the R388 plasmid (51) into human cultured cells via a conjugation-like mechanism. Indeed, the bacterial strains used in these experiments harbored a T4SS, which was required for the transport of plasmids from the bacterium to the host cell, and this DNA transfer was disrupted in B. henselae strains with a mutated virB region. Interestingly, host cell division was required for expression of the transgene, suggesting that the bacterium is unable to utilize the host nuclear import pathways and, instead, relies on breakdown of the host nuclear envelope. This B. henselae-human cell DNA transfer produced stable transgenic cell lines, indicating the integration of the transferred sequences in the host cell genome. Whereas the Bartonella T4SS is known to transport effector proteins essential for virulence into the host cells (52), its apparent ability to transfer plasmid DNA has no demonstrated role in the infection process and may represent the relic of an ancestral function.
POTENTIAL MECHANISMS OF BACTERIUM-TO-EUKARYOTE HGT
DNA transfer into bacterial cells is known to occur via three different mechanisms: transformation (uptake of free DNA in solution), bacteriophage-mediated transduction (i.e., both generalized and specialized transduction), and plasmid-mediated transfer (i.e., conjugation, which usually requires close contact between donor and recipient cells). But are these mechanisms applicable to HGT? Potential HGT pathways may be inferred from studies of natural and experimental bacterium-to eukaryote gene transfer systems. Although yeast cells have been suggested to acquire exogenous DNA under conditions close to their natural environment (53), there are no known naturally occurring mechanisms of DNA uptake in eukaryotic cells. In a study of HGT between Wolbachia and Aedes aegypti (54), bacteriophage sequences were found close to the transferred genes, suggesting the role of bacteriophages as HGT vectors. It has also been suggested that some viruses, in particular giant viruses, may mediate transfer of DNA from bacteria to eukaryotes, but this pathway of HGT has not been confirmed experimentally (12).
Notably, most bacteria possessing the ability to transfer DNA to eukaryotic host cells belong to the Alphaproteobacteria class (55). Most of these bacterial species show a degree of interaction with eukaryotic hosts, from pathogenic or symbiotic lifestyles, e.g., Agrobacterium and Rhizobium, to optional or obligate parasitism of intracellular bacteria, e.g., Bartonella and Wolbachia. In these species, the “mobilome,” i.e., the pool of plasmids containing shared genetic information, plays a prominent role, which most likely underlies high genome plasticity and gene mobility between bacteria as well as the ability of these bacteria to transfer DNA to eukaryotic cells.
In known natural and experimental bacterium-to-eukaryote DNA transfer systems, i.e., bacterium-yeast conjugation, Bartonella-mediated transformation of animal cells, and Agrobacterium/Rhizobium-mediated genetic transformation of diverse eukaryotes, the transport of DNA from the bacterial cell to the recipient cell cytoplasm depends on conjugation-like mechanisms mediated by the bacterial T4SS. T4SSs are specialized molecular superstructures able to transport protein and DNA molecules between donor bacteria and a variety of recipient cells (56, 57). They are encoded by many bacterial species and are often involved in conjugation, i.e., transfer of genetic information between bacterial cells of the same or closely related species. However, T4SSs are also known to mediate macromolecular transport of DNA and/or proteins from bacteria to cells of their eukaryotic hosts. In fact, to date, T4SS represents the only demonstrated mechanism of transfer of genetic material to eukaryotic cells from bacteria in nature or under laboratory conditions. Although the export of macromolecules across the bacterial membranes and cell walls through the T4SS is well understood (57, 58), how the transported protein or DNA molecule passes across the eukaryotic recipient cell wall and membrane remains obscure. By analogy to type III secretion systems (T3SSs) (59), the T4SS pilus itself might pierce the host cell barriers to inject macromolecules directly into the host cytoplasm, but this mechanism has never been directly observed. The diameter of this channel is compatible with the size of transported macromolecules (60), and two studies have shown that DNA can be transferred by conjugation between bacterial cells without direct cell-to-cell contact, suggesting that DNA transits through the F pilus lumen (61, 62). Alternatively, the exported macromolecules first may be deposited at the surface of the recipient cell by the T4SS and then internalized by a separate mechanism that may involve host receptors and/or endocytosis. Indeed, studies of direct transformation of yeast cells have shown that once DNA molecules are placed at the surface of the host cell membrane, they may become internalized (e.g., reference 63).
Our knowledge about the molecular reactions that occur after the entry of the transferred DNA into the host cell cytoplasm and lead to the transgene expression and integration derives largely from the studies of Agrobacterium-mediated genetic transformation, which indicate that they rely on interactions of the transferred bacterial DNA and proteins with host factors. Specifically, in a eukaryotic cell, the incoming single-stranded DNA molecule, which represents a mobile T-DNA or a conjugative plasmid, should be imported into the cell nucleus before further processing. Nuclear uptake of the Agrobacterium T-DNA and its associated proteins is mediated by the host nuclear import machinery (35). In addition, it was suggested that, in some cases, the host cell division could be required for expression or integration of the invading DNA, thereby circumventing the necessity for active nuclear import and allowing passive entry in the nucleus following disruption of the nuclear envelope (50). Inside the nucleus, replication of the DNA, imported as a single-stranded molecule via the bacterial T4SS, is thought to occur before plasmid circularization or before integration (64–66) and is likely mediated by the host DNA replication machinery. In the case of Agrobacterium, the T-DNA circularization has been shown to occur, although it remains unclear whether these circles function as intermediates of integration (64–66). Furthermore, the single-stranded T-DNA molecule is converted to a double-stranded molecule before integration (67, 68), whereas the existence of other pathways of integration cannot be excluded. Finally, the host double-strand break (DSB) DNA repair pathways play a crucial role in integration of the transferred sequences (69).
EVOLUTIONARY IMPLICATIONS OF HGT
A few decades ago, the only demonstrated bacterium-to-eukaryote gene transfer was represented by plant genetic transformation mediated by Agrobacterium. Subsequent studies showed that this capability could be observed in other related bacterial species, such as many Rhizobiaceae, including R. etli, E. coli, and Bartonella, and with a wider range of host species from all eukaryotic taxa. In addition, the ever-increasing amount of genomic data from both prokaryotes and eukaryotes led to the discovery of HGT genomic signatures in a broad spectrum of eukaryote species. Globally, the influence of bacterium-to-eukaryote HGT may have been more important for evolution of eukaryotes than previously thought, to the extent that HGT has been proposed to underlie the emergence of several lineages of eukaryotic organisms as opposed to the idea of all eukaryotes descending from a single universal ancestor (7).
It is important to note that the presence of bacterial HGT signatures in eukaryotic genomes does not depend solely on the ability of bacteria to transfer DNA into the host cell. Instead, four additional major conditions should be met. First, the transferred DNA must become integrated into the host genome. Second, the foreign sequence must not be lost after rearrangements of the genome during subsequent cell divisions. Third, for multicellular eukaryotes, the transformed cell must either be fixed in the germline for genetic modification of animals and plant germlines or regenerate into a viable organism when asexual reproduction is possible, for example, via cell dedifferentiation in plants. Finally, the integrated sequence must be preserved in the course of evolution, which is more likely to occur if the acquired gene confers a selective advantage or is at least neutral rather than deleterious. Thus, transient transformation events, which, by definition, are not retained in the genome, probably occur at a much higher rate than is suggested by the HGT signatures discovered in eukaryote genomes. Yet, this phenomenon of “transient expression,” which is well known in the Agrobacterium-plant interaction, might play a role in promoting HGT. For example, hypothetically, transient genetic transformation could express putative effector proteins, which are analogous to effectors translocated from many pathogenic or symbiotic bacteria to their eukaryotic hosts, which in turn will facilitate subsequent rounds of bacterial infection.
The very wide range of eukaryotic cells that can be transformed by Agrobacterium suggests that the DNA transfer involves fundamental biological processes common to most, if not all, eukaryotes and is not dependent on host species-specific factors (37). Indeed, in addition to numerous plant species that can be transformed by Agrobacterium either naturally or under laboratory conditions, other, evolutionarily distant eukaryotes such as yeast (70, 71) and many other fungi (72, 73) and arachnid (74) and human cultured (75) cells are amenable to Agrobacterium-mediated transformation. Moreover, under laboratory conditions, DNA transfer via conjugation-related mechanisms is possible between other bacteria, e.g., E. coli, and several eukaryote species. Thus, the combined potential of different bacterial species to modify genetically cells of virtually all eukaryotes supports the notion of HGT as a widespread mechanism in evolution.
POTENTIAL APPLICATIONS
Besides its importance for understanding the evolution of modern eukaryotes, the bacterium-to-eukaryote gene transfer has a unique and highly significant application potential, which lies mainly in two major areas, research and biotechnology. Research applications mainly aim at discovering new protein functions and cellular pathways by expressing specific genes of interest, delivering gene-targeting systems, such as CRISPR/Cas9 (76), or insertional mutagenesis of genomes of interest, for example, by generating T-DNA insertion mutant libraries of the model plant Arabidopsis thaliana (77). Biotechnological applications aim at expressing traits of interest, such as pathogen and abiotic stress resistance genes, genetically engineered pathways for production of biofuels and pharmaceuticals, or generation of desired phenotypes, e.g., color and fragrance of flowers and fruits in agriculture or restoration of normal cellular functions in gene therapy. To achieve these general goals, it is important to adapt and optimize different bacterial DNA transfer systems—or even discover new bacterial species capable of DNA transfer to eukaryotes—for use as vectors for genetic transformation of specific eukaryotic cells or organisms, allowing development of highly efficient “custom-tailored” DNA transfer tools for each specific application.
In plants, Agrobacterium-mediated genetic transformation already is efficient for some species, but many other plant species or cultivars, especially those of agronomical importance, are still considered “recalcitrant to transformation” and might become more amenable to gene transfer by a different bacterial vector, for example, belonging to the rhizobial group, with a more appropriate natural host range. Indeed, the early steps of plant genetic transformation rely on close interactions between the bacterium and plant cells, which may be more efficient between plant-associated rhizobia and their hosts than Agrobacterium interactions with the same host species. The same approach also may lead to improving the efficiency of transformation of fungal and other eukaryotic species. In fact, the Agrobacterium-mediated transformation has become a technique of choice for genetic modification of different species of fungi (73); furthermore, it was also suggested that methods based on bacterium-yeast cell conjugation, using, for example, E. coli, could be extended to other species of fungi (78). Obviously, these approaches could be adapted to other eukaryotic cells, such as animal or algal cells.
In animals, two main types of vectors are employed to introduce DNA of interest into human cells for gene therapy: biologicals, such as viruses or bacteria, and biomaterials (79). To date, viruses represent the overwhelmingly predominant vector for use in gene therapy; yet, our increasing knowledge about bacterial DNA transfer systems positions bacteria as a promising alternative for viral vectors (80). Bacterial vectors possess specific features that could be advantageous under specific circumstances: for example, bacterial vectors may be introduced into a tissue for transformation and then easily eliminated by application of antibiotics; many bacteria remain extracellular during DNA transfer, thereby avoiding transfer of DNA sequences other than the gene of interest; and some bacterial strains can target a specific cell type or be engineered for that purpose (80).
Conversely, it is also necessary to investigate and understand potential implications of natural cases of bacterium-to-eukaryote DNA transfer in development of animal and human diseases, such as cancer. Indeed, it is estimated that about 20% of cancers are caused by bacterial or viral infection (81). For instance, one of the best-characterized cases of cancer caused by or associated with bacterial infection is the induction of gastric carcinoma by Helicobacter pylori (82). In most cases, it is thought that carcinogenesis results from stress, e.g., tissue inflammation, caused by the infection process (83). However, it has been also proposed that the transfer and insertion of bacterial DNA sequences into the host cell genome may represent another and more specific cause of cancer development (84, 85). If this hypothesis is confirmed, our accumulating knowledge of bacterium-eukaryote DNA transfer will represent an invaluable tool for providing novel insights into early stages of carcinogenesis.
CONCLUSION
Transfer of genetic information from bacteria to eukaryote cells, once believed to occur exclusively during infection of plants by Agrobacterium, probably occurs in many other bacterium-host cell interactions, which include a wide variety of combinations of donor bacterial species and recipient eukaryote species, at least under laboratory conditions. Studies of these gene transfer systems are critical for understanding their potential ecological and evolutionary significance as well as for their utilization for development of new biological tools for fundamental and applied purposes.
ACKNOWLEDGMENTS
Due to space constraints, we relied largely on citing review articles, and we apologize to our colleagues whose original publications, therefore, have not been cited.
The work in the V.C. laboratory is supported by grants from NIH, NSF, NIFA/USDA, BARD, and BSF.
Footnotes
Citation Lacroix B, Citovsky V. 2016. Transfer of DNA from bacteria to eukaryotes. mBio 7(4):e00863-16. doi:10.1128/mBio.00863-16.
REFERENCES
- 1.Keeling PJ, Palmer JD. 2008. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet 9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 2.Akiba T, Koyama K, Ishiki Y, Kimura S, Fukushima T. 1960. On the mechanism of the development of multiple-drug-resistant clones of Shigella. Jpn J Microbiol 4:219–227. doi: 10.1111/j.1348-0421.1960.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 3.Eisen JA. 2000. Horizontal gene transfer among microbial genomes: new insights from complete genome analysis. Curr Opin Genet Dev 10:606–611. doi: 10.1016/S0959-437X(00)00143-X. [DOI] [PubMed] [Google Scholar]
- 4.Koonin EV, Makarova KS, Aravind L. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol 55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beiko RG, Harlow TJ, Ragan MA. 2005. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci U S A 102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archibald JM. 2015. Endosymbiosis and eukaryotic cell evolution. Curr Biol 25:R911–R921. doi: 10.1016/j.cub.2015.07.055. [DOI] [PubMed] [Google Scholar]
- 7.Syvanen M. 2012. Evolutionary implications of horizontal gene transfer. Annu Rev Genet 46:341–358. doi: 10.1146/annurev-genet-110711-155529. [DOI] [PubMed] [Google Scholar]
- 8.Ku C, Nelson-Sathi S, Roettger M, Sousa FL, Lockhart PJ, Bryant D, Hazkani-Covo E, McInerney JO, Landan G, Martin WF. 2015. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524:427–432. doi: 10.1038/nature14963. [DOI] [PubMed] [Google Scholar]
- 9.Schönknecht G, Weber AP, Lercher MJ. 2014. Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. Bioessays 36:9–20. doi: 10.1002/bies.201300095. [DOI] [PubMed] [Google Scholar]
- 10.Fitzpatrick DA. 2012. Horizontal gene transfer in fungi. FEMS Microbiol Lett 329:1–8. doi: 10.1111/j.1574-6968.2011.02465.x. [DOI] [PubMed] [Google Scholar]
- 11.Eichinger L, Pachebat JA, Glöckner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, Tunggal B, Kummerfeld S, Madera M, Konfortov BA, Rivero F, Bankier AT, Lehmann R, Hamlin N, Davies R, Gaudet P, Fey P, Pilcher K, Chen G, Saunders D, Sodergren E, Davis P, Kerhornou A, Nie X, Hall N, Anjard C, Hemphill L, Bason N, Farbrother P, Desany B, Just E, Morio T, Rost R, Churcher C, Cooper J, Haydock S, van Driessche N, Cronin A, Goodhead I, Muzny D, Mourier T, Pain A, Lu M, Harper D, Lindsay R, Hauser H, James K, Quiles M, Madan Babu M, Saito T, Buchriser C, Wardroper A, Felder M, Thangavelu M, Johnson D, Knights A, Loulseged H, Mungall K, Oliver K, Price C, Quail M, Urushihara H, Hernandez J, Rabbinowitsch E, Steffen D, Sanders M, Ma J, Kohara Y, Sharp S, Simmonds M, Spiegler S, Tivey A, Sugano S, White B, Walker D, Woodward J, Winckler T, Tanaka Y, Shaulsky G, Schleicher M, Weinstock G, Rosenthal A, Cox EC, Chisholm RL, Gibbs R, Loomis WF, Platzer M, Kay RR, Williams J, Dear PH, Noegel AA, Barrell B, Kuspa A. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schönknecht G, Chen WH, Ternes CM, Barbier GG, Shrestha RP, Stanke M, Bräutigam A, Baker BJ, Banfield JF, Garavito RM, Carr K, Wilkerson C, Rensing SA, Gagneul D, Dickenson NE, Oesterhelt C, Lercher MJ, Weber AP. 2013. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339:1207–1210. doi: 10.1126/science.1231707. [DOI] [PubMed] [Google Scholar]
- 13.Gojković Z, Knecht W, Zameitat E, Warneboldt J, Coutelis JB, Pynyaha Y, Neuveglise C, Møller K, Löffler M, Piskur J. 2004. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol Genet Genomics 271:387–393. doi: 10.1007/s00438-004-0995-7. [DOI] [PubMed] [Google Scholar]
- 14.Hall C, Brachat S, Dietrich FS. 2005. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot Cell 4:1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rolland T, Neuvéglise C, Sacerdot C, Dujon B. 2009. Insertion of horizontally transferred genes within conserved syntenic regions of yeast genomes. PLoS One 4:e6515. doi: 10.1371/journal.pone.0006515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardiner DM, Kazan K, Manners JM. 2013. Cross-kingdom gene transfer facilitates the evolution of virulence in fungal pathogens. Plant Sci 210:151–158. doi: 10.1016/j.plantsci.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Jaramillo VD, Sukno SA, Thon MR. 2015. Identification of horizontally transferred genes in the genus Colletotrichum reveals a steady tempo of bacterial to fungal gene transfer. BMC Genomics 16:2. doi: 10.1186/1471-2164-16-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klosterman SJ, Subbarao KV, Kang S, Veronese P, Gold SE, Thomma BP, Chen Z, Henrissat B, Lee YH, Park J, Garcia-Pedrajas MD, Barbara DJ, Anchieta A, de Jonge R, Santhanam P, Maruthachalam K, Atallah Z, Amyotte SG, Paz Z, Inderbitzin P, Hayes RJ, Heiman DI, Young S, Zeng Q, Engels R, Galagan J, Cuomo CA, Dobinson KF, Ma LJ. 2011. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog 7:e1002137. doi: 10.1371/journal.ppat.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunning Hotopp JC. 2011. Horizontal gene transfer between bacteria and animals. Trends Genet 27:157–163. doi: 10.1016/j.tig.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunning Hotopp JC, Clark ME, Oliveira DC, Foster JM, Fischer P, Muñoz Torres MC, Giebel JD, Kumar N, Ishmael N, Wang S, Ingram J, Nene RV, Shepard J, Tomkins J, Richards S, Spiro DJ, Ghedin E, Slatko BE, Tettelin H, Werren JH. 2007. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 21.Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, Disbennett K, Pfannkoch C, Sumin N, Sutton GG, Viswanathan LD, Walenz B, Goodstein DM, Hellsten U, Kawashima T, Prochnik SE, Putnam NH, Shu S, Blumberg B, Dana CE, Gee L, Kibler DF, Law L, Lindgens D, Martinez DE, Peng J, Wigge PA, Bertulat B, Guder C, Nakamura Y, Ozbek S, Watanabe H, Khalturin K, Hemmrich G, Franke A, Augustin R, Fraune S, Hayakawa E, Hayakawa S, Hirose M, Hwang JS, Ikeo K, Nishimiya-Fujisawa C, Ogura A, Takahashi T, Steinmetz PR, Zhang X, Aufschnaiter R, Eder M-K, Gorny A-K, Salvenmoser W, Heimberg AM, Wheeler BM, Peterson KJ, Böttger A, Tischler P, Wolf A, Gojobori T, Remington KA, Strausberg RL, Venter JC, Technau U, Hobmayer B, Bosch TCG, Holstein TW, Fujisawa T, Bode HR, David CN, Rokhsar DS, Steele RE. 2010. The dynamic genome of Hydra. Nature 464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gladyshev EA, Meselson M, Arkhipova IR. 2008. Massive horizontal gene transfer in bdelloid rotifers. Science 320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 23.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 24.Stanhope MJ, Lupas A, Italia MJ, Koretke KK, Volker C, Brown JR. 2001. Phylogenetic analyses do not support horizontal gene transfers from bacteria to vertebrates. Nature 411:940–944. doi: 10.1038/35082058. [DOI] [PubMed] [Google Scholar]
- 25.Yue J, Hu X, Sun H, Yang Y, Huang J. 2012. Widespread impact of horizontal gene transfer on plant colonization of land. Nat Commun 3:1152. doi: 10.1038/ncomms2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue J, Hu X, Huang J. 2014. Origin of plant auxin biosynthesis. Trends Plant Sci 19:764–770. doi: 10.1016/j.tplants.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Matveeva TV, Lutova LA. 2014. Horizontal gene transfer from Agrobacterium to plants. Front Plant Sci 5:326. doi: 10.3389/fpls.2014.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White FF, Garfinkel DJ, Huffman GA, Gordon MP, Nester EW. 1983. Sequences homologous to Agrobacterium rhizogenes T-DNA in the genomes of uninfected plants. Nature 301:348–350. doi: 10.1038/301348a0. [DOI] [Google Scholar]
- 29.Aoki S, Kawaoka A, Sekine M, Ichikawa T, Fujita T, Shinmyo A, Syono K. 1994. Sequence of the cellular T-DNA in the untransformed genome of Nicotiana glauca that is homologous to ORFs 13 and 14 of the Ri plasmid and analysis of its expression in genetic tumours of N. glauca x N. langsdorffii. Mol Gen Genet 243:706–710. [DOI] [PubMed] [Google Scholar]
- 30.Furner IJ, Huffman GA, Amasino RM, Garfinkel DJ, Gordon MP, Nester EW. 1986. An Agrobacterium transformation in the evolution of the genus Nicotiana. Nature 319:422–427. doi: 10.1038/319422a0. [DOI] [Google Scholar]
- 31.Intrieri MC, Buiatti M. 2001. The horizontal transfer of Agrobacterium rhizogenes genes and the evolution of the genus Nicotiana. Mol Phylogenet Evol 20:100–110. doi: 10.1006/mpev.2001.0927. [DOI] [PubMed] [Google Scholar]
- 32.Matveeva TV, Bogomaz DI, Pavlova OA, Nester EW, Lutova LA. 2012. Horizontal gene transfer from genus Agrobacterium to the plant Linaria in nature. Mol Plant Microbe Interact 25:1542–1551. doi: 10.1094/MPMI-07-12-0169-R. [DOI] [PubMed] [Google Scholar]
- 33.Kyndt T, Quispe D, Zhai H, Jarret R, Ghislain M, Liu Q, Gheysen G, Kreuze JF. 2015. The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: an example of a naturally transgenic food crop. Proc Natl Acad Sci U S A 112:5844–5849. doi: 10.1073/pnas.1419685112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gelvin SB. 2003. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacroix B, Citovsky V. 2013. The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int J Dev Biol 57:467–481. doi: 10.1387/ijdb.130199bl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghai J, Das A. 1989. The virD operon of Agrobacterium tumefaciens Ti plasmid encodes a DNA-relaxing enzyme. Proc Natl Acad Sci U S A 86:3109–3113. doi: 10.1073/pnas.86.9.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lacroix B, Tzfira T, Vainstein A, Citovsky V. 2006. A case of promiscuity: Agrobacterium’s endless hunt for new partners. Trends Genet 22:29–37. doi: 10.1016/j.tig.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Buchanan-Wollaston V, Passiatore JE, Cannon F. 1987. The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature 328:172–175. doi: 10.1038/328172a0. [DOI] [Google Scholar]
- 39.Hooykaas PJJ, Klapwijk PM, Nuti MP, Schilperoort RA, Rorsch A. 1977. Transfer of the Agrobacterium tumefaciens Ti plasmid to avirulent agrobacteria and to Rhizobium ex planta. J Gen Microbiol 98:477–484. doi: 10.1099/00221287-98-2-477. [DOI] [Google Scholar]
- 40.Broothaerts W, Mitchell HJ, Weir B, Kaines S, Smith LM, Yang W, Mayer JE, Roa-Rodríguez C, Jefferson RA. 2005. Gene transfer to plants by diverse species of bacteria. Nature 433:629–633. doi: 10.1038/nature03309. [DOI] [PubMed] [Google Scholar]
- 41.Wendt T, Doohan F, Winckelmann D, Mullins E. 2011. Gene transfer into Solanum tuberosum via Rhizobium spp. Transgenic Res 20:377–386. doi: 10.1007/s11248-010-9423-4. [DOI] [PubMed] [Google Scholar]
- 42.Wendt T, Doohan F, Mullins E. 2012. Production of Phytophthora infestans-resistant potato (Solanum tuberosum) utilising Ensifer adhaerens OV14. Transgenic Res 21:567–578. doi: 10.1007/s11248-011-9553-3. [DOI] [PubMed] [Google Scholar]
- 43.Zuniga-Soto E, Mullins E, Dedicova B. 2015. Ensifer-mediated transformation: an efficient non-Agrobacterium protocol for the genetic modification of rice. Springerplus 4:600. doi: 10.1186/s40064-015-1369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lacroix B, Citovsky V. 2016. A functional bacterium-to-plant DNA transfer machinery of Rhizobium etli. PLoS Pathog 12:e1005502. doi: 10.1371/journal.ppat.1005502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinemann JA, Sprague JF Jr. 1989. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 340:205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- 46.Hayman GT, Bolen PL. 1993. Movement of shuttle plasmids from Escherichia coli into yeasts other than Saccharomyces cerevisiae using trans-kingdom conjugation. Plasmid 30:251–257. doi: 10.1006/plas.1993.1056. [DOI] [PubMed] [Google Scholar]
- 47.Inomata K, Nishikawa M, Yoshida K. 1994. The yeast Saccharomyces kluyveri as a recipient eukaryote in transkingdom conjugation: behavior of transmitted plasmids in transconjugants. J Bacteriol 176:4770–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waters VL. 2001. Conjugation between bacterial and mammalian cells. Nat Genet 29:375–376. doi: 10.1038/ng779. [DOI] [PubMed] [Google Scholar]
- 49.Karas BJ, Diner RE, Lefebvre SC, McQuaid J, Phillips AP, Noddings CM, Brunson JK, Valas RE, Deerinck TJ, Jablanovic J, Gillard JT, Beeri K, Ellisman MH, Glass JI, Hutchison CA III, Smith HO, Venter JC, Allen AE, Dupont CL, Weyman PD. 2015. Designer diatom episomes delivered by bacterial conjugation. Nat Commun 6:6925. doi: 10.1038/ncomms7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schröder G, Schuelein R, Quebatte M, Dehio C. 2011. Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae. Proc Natl Acad Sci U S A 108:14643–14648. doi: 10.1073/pnas.1019074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernández-González E, de Paz HD, Alperi A, Agúndez L, Faustmann M, Sangari FJ, Dehio C, Llosa M. 2011. Transfer of R388 derivatives by a pathogenesis-associated type IV secretion system into both bacteria and human cells. J Bacteriol 193:6257–6265. doi: 10.1128/JB.05905-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siamer S, Dehio C. 2015. New insights into the role of Bartonella effector proteins in pathogenesis. Curr Opin Microbiol 23:80–85. doi: 10.1016/j.mib.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 53.Mitrikeski PT. 2013. Yeast competence for exogenous DNA uptake: towards understanding its genetic component. Antonie Van Leeuwenhoek 103:1181–1207. doi: 10.1007/s10482-013-9905-5. [DOI] [PubMed] [Google Scholar]
- 54.Klasson L, Kambris Z, Cook PE, Walker T, Sinkins SP. 2009. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genomics 10:33. doi: 10.1186/1471-2164-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rämö P, Drewek A, Arrieumerlou C, Beerenwinkel N, Ben-Tekaya H, Cardel B, Casanova A, Conde-Alvarez R, Cossart P, Csúcs G, Eicher S, Emmenlauer M, Greber U, Hardt WD, Helenius A, Kasper C, Kaufmann A, Kreibich S, Kühbacher A, Kunszt P, Low SH, Mercer J, Mudrak D, Muntwiler S, Pelkmans L, Pizarro-Cerda J, Podvinec M, Pujadas E, Rinn B, Rouilly V, Schmich F, Siebourg-Polster J, Snijder B, Stebler M, Studer G, Szczurek E, Truttmann M, von Mering C, Vonderheit A, Yakimovich A, Bühlmann P, Dehio C. 2014. Simultaneous analysis of large-scale RNAi screens for pathogen entry. BMC Genomics 15:1162. doi: 10.1186/1471-2164-15-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhatty M, Laverde Gomez JA, Christie PJ. 2013. The expanding bacterial type IV secretion lexicon. Res Microbiol 164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christie PJ, Whitaker N, González-Rivera C. 2014. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta 1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kado CI. 2000. The role of the T-pilus in horizontal gene transfer and tumorigenesis. Curr Opin Microbiol 3:643–648. doi: 10.1016/S1369-5274(00)00154-5. [DOI] [PubMed] [Google Scholar]
- 61.Harrington LC, Rogerson AC. 1990. The F pilus of Escherichia coli appears to support stable DNA transfer in the absence of wall-to-wall contact between cells. J Bacteriol 172:7263–7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Babić A, Lindner AB, Vulić M, Stewart EJ, Radman M. 2008. Direct visualization of horizontal gene transfer. Science 319:1533–1536. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- 63.Kawai S, Pham TA, Nguyen HT, Nankai H, Utsumi T, Fukuda Y, Murata K. 2004. Molecular insights on DNA delivery into Saccharomyces cerevisiae. Biochem Biophys Res Commun 317:100–107. doi: 10.1016/j.bbrc.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 64.Singer K, Shiboleth YM, Li J, Tzfira T. 2012. Formation of complex extrachromosomal T-DNA structures in Agrobacterium tumefaciens-infected plants. Plant Physiol 160:511–522. doi: 10.1104/pp.112.200212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang Z, Tzfira T. 2013. In vivo formation of double-stranded T-DNA molecules by T-strand priming. Nat Commun 4:2253. doi: 10.1038/ncomms3253. [DOI] [PubMed] [Google Scholar]
- 66.Dafny-Yelin M, Levy A, Dafny R, Tzfira T. 2015. Blocking T-strand conversion to double-stranded intermediates by overexpression of yeast DNA replication factor A. Plant Physiol 167:153–163. doi: 10.1104/pp.114.250639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tzfira T, Frankmen L, Vaidya M, Citovsky V. 2003. Site-specific integration of Agrobacterium T-DNA via double-stranded intermediates. Plant Physiol 133:1011–1023. doi: 10.1104/pp.103.032128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chilton MD, Que Q. 2003. Targeted integration of T-DNA into the tobacco genome at double-strand breaks: new insights on the mechanism of T-DNA integration. Plant Physiol 133:956–965. doi: 10.1104/pp.103.026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tzfira T, Li J, Lacroix B, Citovsky V. 2004. Agrobacterium T-DNA integration: molecules and models. Trends Genet 20:375–383. doi: 10.1016/j.tig.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 70.Piers KL, Heath JD, Liang X, Stephens KM, Nester EW. 1996. Agrobacterium tumefaciens-mediated transformation of yeast. Proc Natl Acad Sci U S A 93:1613–1618. doi: 10.1073/pnas.93.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bundock P, Hooykaas PJ. 1996. Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc Natl Acad Sci U S A 93:15272–15275. doi: 10.1073/pnas.93.26.15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Groot MJ, Bundock P, Hooykaas PJ, Beijersbergen AG. 1998. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol 16:839–842. (Erratum, 16:1074.) doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- 73.Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF. 2005. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr Genet 48:1–17. doi: 10.1007/s00294-005-0578-0. [DOI] [PubMed] [Google Scholar]
- 74.Machado-Ferreira E, Balsemão-Pires E, Dietrich G, Hojgaard A, Vizzoni VF, Scoles G, Bell-Sakyi L, Piesman J, Zeidner NS, Soares CA. 2015. Transgene expression in tick cells using Agrobacterium tumefaciens. Exp Appl Acarol 67:269–287. doi: 10.1007/s10493-015-9949-5. [DOI] [PubMed] [Google Scholar]
- 75.Kunik T, Tzfira T, Kapulnik Y, Gafni Y, Dingwall C, Citovsky V. 2001. Genetic transformation of HeLa cells by Agrobacterium. Proc Natl Acad Sci U S A 98:1871–1876. doi: 10.1073/pnas.041327598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S. 2013. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 77.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 78.Moriguchi K, Yamamoto S, Ohmine Y, Suzuki K. 2016. A fast and practical yeast transformation method mediated by Escherichia coli based on a trans-kingdom conjugal transfer system: just mix two cultures and wait one hour. PLoS One 11:e0148989. doi: 10.1371/journal.pone.0148989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hill AB, Chen M, Chen CK, Pfeifer BA, Jones CH. 2016. Overcoming gene-delivery hurdles: physiological considerations for nonviral vectors. Trends Biotechnol 34:91–105. doi: 10.1016/j.tibtech.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Llosa M, Schröder G, Dehio C. 2012. New perspectives into bacterial DNA transfer to human cells. Trends Microbiol 20:355–359. doi: 10.1016/j.tim.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 81.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. 2012. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol 13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 82.Khatoon J, Rai RP, Prasad KN. 2016. Role of Helicobacter pylori in gastric cancer: updates. World J Gastrointest Oncol 8:147–158. doi: 10.4251/wjgo.v8.i2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cover TL. 2016. Helicobacter pylori diversity and gastric cancer risk. mBio 7:e01869-15. doi: 10.1128/mBio.01869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson KM, Sieber KB, Dunning Hotopp JC. 2013. A review of bacteria-animal lateral gene transfer may inform our understanding of diseases like cancer. PLoS Genet 9:e1003877. doi: 10.1371/journal.pgen.1003877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson KM, Dunning Hotopp JC. 2014. Mobile elements and viral integrations prompt considerations for bacterial DNA integration as a novel carcinogen. Cancer Lett 352:137–144. doi: 10.1016/j.canlet.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]