ABSTRACT
Cell density-dependent regulation of gene expression in Xylella fastidiosa that is crucial to its switching between plant hosts and insect vectors is dependent on RpfF and its production of 2-enoic acids known as diffusible signal factor (DSF). We show that X. fastidiosa produces a particularly large variety of similar, relatively long-chain-length 2-enoic acids that are active in modulating gene expression. Both X. fastidiosa itself and a Pantoea agglomerans surrogate host harboring X. fastidiosa RpfF (XfRpfF) is capable of producing a variety of both saturated and unsaturated free fatty acids. However, only 2-cis unsaturated acids were found to be biologically active in X. fastidiosa. X. fastidiosa produces, and is particularly responsive to, a novel DSF species, 2-cis-hexadecanoic acid that we term XfDSF2. It is also responsive to other, even longer 2-enoic acids to which other taxa such as Xanthomonas campestris are unresponsive. The 2-enoic acids that are produced by X. fastidiosa are strongly affected by the cellular growth environment, with XfDSF2 not detected in culture media in which 2-tetradecenoic acid (XfDSF1) had previously been found. X. fastidiosa is responsive to much lower concentrations of XfDSF2 than XfDSF1. Apparently competitive interactions can occur between various saturated and unsaturated fatty acids that block the function of those agonistic 2-enoic fatty acids. By altering the particular 2-enoic acids produced and the relative balance of free enoic and saturated fatty acids, X. fastidiosa might modulate the extent of DSF-mediated quorum sensing.
IMPORTANCE
X. fastidiosa, having a complicated lifestyle in which it moves and multiplies within plants but also must be vectored by insects, utilizes DSF-based quorum sensing to partition the expression of traits needed for these two processes within different cells in this population based on local cellular density. The finding that it can produce a variety of DSF species in a strongly environmentally context-dependent manner provides insight into how it coordinates the many genes under the control of DSF signaling to successfully associate with its two hosts. Since the new DSF variant XfDSF2 described here is much more active than the previously recognized DSF species, it should contribute to plant disease control, given that the susceptibility of plants can be greatly reduced by artificially elevating the levels of DSF in plants, creating “pathogen confusion,” resulting in lower virulence.
INTRODUCTION
The xylem-limited plant pathogen Xylella fastidiosa causes serious diseases of several important agricultural crops, including Pierce’s disease (PD) of grapevine and variegated chlorosis in citrus (CVC) (1, 2). X. fastidiosa is obligately transmitted from one plant to another by xylem sap-feeding insects. Like related Xanthomonas species, X. fastidiosa utilizes one or more signal molecules known as diffusible signaling factor (DSF) to regulate its behavior in a cell density-dependent manner (3, 4).
Previous studies implicated DSF-mediated cell-cell signaling in host switching by X. fastidiosa. Such signaling is apparently a cue that enables a subset of cells inside the plant to become preadapted to acquisition and transmission by insects to new host plants once a sufficiently high level of DSF is experienced. Acquisition of cells by insect vectors is strongly dependent on their ability to adhere to the walls of the insect’s foregut. DSF-mediated signaling regulates the transition from a nonadhesive, motile phenotype that allows systemic plant colonization to more adhesive cells that can form biofilms in insects and colonize insects (reviewed in reference 5). DSF induces the expression of many genes in X. fastidiosa (6, 7) including hxfA and hxfB that encode hemagglutinin-like proteins that are involved in cell-cell aggregation and biofilm formation (8). Since attachment-promoting traits in X. fastidiosa are incompatible with its movement inside the plant, DSF-producing grape plants were successfully employed to control PD by trapping the pathogen in a phenotype inconsistent with movement in plants, causing them to remain localized near the point of inoculation (9).
The DSF molecules that have been characterized are typically 2-cis enoic acids with a chain length of 12 to 14 carbons (10). To date, eight active DSF molecules have been reported in a variety of bacterial species. DSF (2-cis-11-methyldodecenoic acid), BDSF (2-cis-dodecenoic acid), CDSF [(2-cis,5-cis)-11-methyldodecadienoic acid], IDSF (2-cis-10-methyl-dodecenoic acid; also called DSF-II), 2-cis-9-methyldecenoic acid, and 2-cis-undecenoic acid were isolated from cultures of Xanthomonas campestris pv. campestris (11, 12, 13). DSF, BDSF, and CDSF were also isolated from Xanthomonas oryzae pv. oryzae (14).
BDSF was originally isolated from Burkholderia cenocepacia (15), while CDSF was originally reported to be produced by several Burkholderia species that were also reported to produce BDSF (16). DSF was found to be produced also by Burkholderia multivorans (16). 2-cis-Decenoic acid has been found in Pseudomonas aeruginosa (17). We previously isolated X. fastidiosa DSF (XfDSF) (2-cis-tetradecenoic acid) from a grape strain of X. fastidiosa (18). A saturated acid molecule, (12-methyltetradecenoic acid) isolated from an X. fastidiosa CVC strain was proposed to be a DSF molecule (19), but it has not been shown to be biologically active.
DSFs are synthesized by RpfF, a unique crotonase that has both 3-hydroxyacyl-acyl carrier protein (ACP) dehydratase and thioesterase activity (20). RpfF first catalyzes the formation of a double bond between carbons 2 and 3 of a 3-hydroxyacyl moiety and then hydrolyzes the thioester bond with ACP to release a free acid. 13C-labeling experiments demonstrated that glucose acts as a substrate to provide a carbon element for DSF biosynthesis (21). Once DSF reaches a threshold concentration outside the cell, it activates its cognate receptor, RpfC, a hybrid membrane sensor kinase that phosphorylates the intracellular response regulator RpfG. RpfG then converts the intercellular signal into an intracellular signal through its cyclic di-GMP phosphodiesterase activity (22), which in turn, alters the expression of target genes (7, 23). In a previous study (24), we demonstrated that the DSF sensing mechanism in X. fastidiosa, unlike in X. campestris, is RpfF dependent; an rpfF deletion mutant could not sense externally applied DSF, while a strain harboring an rpfF variant (designated rpfF*) in which DSF synthesis was blocked via substitution of two glutamic acid residues with alanine residues (E141A and E161A), was able to sense and respond to DSF. This strain was the basis for an X. fastidiosa-based DSF sensor we designate the XfDSF-biosensor strain.
The composition of the mixed DSF signals produced by X. oryzae was shown to be influenced by the composition of the culture media in which it grew (12, 14). In addition, while DSF or BDSF production has not been observed in X. fastidiosa, replacement of the native RpfF of X. campestris with that of X. fastidiosa (XfRpfF) conferred production of DSF and BDSF as well as abundant XfDSF (20). Therefore, we hypothesized that XfRpfF, while exhibiting some degree of specificity in the production of the longer-chain XfDSF, is relatively promiscuous compared to that of RpfF from X. campestris, and its products will be strongly influenced by the host in which it is being expressed and its environment. This suggests that a different compositional mixture of known DSF species and perhaps novel DSF species might be produced by XfRpfF when grown under different conditions, such as in the xylem of host plants.
RESULTS
XfRpfF produces a collection of free fatty acids.
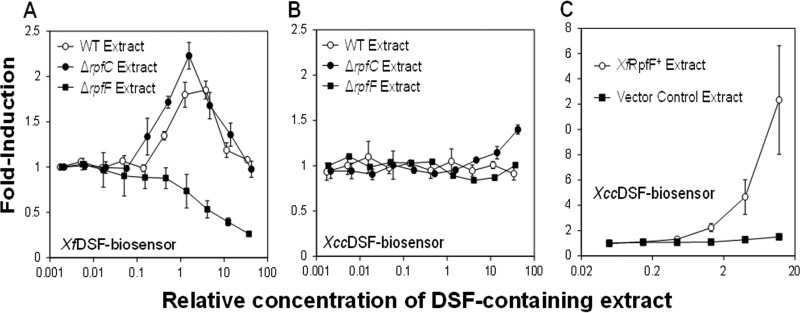
In a previous study (18), we isolated and characterized a 14-carbon DSF species 2-cis-tetradecenoic acid (XfDSF) that was produced and recognized by X. fastidiosa. In this study, XfDSF was produced in the surrogate host Pantoea agglomerans 299R harboring XfRpfF, and its isolation was guided by an X. campestris-based DSF-biosensor (4), which we designate the XccDSF-biosensor strain (Xcc stands for X. campestris pv. campestris) here. XfDSF production in X. fastidiosa was then confirmed in an rpfC mutant strain that overproduces DSF (6, 18). Since the XccDSF-biosensor strain is more responsive to the DSF produced by X. campestris than to the XfDSF produced by X. fastidiosa (18, 24), we hypothesized that other DSF molecules to which X. campestris would be unresponsive are produced by X. fastidiosa. In order to address this hypothesis, DSF production by a wild-type (WT) strain of X. fastidiosa, a ΔrpfC mutant which is an overproducer of DSF (6), and a ΔrpfF mutant blocked in DSF production was assessed using both the XccDSF-biosensor strain and the XfDSF-biosensor strain. While the XfDSF-biosensor strain was activated by DSF-containing extracts of cultures of the WT and the ΔrpfC mutant representing as little as 0.42 and 0.17 the concentration present in the original culture media, respectively (Fig. 1A), the XccDSF-biosensor strain was activated only by the DSF-containing extract of the ΔrpfC mutant that was 14-fold higher than that in the original culture media (Fig. 1B). No DSF could be detected by the XccDSF-biosensor strain in extracts of cultures of the WT strain even at high concentrations (Fig. 1B). Since the XfDSF-biosensor strain responds to XfDSF at 10-fold-lower concentrations than those perceived by the XccDSF-biosensor strain (18), the greater responsiveness of the XfDSF-biosensor strain to extracts of cultures of X. fastidiosa than that of the XccDSF-biosensor strain suggested that DSF species that were not recognized by X. campestris were present in these extracts. While the XccDSF-biosensor strain was largely unresponsive to extracts of X. fastidiosa cultures, a strong response to extracts of cultures of P. agglomerans harboring XfRpfF was seen (Fig. 1C), suggesting that the cellular environment of XfRpfF determines the quantities and/or properties of the DSF species it produces. This crude extract was toxic to the XfDSF-biosensor strain (not shown) and therefore could not be tested for active molecules in the Xylella system.
FIG 1 .
(A) Induction of the Xylella fastidiosa-based DSF biosensor strain (XfDSF-biosensor strain) and (B) the Xanthomonas campestris-based DSF biosensor strain (XccDSF-biosensor strain) by various concentrations of DSF-containing culture extracts of Xylella fastidiosa strain. (C) Induction of the XccDSF-biosensor strain by extracts of Pantoea agglomerans 299R expressing XfRpfF or harboring a vector only. The extract concentration reflects the volume of culture supernatant that would have been extracted with ethyl acetate to yield that added to 1 volume of assay medium.
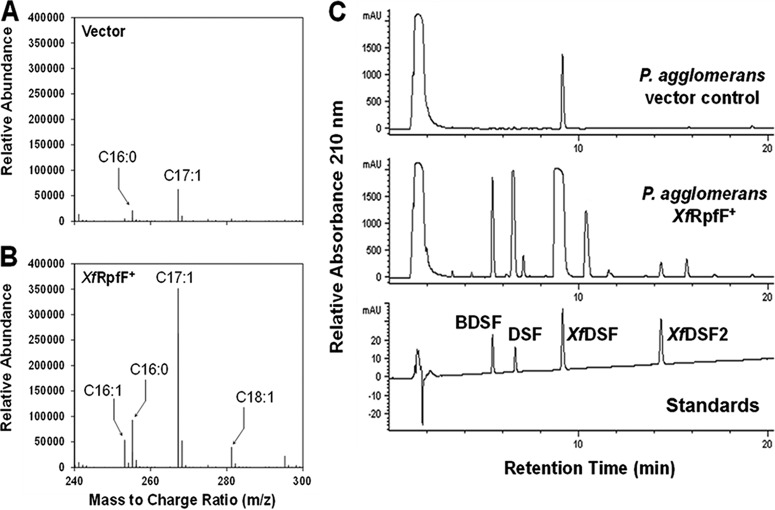
DSF-containing extracts of WT X. fastidiosa and the ΔrpfC and ΔrpfF mutants, as well as P. agglomerans expressing XfRpfF or harboring an empty vector, were analyzed by electrospray ionization mass spectrometry (ESI-MS) in a negative-ion mode to identify fatty acids that were expected to serve as signaling molecules. Free ionized fatty acids were identified by their mass-to-charge ratio (m/z). The accumulation of several saturated and unsaturated fatty acids was strongly dependent on XfRpfF in both X. fastidiosa and P. agglomerans (Table 1; Fig. 2A and B). Five unsaturated fatty acids detected in P. agglomerans were apparently XfRpfF-dependent, being 5-fold or more abundant in strains harboring XfRpfF than in the vector control. These molecules could be tentatively identified as enoic acids from their m/z values: 225.185 (C14:1), 253.217 (C16:1), 267.232 (C17:1), 281.248 (C18:1), and 295.263 (C19:1) (Table 1). In addition, molecules with an m/z corresponding to DSF (m/z = 211.169) and BDSF (m/z = 197.154) were also found only in cells harboring XfRpfF (Table 1), but their overall abundance and enrichment in the strain harboring XfRpfF compared to the control strain was low. Three of these DSF-like molecules, C16:1, C17:1, and C18:1, were also found to be RpfF dependent in extracts of X. fastidiosa cultures, being in higher abundance in extracts of the ΔrpfC mutant than in the WT strain (Table 1) and greatly reduced in abundance in a ΔrpfF mutant. Interestingly, XfDSF (C14:1) was not among the molecules produced by the WT X. fastidiosa strain, perhaps since the culture medium (PD3) used in this study was different than the periwinkle wilt GelRite medium (PWG) used previously (18). A low apparent abundance of C14:1 in extracts of the ΔrpfC X. fastidiosa strain (Table 1) probably accounts for the weak responsiveness of the XccDSF-biosensor strain to culture extracts of this strain, and suggested that one or more of the other putative enoic acids contributed to the responsiveness of the XfDSF-biosensor strain (Fig. 1B).
TABLE 1 .
Abundance of putative fatty acid-derived molecular ionsa
| Mass/charge (m/z)b | Formulab | Predicted molecule | Abundance of putative fatty acid-derived molecular ions inc: |
||||
|---|---|---|---|---|---|---|---|
|
P. agglomerans
|
X. fastidiosa strains |
||||||
| XfRpfF+ | Vector | WT | ΔrpfC | ΔrpfF | |||
| 197.154 | C12H21O2 | BDSF (C12:1) | 1,460 | 635 | 0 | 0 | 0 |
| 211.169 | C13H23O2 | DSF (C13:1) | 1,724 | 683 | 0 | 0 | 0 |
| 225.185 | C14H25O2 | XfDSF (C14:1) | 4,165 | 469 | 0 | 421 | 0 |
| 239.201 | C15H27O2 | C15:1 | 0 | 0 | 858 | 0 | 0 |
| 253.216 | C16H29O2 | XfDSF2 (C16:1) | 51,594 | 5,269 | 258,017 | 394,650 | 13,967 |
| 251.201 | C16H27O2 | C16:2 | 130 | 0 | 0 | 0 | 0 |
| 267.232 | C17H31O2 | C17:1 | 351,331 | 62,930 | 1,855 | 6,046 | 0 |
| 281.248 | C18H33O2 | C18:1 | 39,796 | 4,268 | 23,281 | 57,148 | 915 |
| 295.263 | C19H35O2 | C19:1 | 22,064 | 295 | 363 | 0 | 0 |
| 309.279 | C20H37O2 | C20:1 | 0 | 0 | 307 | 0 | 0 |
| 199.169 | C12H23O2 | C12:0 | 1,481 | 29 | 8,419 | 1,378 | 2,371 |
| 213.185 | C13H25O2 | C13:0 | 0 | 0 | 0 | 645 | 715 |
| 227.201 | C14H27O2 | C14:0 | 6,080 | 1935 | 0 | 0 | 0 |
| 241.216 | C15H29O2 | CVC-DSF (C15:0) | 213 | 35 | 7,225 | 4,009 | 0 |
| 255.232 | C16H31O2 | C16:0 | 92,927 | 20,284 | 108,892 | 216,303 | 6,069 |
| 269.248 | C17H33O2 | C17:0 | 4,305 | 754 | 1994 | 16,567 | 0 |
| 283.263 | C18H35O2 | C18:0 | 1,896 | 608 | 22,145 | 71,704 | 830 |
| 297.279 | C19H37O2 | C19:0 | 0 | 0 | 0 | 0 | 0 |
| 311.295 | C20H39O2 | C20:0 | 0 | 0 | 543 | 8,187 | 0 |
Abundance of putative fatty acid-derived molecular ions resolved by electrospray ionization-mass spectrometry (ESI-MS) analysis of DSF-containing culture extracts of P. agglomerans and X. fastidiosa strains.
The m/z values and chemical formulas are of ionized molecules.
Abundance of putative fatty acid-derived molecular ions (in relative arbitrary counts) resolved by ESI-MS analysis of DSF-containing culture extracts of P. agglomerans expressing vector only or expressing XfRpfF (XfRpfF+) and three X. fastidiosa strains, the wild-type X. fastidiosa strain and rpfF and rpfC deletion mutant strains.
FIG 2 .
Electrospray ionization mass spectra (ESI-MS) of DSF-containing culture extracts of Pantoea agglomerans 299R harboring a vector only (A) or expressing XfRpfF (B). (C) High-performance liquid chromatographs (HPLC) analysis of Pantoea agglomerans 299R harboring a vector only (top panel) or expressing XfRpfF (middle panel) or solutions (250 µM) of synthetic standards.
It should be noted that fatty acids with identical m/z values could have different unsaturated sites or could be in a trans conformation rather than a cis conformation. For example, C16:1 and C18:1 could represent not only DSF-like molecules with cis unsaturated bonds at carbon atom 2 but also components of the bacterial membrane typically having unsaturated bonds in the middle of the aliphatic chain (reviewed in reference 25). However, given that the relative abundance of ions corresponding to C16:1 or C18:1 were 18- to 62-fold higher in extracts of the X. fastidiosa WT and ΔrpfC strains compared to that in the ΔrpfF strain, DSF-like 2-cis-enoic acid molecules apparently dominate these extracts with other, presumably membrane-derived molecules that are uncommon.
XfRpfF produces 2-cis-hexadecenoic acid.
In order to determine the structure and conformation of the XfRpfF-dependent enoic acids discovered by ESI-MS, we employed reverse-phase high-performance liquid chromatography (HPLC) after separation on a C18 column, using selected synthetic 2-cis enoic acids as standards. Eight XfRpfF-dependent molecules produced by P. agglomerans could be detected (Fig. 2C), while X. fastidiosa extracts did not contain enough material for such an analysis (not shown). Four compounds in extracts of P. agglomerans cultures coeluted with synthetic standards: BDSF (retention time [RT] = 5.5 min), DSF (RT = 7.1 min), XfDSF (RT = 9.2 min), and 2-cis-hexadecenoic acid (RT = 14.3 min) (Fig. 2C). HPLC analysis enabled the relative concentrations of these molecules to be more readily determined than ESI-MS did. Of these four molecules, XfDSF was present in the highest concentration (50 µM), while DSF, BDSF, and 2-cis-hexadecenoic acid were equally abundant. The other four molecules were isolated and tested for activity with the XfDSF-biosensor strain (see Fig. S1 in the supplemental material). Only the compound with a retention time of 10.5 min activated the biosensor, and its analysis by ESI-MS revealed a molecule with an m/z of 251.20, corresponding to a 16-carbon enoic acid with two unsaturated sites. Nucleic magnetic resonance (NMR) analysis confirmed that one unsaturated site is at position 2 and in the cis conformation, but the position and conformation of the second unsaturated site could not be determined (data not shown).
Analysis of extracts of X. fastidiosa cultures and synthetic standard molecules using gas chromatography-mass spectrometry (GC-MS) was used to support the presence of 2-cis-hexadecenoic acid (C16:1) in the extract of the WT strain (see Fig. S2 in the supplemental material). Extracts of the WT and mutant strains were treated with boron trifluoride diethyl etherate (BF3 ⋅ OEt2) to esterify fatty acid derivatives, making them easier to detect by this method. While a peak with the same retention time as that of the synthetic C16:1 was observed in both the WT and ΔrpfC mutant extracts, the mass signature for C16:1 was below the detection limit at the concentrations found in the extracts. Other DSF-like molecules were not identified by GC-MS and were most likely present at concentrations too low to detect using this chromatography method.
Interestingly, small amounts of a compound that coeluted with XfDSF (RT = 9.2 min) were detected in HPLC analysis of extracts of the P. agglomerans control strain harboring only the empty vector (Fig. 2C). DSF signaling has not been reported in this organism. A search for orthologous proteins to XfRpfF in the draft genome of P. agglomerans 299R (26) revealed the presence of gene F385_3254, encoding a protein with 37% identity to that of X. fastidiosa RpfF (see Fig. S3 in the supplemental material). The XccDSF-biosensor strain exhibited a weak response to this extract when present in relatively high concentrations (at higher concentrations than those used in Fig. 1C), further suggesting that P. agglomerans 299R itself produces a small amount of a fatty acid species capable of DSF signaling activity.
XfRpfF-dependent unsaturated fatty acids are biologically active.
Since all demonstrably active DSF species detected thus far are 2-cis-unsaturated fatty acids (10, 11, 14, 15, 18), we posited that the XfRpfF-dependent species C14:1, C16:1, C17:1, C18:1, and C19:1 would be 2-cis-tetradecenoic acid, 2-cis-hexadecenoic acid, 2-cis-heptadecenoic acid, 2-cis-octadecenoic acid, and 2-cis-nonadecanoic acid, respectively. Therefore, we tested the biological activity of a set of synthetic 2-cis enoic acids ranging in length from 2-cis-octenoic acid (C8:1) to 2-cis-eicosenoic acid (C20:1) using both the XfDSF-biosensor strain and the XccDSF-biosensor strain. While the XccDSF-biosensor strain responded to 2-cis-enoic acids that varied in length from 10 to 14 carbons, the XfDSF-biosensor strain responded to molecules with chain lengths of 12 to 18 carbons. 2-cis-Enoic acids with chain lengths of 8 to 11 carbons were toxic to X. fastidiosa but did not influence the growth of X. campestris (Table 2). While X. fastidiosa responded with the greatest sensitivity and intensity to 2-cis-hexadecenoic and 2-cis-heptadecenoic acids, X. campestris was much more responsive to DSF and 2-cis-tridecenoic acid. X. fastidiosa did not respond to any 14-carbon enoic acids with unsaturated sites other than at carbon atom 2 or those with a trans conformation rather than a cis conformation (Table 2; Fig. 3). It did respond, however, although only at higher concentrations, to 9-cis-hexadecenoic acid (Table 2; Fig. 3), but the activity of the XfDSF-biosensor strain in response to a given concentration of this compound was only about half as great as to an equivalent concentration of 2-cis-hexadecenoic acid (Fig. 3). Since 2-cis-hexadecenoic acid was a natural product of XfRpfF in X. fastidiosa (Table 1; see Fig. 2S in the supplemental material) and since X. fastidiosa is particularly responsive to it (Table 2; Fig. 4A), it is a novel X. fastidiosa DSF which we term XfDSF2.
TABLE 2 .
Activity of various unsaturated fatty acids as signal molecules in Xylella fastidiosa and Xanthomonas campestris
| Chain length | Orientation | Unsaturated site | Molecule name |
XfDSF-biosensor strain |
XccDSF-biosensor strain |
||
|---|---|---|---|---|---|---|---|
| Response and/or minimum concn detected (µM) | Fold induction | Response or minimum concn detected (µM) | Fold induction | ||||
| 8 | cis | 2 | 2-cis-Octenoic acid | Toxic 30 | No response | ||
| 9 | cis | 2 | 2-cis-Nonenoic acid | Toxic 1 | No response | ||
| 10 | cis | 2 | 2-cis-Decenoic acid | Toxic 2 | 10 | 4.1 | |
| 11 | cis | 2 | 2-cis-Undecenoic acid | Toxic 3 | 1 | 12.4 | |
| 12 | cis | 2-cis-Dodecenoic acid (BDSF) | 3 | 3.2 | 0.5 | 12.4 | |
| 12 | trans | 2 | 2-trans-Dodecenoic acid | Toxic 6 | 3 | 7.9 | |
| 13 | cis | 2 | 2-cis-Tridecenoic acid | Toxic | 0.1 | 17.9 | |
| 13 | cis | 2 | 2-cis-11-Methyldodecenoic acid (DSF) | 3 | 17.5 | 0.05 | 17.9 |
| 14 | cis | 2 | 2-cis-Tetradecenoic acid (XfDSF) | 1 | 3.3 | 7 | 4.8 |
| 14 | cis | 5 | 5-cis-Tetradecenoic acid | No response | No response | ||
| 14 | cis | 6 | 6-cis-Tetradecenoic acid | No response | No response | ||
| 14 | cis | 9 | 9-cis-Tetradecenoic acid (myristoleic acid) | No response | No response | ||
| 14 | 0 | Tetradecenoic acid (myristic acid) | No response | ||||
| 15 | cis | 2 | 2-cis-Pentadecenoic acid | 10 | 4.2 | No response | |
| 15 | 0 | 12-Methyltetradecanoic acid (CVC-DSF) | No response | No response | |||
| 16 | cis | 2 | 2-cis-Hexadecenoic acid (XfDSF2) | 0.15 | 8.9 | No response | |
| 16 | 0 | Hexadecanoic acid | No response | No response | |||
| 16 | cis | 9 | 9-cis-Hexadecenoic acid (palmitoleic acid) | 1 | 4.6 | No response | |
| 16 | trans | 9 | 9-trans-Hexadecenoic acid (palmitelaidic acid) | No response | No response | ||
| 17 | cis | 2 | 2-cis-Heptadecenoic acid | 0.3 | 8.6 | No response | |
| 18 | cis | 2 | 2-cis-Octadecenoic acid | 1 | 6.5 | No response | |
| 19 | cis | 2 | 2-cis-Nonadecenoic acid | No response | No response | ||
| 20 | cis | 2 | 2-cis-Eicodecenoic acid | No response | No response | ||
FIG 3 .
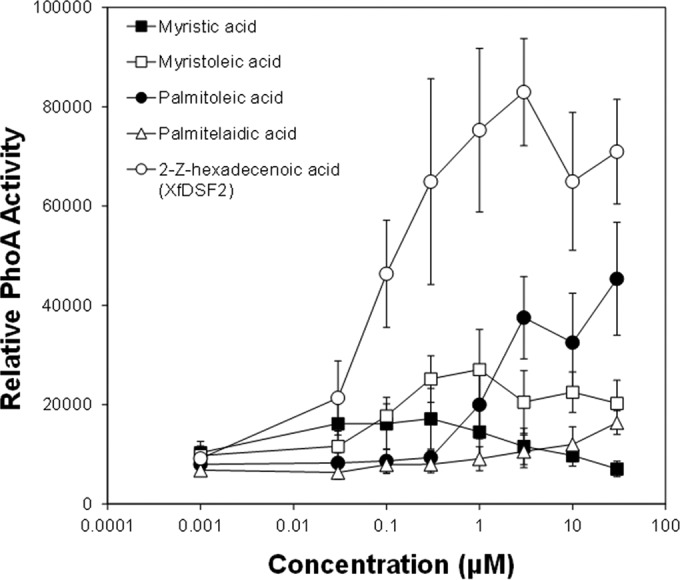
Alkaline phosphatase activity exhibited by the Xylella fastidiosa-based DSF biosensor strain (XfDSF-biosensor strain) in cultures exposed to different concentrations of myristic acid (tetradecanoic acid), myristoleic acid (9-cis-tetradecenoic acid), palmitoleic acid (9-cis-hexadecenoic acid), palmitelaidic acid (9-trans-hexadecenoic acid), and 2-cis-hexadecenoic acid (XfDSF) shown on the abscissa after 96-h incubation. The error bars represent the standard errors of the means.
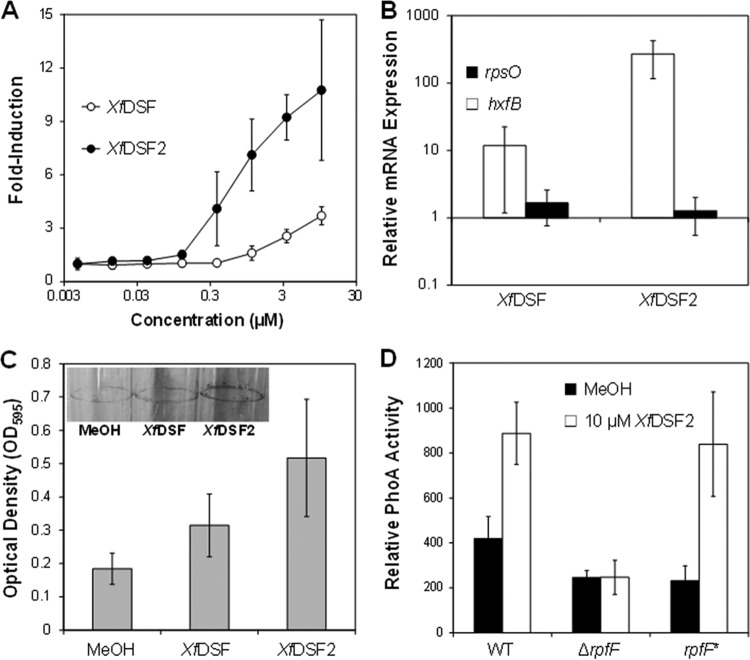
FIG 4 .
(A) Dose-dependent induction of the Xylella fastidiosa-based DSF biosensor (XfDSF-biosensor strain) by various concentrations of XfDSF and XfDSF2. PhoA activity was measured at 72 h. (B) qRT-PCR analysis of hxfB expression as well as expression of the housekeeping gene rpsO in the Xylella fastidiosa rpfF* strain after 72 h or growth in PD3 broth supplemented with 10 µM XfDSF or 10 µM XfDSF2. rpoD and rpsO were used endogenous control genes. Values shown are the ratios of transcript abundance relative to that of cells exposed to a similar volume of MeOH alone. (C) Biofilm formation at the liquid-air interface of shaken glass tubes by the Xylella fastidiosa rpfF* mutant strain after 24 h of growth in PD3 in broth supplemented with 10 µM XfDSF, 10 µM XfDSF2, or MeOH alone as measured by a crystal violet assay. (D) Induction of the hxfA′::phoA transcriptional fusion in Xylella fastidiosa WT, ΔrpfF, and rpfF* strains by 10 µM XfDSF2 as determined by alkaline phosphatase activity.
XfDSF2 is more active as a signaling molecule than XfDSF.
X. fastidiosa responded to lower concentrations of XfDSF2 than to XfDSF (minimum detected concentrations of 0.15 µM versus 1.0 µM, respectively) as measured by the promoter activity of the hxfA gene in the XfDSF-biosensor strain (Table 2; Fig. 4A). Furthermore, above these threshold levels, the alkaline phosphatase activity exhibited by the XfDSF-biosensor strain at a given concentration was much higher in the presence of XfDSF2 than in the presence of XfDSF (Fig. 4A). The higher ability of XfDSF2 to induce gene expression in X. fastidiosa was confirmed in measurements of the expression of the hxfB gene, encoding another hemagglutinin-like protein that is negatively controlled by XfRpfF but located elsewhere in the genome. Expression of hxfA in the X. fastidiosa rpfF* mutant upon exposure to 10 µM XfDSF2 was more than 10-fold higher than when exposed to 10 µM XfDSF (Fig. 4B). Given that XfDSF stimulates biofilm formation and increases the attachment of cells to the liquid-air interface of shaken cultures in glass tubes (15), we compared the apparent adhesiveness of cells of the rpfF* mutant in the presence of 10 µM XfDSF or XfDSF2 grown in this way. After as little as 24 h of incubation, the number of cells attached to the glass was 2.8- and 1.7-fold higher in the presence of XfDSF2 and XfDSF1, respectively, than in the control (Fig. 4C), indicating that XfDSF2 more effectively induces the transition of planktonic to sessile cells. We previously also reported that expression of hxfA in the X. fastidiosa ΔrpfF deletion mutant was unresponsive to XfDSF (20); this mutant also did not respond to XfDSF2, while a high level of induction of hxfA was observed in the rpfF* mutant (Fig. 4D).
Saturated fatty acids inhibit induction of the XfDSF2-dependent hxfA promoter.
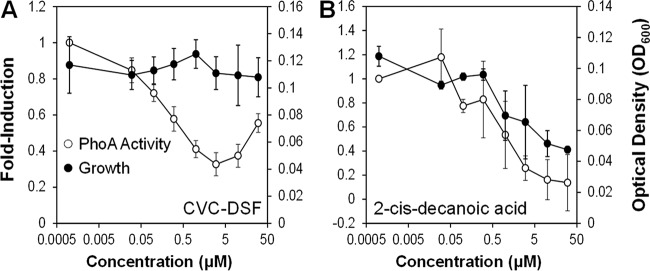
While saturated fatty acids conferred no induction of hxfA in the XfDSF-biosensor strain (Table 2), some fatty acids, such as tetradecanoic acid (myristic acid) and CVC-DSF, suppressed the basal activity of the XfDSF-biosensor strain in a dose-dependent manner without affecting its growth (Fig. 5A). This phenomenon was distinct from that seen with short-chain 2-cis-enoic acids, such as 2-cis-decenoic acid that both inhibited hxfA activity and suppressed growth of the XfDSF-biosensor strain in a dose-dependent manner (Fig. 5B). The latter molecules were therefore termed “inhibitory fatty acids” to distinguish them from the “antagonistic” saturated fatty acids that merely reduced the expression of DSF-dependent genes in X. fastidiosa. Not only did CVC-DSF reduce the basal expression of the hxfA promoter in the XfDSF-biosensor strain (Table 2), it also reduced the responsiveness of X. fastidiosa to DSF species such as XfDSF2. The activity of the XfDSF-biosensor strain in the presence of 1 µM XfDSF2 decreased proportionally as the concentration of CVC-DSF added to cultures increased (Fig. 6). At relatively high concentrations of CVC-DSF (10 µM), no apparent induction of the hxfA promoter was observed, even in the presence of sufficient XfDSF2 to elicit strong induction of hxfA. At equal molar concentrations of XfDSF2 and CVC-DSF, the apparent expression of the hxfA promoter, as measured by the alkaline phosphatase activity exhibited by the XfDSF-biosensor strain, was reduced about 50% (Fig. 6). Induction of the hxfA promoter by various agonistic fatty acids, including XfDSF, XfDSF2, and 9-cis-hexadecenoic acid, could all be suppressed in the presence of saturated fatty acids such as hexadecanoic acid (palmitic acid). In all cases, the addition of palmitic acid to cultures containing one of these various DSF species decreased the alkaline phosphatase activity exhibited by the XfDSF-biosensor strain in a dose-dependent manner (Fig. 7). As was seen with mixtures of CVC-DSF and XfDSF2, equal molar concentrations of palmitic acid added to culture media with XfDSF2 reduced the apparent expression of the hxfA promoter about 50%. Likewise, induction of hxfA by XfDSF was reduced about 50% when an equal molar concentration of palmitic acid was added to cultures (Fig. 7). Curiously, even though 9-cis-hexadecenoic acid (palmitoleic acid) was able to induce hxfA expression at relatively high concentrations, it also reduced the induction of hxfA in the presence of XfDSF2 by ca. 3-fold when added at equal molar concentrations to culture media (Fig. 7). Thus, the regulation of genes dependent on XfRpfF and thus, DSF-mediated signaling in X. fastidiosa, appears to be conferred by the presence of a variety of similar, relatively long-chain unsaturated fatty acids. Furthermore, apparently competitive interactions can occur between various saturated and unsaturated fatty acids that are not effective inducers of DSF-dependent gene expression to block the function of those unsaturated fatty acids that can successfully interact with DSF receptors to modulate gene expression.
FIG 5 .
Suppression of the activity of the alkaline phosphatase activity exhibited by the Xylella fastidiosa-based DSF biosensor (XfDSF-biosensor strain) by different concentrations of CVC-DSF (12-methyltetradecanoic acid) (A) and 2-cis-decanoic acid (B). PhoA activity was measured after 96 h of incubation and is shown as the proportion of that exhibited by control cultures to which no test material was added. Cell concentration (OD600) of cultures after incubation for 96 h (closed circles) is shown on the right ordinate.
FIG 6 .
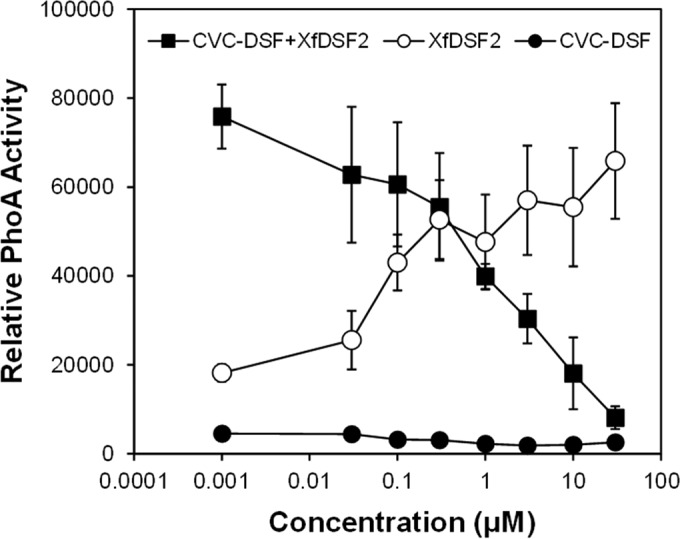
Alkaline phosphatase activity exhibited by the Xylella fastidiosa-based DSF biosensor (XfDSF-biosensor strain) in cultures exposed to various concentrations of 2-cis-hexadecenoic acid (XfDSF2), 12-methyltetradecanoic acid (CVC-DSF), or both 1 µM 2-cis-hexadecenoic acid and various concentrations of 12-methyltetradecanoic acid after 96-h incubation. The error bars represent the standard errors of the means.
FIG 7 .

Alkaline phosphatase activity exhibited by the XfDSF-biosensor strain in cultures exposed to various concentrations of fatty acids. The cultures were exposed to 9-cis-hexadecenoic acid (palmitoleic acid) or 2-cis-hexadecenoic acid (XfDSF2) or to both 3 µM 9-cis-hexadecenoic acid and various concentrations of hexadecanoic acid (palmitic acid), 3 µM 2-cis-tetradecenoic acid (XfDSF) and various concentrations of hexadecanoic acid), 1 µM 2-cis-hexadecenoic acid (XfDSF2) and various concentrations of hexadecanoic acid, 1 µM 2-cis-hexadecenoic acid and various concentrations of 9-cis-hexadecanoic acid shown on the abscissa after 96-h incubation. The error bars represent the standard errors of the means.
DISCUSSION
Structurally confirmed members of the DSF family are all fatty acids that are unsaturated at carbon atom 2, a feature shown to be important for signaling in Xanthomonas and Burkholderia species (10, 20). X. fastidiosa RpfF, like the DSF synthases of other bacterial species, is a unique member of the corotonase family that produce 2-cis enoic acids. In this work we present evidence that X. fastidiosa RpfF produces and responds to exceptionally long-chain DSF species such as 2-cis-hexdecenoic acid, that we term XfDSF2. Some evidence from mass spectrometry suggests that X. fastidiosa RpfF might produce even longer 2-cis enoic acids of 17 and 18 carbons. However, while X. fastidiosa is quite responsive to these two enoic acids (Table 2), insufficient amounts were recovered from culture media to confirm their levels.
While short- and medium-chain-length DSF species, such as BDSF and XfDSF, can be predicted from their partition coefficient (log Kow = 4.78 and 5.77, respectively, where Kow is the octanol-water partition coefficient; http://www.chemspider.com) to be sufficiently soluble in water (23 µM and 4.2 µM, respectively) to account for their ability to act as signaling molecules at concentrations near 1 µM, XfDSF2 is more hydrophobic and is estimated to have a solubility of only about 0.5 µM (log Kow = 6.58). However, the X. fastidiosa DSF-biosensor strain exhibited a progressive dose-dependent response when XfDSF2 was added to culture media at concentrations greater than 0.5 µM (Fig. 3 and 4A), indicating that it was bioavailable. These observations raise the question as to how such an apparently insoluble signal molecule such as XfDSF2 can be acquired by cells and subsequently interact with the DSF sensor RpfC. We hypothesize that since DSF species, particularly XfDSF2, are hydrophobic, they would interact with hydrophobic matrices such as bacterial membranes. Exogenously supplied DSF might thus be inserted into membranes where they are then distributed from one cell to another by contact or by sharing outer membrane vesicles shed by cells. While X. fastidiosa is a prolific producer of outer membrane vesicles (27), further work will be needed to address whether such vesicles play a role in DSF-mediated cell-cell signaling, as has been suggested for PQS (Pseudomonas quinolone signal) in Pseudomonas aeruginosa (28). Since X. fastidiosa lives exclusively within either xylem vessels or the mouthparts of insect vectors, both sites of rapid fluid flow, an extracellular signal molecule would be expected to be removed from this habitat if it were freely soluble. The relatively insoluble enoic acids such as XfDSF and XfDSF2 to which X. fastidiosa is most responsive are therefore less likely to be lost from the producing cells.
XfRpfF is a promiscuous enzyme that has the potential to produce a variety of DSF molecules. As has been suggested from other studies of X. campestris (14), the products of X. fastidiosa RpfF are very dependent on the host environment in which it is expressed. For instance, XfDSF was isolated from X. fastidiosa grown on PWG in a previous study (18), but when X. fastidiosa was grown on PD3 plates in this study, we could not detect XfDSF in the extracts, indicating that the environment in which XfRpfF functions determines the DSF species that it produces. This suggests that the dominant DSF species produced by bacteria in their natural habitat might be different than those produced in a given in vitro environment. Presumably such patterns of DSF production maximizes fitness by enabling appropriate responses to a given habitat.
It was reported (29) that in Stenotrophomonas maltophilia, RpfF plays a role in the biosynthesis of eight fatty acids, two of which are saturated fatty acids while six are unsaturated fatty acids (all six with double bonds at position 2). Similarly, we report here that the accumulation of several saturated and unsaturated fatty acids is dependent upon RpfF. Since RpfF is a bifunctional crotonase having both dehydratase and thioesterase activities (20), this observation can be explained if some of its substrates are cleaved without being first dehydrated. In Burkholderia cenocepacia, the RpfF homolog has been reported to catalyze the in vitro cleavage of acyl-ACP thioester bonds to yield a holo-ACP and a free fatty acid, a process independent of its dehydratase activity (20). Since the thioester bond is required for the formation of the unsaturated bond at carbon atom 2 (30), thioester cleavage would abort the dehydratase reaction and result in the release and accumulation of free saturated fatty acids. As such molecules are produced in vivo in X. fastidiosa, we hypothesize that they play a role in modulating cell-cell signaling. We report here that CVC-DSF and other saturated fatty acids that do not activate hxfA or other DSF-responsive genes in X. fastidiosa antagonize DSF-mediated signaling in X. fastidiosa. This antagonism is apparently not associated with any toxicity to the cells and thus growth inhibition. Instead, such saturated molecules appear to compete directly with 2-enoic acids for DSF receptors such as RpfC, since the responses to various enoic acids were reduced in a dose-dependent manner by an equal concentration of such molecules. This signaling antagonism did not appear to be very specific, as the response to all enoic acids investigated could be blocked by a given saturated fatty acid (Fig. 6 and 7), and a given saturated fatty acid, such as palmitic acid, could block the response to more than one enoic acid, such as XfDSF, XfDSF2, and palmitoleic acid (Fig. 7).
It is tempting to speculate that the role of RpfB, which has been shown to act as a fatty acyl-coenzyme A (CoA) ligase that counteracts RpfF thioesterase activity by catalyzing the uptake and activation of free fatty acids to yield acyl-CoAs (31), is to alter the relative abundance of fatty acids in the cell. Not only would RpfB restore fitness to cells by restoring some ability to synthesize membrane lipids in cells in which RpfF was active, but it could also play a role in modulating the composition of free fatty acids present in the cell if they were a selective substrate for RpfF. It was recently reported that in X. campestris, RpfB increased DSF and BDSF turnover (32). That is, by altering the relative balance of free enoic and saturated fatty acids in X. fastidiosa DSF-mediated signaling, RpfB could modulate the extent of quorum sensing. Given that X. fastidiosa both produces and is responsive to a particularly large range of enoic acids, the suggestion that it may have more than one receptor for such signal molecules (6) offers the possibility of having a contextual response to increasing cell density. For example, it is apparent from this study that different enoic acids are produced under different culture conditions, and thus, they probably also differ under the various conditions experienced in plant and insect hosts. Furthermore, the output of quorum sensing might also be modulated by the activity of RpfB (33). It is intriguing that unlike other bacteria capable of DSF-based cell signaling such as X. campestris, rpfB is located elsewhere from rpfF in the genome, suggesting that its expression might be independent of that of rpfF, unlike in other bacteria in which they are found together in an operon. The fact that RpfB mutants of X. fastidiosa are more deficient in traits enabling insect colonization and transmission to new host plants than virulence to plants (34) supports the conjecture that RpfB modulates the abundance of DSF species to modulate gene expression.
The complex lifestyle of X. fastidiosa in which the traits that are required for it to move and multiply within the xylem vessels of plants are incompatible with those required for its acquisition from plants by insect vectors may also have led to its apparently more versatile DSF-mediated cell signaling system. As a xylem-limited colonist of plants, X. fastidiosa would encounter relatively few other bacteria in this habitat, since the endophytic population size of most plants is quite low (35). As such, X. fastidiosa may uncommonly encounter other bacteria that produce fatty acid signal molecules. In contrast, species such as X. campestris which has a prominent epiphytic stage on plants might commonly encounter DSF-producing bacteria. Because of this putative chemical isolation, X. fastidiosa may not have been under pressure to restrict the interactions of its DSF receptors only to the various DSF species that it produces. Furthermore, as noted above, it might need to be able to flexibly interact with a suite of signal molecules to enable appropriate varied responses to different environmental conditions. While there has not been extensive study of the diversity of bacteria that produce DSF-like signal molecules, an increasing list of bacterial species capable of producing DSF (36, 37, 38) and the finding here that the common plant epiphyte P. agglomerans can apparently produce at least some XfDSF (Fig. 2C) and harbors an apparently functional homolog to X. fastidiosa RpfF suggest that DSF-mediated signaling may be more common than previously thought. Various enoic acids thus have the potential to participate widely in interspecies interactions, a topic worthy of further investigation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 3. The XfDSF-biosensor strain (previously designated X. fastidiosa rpfF*-XfHA biosensor [24]) consists of the rpfF* mutant (E141A E161A) that exhibits blocked DSF synthesis but can still sense externally applied DSF) harboring pXfHA (hxfA′::phoA). The X. campestris-based DSF-biosensor strain (XccDSF-biosensor) (4) consists of strain 8523 (rpfF mutant) harboring pKLN55 (engXCA′::gfp). P. agglomerans harboring plasmid pVSP61-rpfF (18) and X. campestris and P. agglomerans cells were grown on King’s B medium (KB) (39). Inoculum of X. fastidiosa was grown on periwinkle wilt GelRite medium (PWG medium) plates for 5 to 7 days before transfer to PD3 broth (40). Antibiotics were added to a final concentration of 50 µg ml−1 for kanamycin and 15 µg ml−1 for gentamicin. All cultures were grown on 28°C in the dark.
TABLE 3 .
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant genotype or characteristic(s) | Reference |
|---|---|---|
| Bacterial strains | ||
| X. fastidiosaTemecula1 | Wild type; ATCC 700964 | |
| Dif7 | X. fastidiosa Temecula1ΔrpfF (markerless) | 6 |
| Rpf* | X. fastidiosa Temecula1 rpfF* (E141A E161A) (Kanr) | 24 |
| MIX3 | X. fastidiosa Temecula1 ΔrpfC (Kanr) | This study |
| 8523 | X. campestris pv. campestris 8004 rpfF::Tn5lac (Kanr) | 3 |
| Pantoeaagglomerans299R | Wild type | 41 |
| XccDSF-biosensor | 8523 bearing pKLN55 | 4 |
| XfDSF-biosensor | MIX2 bearing pXfHA | 18 |
| Plasmids | ||
| pFXFkan | pUC19 Kanr [aph(3′)II] | 24 |
| pVSP61 | pVS1 and pACY184 Ori Kanr | 42 |
| pFXF7 | pFXFkan “rpfG-kanR-rpfF” | This study |
| pVSP61-rpfF | pVSP61 kan′::rpfF (rpfF of X. fastidiosa) | 18 |
| pKLN55 | pVSP61 Xanthomonas campestris pv. campestris engXCA′::gfp | 4 |
| pXfHA | pBBR1MCS-5 hxfA′::phoA | 18 |
Extraction of DSF from bacterial cultures.
DSF was extracted from P. agglomerans strains grown for 48 h in 1 to 3 liters of KB broth that was shaken at 200 rpm at 28°C. The pH of the medium was then adjusted to 4.0 (14), and an equal volume of water-saturated ethyl acetate (EtOAc) was added and mixed for 10 min. The EtOAc fraction was then separated from the medium using a separatory flask and concentrated by evaporation using a Rotavapor R evaporator (Büchi, Switzerland). The dried residues were dissolved in 1 to 3 ml methanol (MeOH).
DSF was extracted from X. fastidiosa strains grown for 2 weeks at 28°C on PD3 agar (150 plates; 3 liters). The medium containing the cells was sliced into 8-mm3 cubes and mixed for 2 h in an equal volume of EtOAc. The EtOAc was decanted by filtration through cheesecloth, and the decanted contents were concentrated in vacuo as described above. The dried residues were dissolved in 3 ml MeOH.
Measurement of DSF biological activity in X. campestris pv. campestris.
DSF, BDSF, CVC-DSF, and myristic acid were purchased from Sigma Aldrich, dissolved in MeOH to a concentration of 100 mM and stored at −20°C. DSF-containing culture extracts or synthetic molecules in MeOH were added in MeOH to wells of Falcon 24-well tissue culture plates (Becton Dickinson, USA), and the MeOH was allowed to escape. An equal volume of MeOH was added to some wells as a negative control. Warm (60°C) KB agar containing kanamycin (2 ml) was then added to each well. Inoculum of the XccDSF-biosensor strain was grown for 2 days on KB containing kanamycin and suspended in 10 mM phosphate buffer (pH 7.4), and the optical density at 600 nm (OD600) was adjusted to 0.1 using a Spectronic 21D spectrophotometer (Milton Roy, USA), and 5-µl drops were spotted onto each well. After 2 days of incubation at 28°C, cells were collected and suspended in 0.2 ml of 10 mM phosphate buffer to a final cell density of 0.2 to 0.3 measured at OD600 in Falcon 96-well tissue culture plates (Becton Dickinson). For each well, both relative fluorescence units (RFU) (excitation wavelength, 485 nm; emission wavelength, 515 nm) and cell density measured as OD600 were recorded using a Synergy 2 plate reader (BioTek, USA), and green fluorescent protein (GFP) fluorescence was normalized as RFU OD600−1.
Measurement of DSF biological activity in X. fastidiosa.
Inoculum of the XfDSF-biosensor strain was grown for 5 or 6 days at 28°C on PWG plates containing gentamicin prior to suspension in PD3 broth containing gentamicin (final OD600 of 0.05) and various amounts of synthetic DSF molecules or DSF-containing culture extracts. MeOH only was used as a control, and its concentration in the medium was always 0.01%. Samples were then distributed (800 µl per well) into 48-well clear tissue culture plates (Becton Dickinson) with six replicates. After 96-h incubation at 28°C without shaking, the alkaline phosphatase activity (PhoA activity) was quantified (43). The plate was centrifuged for 10 min at 2,254 × g in an Eppendorf model 5804 centrifuge (Eppendorf, Germany), the growth medium was removed by aspiration, and the cells were resuspended in 0.4 ml of 10 mM Tris base (pH 8.0) containing 10 mM MgSO4. The cells were pelleted again by centrifuging for 10 min at 2,254 × g, resuspended in 0.4 ml of 1 M Tris base (pH 8.0) containing 0.4 mM ZnCl2, and the OD600 in each well was measured. The cells were then disrupted by adding 10 µl of 0.1% sodium dodecyl sulfate (SDS) and 10 µl chloroform to each well followed by 5 min of shaking (200 rpm) at room temperature. Sixty microliters of 1 M Tris base (pH 8.0) containing 0.4 mM ZnCl2 supplemented with 100 µM fluorescein diphosphate (AnaSpec, USA) stock solution was then added to each well, and fluorescence (excitation wavelength, 485 nm; emission wavelength, 515 nm) was measured at 2-min intervals for 30 min. OD600 and fluorescence were both measured using a Synergy 2 plate reader (Biotek, USA). Enzyme activity was calculated as the rate of increase of fluorescence over time divided by the cell density.
Electrospray ionization mass spectrometry (ESI-MS).
DSF-containing culture extracts were analyzed using an LTQ Orbitrap XL mass spectrometer equipped with an electrospray ionization (ESI) source (Thermo Fisher Scientific, Waltham, MA, USA). Mass spectra were recorded in the negative-ion mode over the m/z range from 100 to 500 using the Orbitrap mass analyzer, in profile format, with a resolution setting of 100,000 (as measured at m/z = 400). Mass spectra were processed using Xcalibur software (version 2.0.7 SP1; Thermo Fisher Scientific).
Synthesis of 2-cis-hexadecenoate.
Still-Gennari reagent (2.00 ml, 8.47 mmol, 1.2 equivalents [equiv]) was added to a solution of 1,4,7,10,13,16-hexaoxacyclooctadecane (18-crown-6) (7.45 g, 4 equiv) in tetrahydrofuran (THF) (60 ml) at room temperature. The mixture was cooled to −78°C, and a solution of potassium hexamethyldisilazide in toluene (17.0 ml of 0.5 M solution, 1.2 equiv) was added dropwise over 5 min. The mixture turned bright orange and was stirred for 45 min. A solution of tetradecanal (1.50 g, 7.06 mmol, 1.0 equiv) in THF (10 ml) was added dropwise over 10 min. The reaction mixture was stirred at −78°C for 3 h. The reaction was quenched by the addition of a saturated solution of NH4Cl (50 ml). The layers were separated, and the aqueous layer was extracted with ethyl acetate (extracted three times with 50 ml). The combined organics were washed with brine (once with 50 ml), the resulting liquid was dried over Na2SO4, and the volatiles were removed in vacuo. The crude oil was purified using flash column chromatography using a 5% to 10% gradient of ethyl acetate in hexane. The ethyl ester was isolated as a colorless oil (835 mg, 41%). The ester (806 mg, 2.85 mmol, 1.0 equiv) was dissolved in THF (10 ml). Solid LiOH (411 mg, 17.1 mmol, 6.0 equiv) was dissolved in H2O (10 ml) and added to the ester solution. The biphasic mixture was stirred vigorously at 60°C for 12 h at which point the reaction was complete by thin layer chromatography. The mixture was acidified with a 10% solution of HCl to a pH of 2. The mixture was extracted with ethyl acetate (three times with 30 ml). The combined organics were washed with brine (once with 20 ml), the resulting liquid was dried over Na2SO4, and 2-cis-hexadecenoic acid was isolated as a white solid (678 mg, 93%).
Reverse-phase HPLC analysis.
DSFs were detected and quantified by reverse-phase high-performance liquid chromatography (HPLC) using an Agilent Technologies 1200 series high-performance liquid 150 chromatography system (Agilent Technologies, USA) as follows. Five microliters of the sample was injected into an HPLC column (Ascentis Express C18 column [150 mm long with an inner diameter of 4.6 mm]; Supelco, USA), which was eluted at a flow rate of 1 ml min−1 with two solvent gradients containing 0.1% trifluoroacetic acid, water (eluent A) or methanol (eluent B), at 50°C. The gradient conditions were as follows: starting at 20% eluent A and 80% eluent B, eluent A was linearly decreased to 10% by increasing the amount of eluent B over 20 min, and the eluent was monitored at 210 nm. DSF concentration in crude extracts was determined based on their peak area and calculated from standard curves generated for each of the DSF species using synthetic forms.
GC-MS.
Gas chromatography-mass spectrometry (GC-MS) analysis of acids present in natural extracts was performed by derivatizing the acids into methyl esters. To confirm the presence of XfDSF2 in natural extracts, a synthetic standard was used. DSFs were detected by GC-MS using an Agilent (HP) model 6890N GC with a 5973 mass selective detector. A vial containing 25.0 µl of each natural extract dissolved in MeOH (as prepared in “Extraction of DSF from bacterial cultures” above) was diluted with 25.0 µl of MeOH. To each vial was added 100.0 µl of a 10% (wt/wt) solution of BF3 ⋅ MeOH (commercially available from Sigma Aldrich). Esterification was carried out for 30 min at 45°C. After the reaction, 150.0 µl and 300.0 µl of hexanes were added to each vial, and the vial was vortexed for 20 s. The organic layer was extracted and dried under a stream of argon gas. Isolated solids were resuspended in 50.0 µl in methyl cyanide (MeCN) (acetonitrile) and analyzed by GC-MS.
To confirm the presence of XfDSF2 in natural extracts, a 5.00 mM stock solution of synthetic 2-cis-hexadecenoic acid in MeOH was prepared. To a vial containing 25.0 µl of each natural extract dissolved in MeOH (as prepared in “Extraction of DSF from bacterial cultures” above) was added 25.0 µl of the 5.00 mM stock solution of synthetic 2-cis-hexadecenoic acid, followed by addition of 100.0 µl of a 10% (wt/wt) solution of BF3 ⋅ MeOH. Esterification was carried out for 30 min at 45°C. After the reaction, 150.0 µl and 300.0 µl of hexanes were added to each vial, and the vial was vortexed for 20 s. The organic layer was extracted and dried under a stream of argon gas. Isolated solids were resuspended in 50.0 µl in MeCN and analyzed by GC-MS.
Both the natural and spiked extracts were compared to a blank sample which was prepared by the use of 50.0 µl of pure MeOH treated as described above. The molecular ion for the methyl ester at 268 m/z was not detected by this ionization method, so the fragments at 237 and 171 m/z were used to identify XfDSF2 in natural extracts.
Quantification of gene expression with real-time PCR.
Gene expression analysis was performed on rpfF* mutant X. fastidiosa strain MIX2 (Table 3) subjected to experimental materials in a procedure similar to that employed for measurement of PhoA activity. Cells were harvested from PWG plates after 7 days of growth and suspended in 120-ml PD3 broth to an OD600 of 0.05. The medium was then divided into 3 equal parts of 40 ml that was then supplemented with 10 µM XfDSF1, 10 µM XfDSF2, or MeOH alone as a control. The samples were then distributed into three 48-well clear tissue culture plates (800 µl per well) and incubated at 28°C without shaking for 3 days. Total RNA was isolated using TRIzol extraction as follows. Cells were collected into a 50-ml Falcon tube and centrifuged for 10 min at 2,254 × g in an Eppendorf model 5804. The pellet was resuspended in 1-ml RNAlater RNA stabilization solution (Ambion, TX, USA) and transferred to a 2-ml Eppendorf tube. The cells were harvested again and resuspended in 1-ml TRIzol reagent (Ambion), mixed vigorously, and incubated until completely lysed at 60°C (ca. 5 min). Chloroform (200 µl) was added and mixed well, and the tubes were allowed to stand on the bench for 5 min before being subjected to 10 min of centrifugation at 20,800 × g. The clear upper phase was then collected (ca. 450 µl) and mixed with 500 µl isopropanol. The tubes were incubated for 30 min at −80°C and then centrifuged for 30 min at 20,800 × g at 4°C. The white pellet containing nucleic acids was washed twice with 70% cold ethanol (EtOH), and the pellet was air dried and suspended in 50 µl nucleic acid-free double-distilled water.
DNA was eliminated using Turbo DNase (Ambion, TX, USA). RNA samples were stored at −80°C, and fresh 1-µg aliquots were used for cDNA synthesis before each analysis, using 3 µg of random hexamers and Superscript II reverse transcriptase (Life Technologies, USA) according to the manufacturer’s instructions. Quantitative PCR was performed in an ABI PRISM 7100 sequence detector system (Applied Biosystems, USA). Detection of PCR products was done by measuring the increase in fluorescence produced upon binding of SYBR green dye (Qiagen, Germany) to double-stranded DNA. Both rpoD and rpsO were used as endogenous control genes to normalize gene expression. The primers for the hxfB, rpoD, and rpsO genes have been previously described (24). To ensure that the threshold cycle (CT) values obtained were from a single PCR product, melting curve analysis was run after each analysis. Relative expression (RQ) was calculated from the threshold cycle (CT) as follows: dCT = CT (target gene) − CT (endogenous control) where dCT is the change in the CT and CT (target gene) is the threshold cycle of the target gene; ddCT = dCT (treatment) − dCT (reference); RQ = 2(−ddCT). RQ values (ratios between two compared samples) are presented as means ± standard deviations from three biological replicates of quantitative reverse transcription-PCR (qRT-PCR) assays performed in triplicate.
Biofilm assay.
The effects of XfDSF and XfDSF2 on the biofilm formation capacity of the X. fastidiosa rpfF* mutant were determined in cells growing in PD3 broth cultures. Cells were grown on PWG plates for 7 days, resuspended in PD3 broth to an OD600 of 0.05, and added (2 ml) to PD3 broth cultures containing a final concentration of 10 µM of either 2-cis-tetradecenoic acid or 2-cis-hexdecenoic acid or an equal volume of MeOH alone as a control. The glass tubes, containing 2 ml of a culture, were shaken (200 rpm) for 24 h in glass tubes at 28°C during which time a visible biofilm formed at the liquid-air interface. The medium, containing unattached cells, was then removed by aspiration, the tubes were washed three times with tap water to remove unattached cells, and the biomass attached to the glass wall was stained with 2 ml of 1% crystal violet (CV) for 10 min. Excess CV was removed by washing the tubes three times with tap water, and the retained CV was dissolved in 1 ml of 95% ethanol and quantified by measuring absorbance at 595 nm (Spectronic 21D spectrophotometer; Milton Roy, USA).
SUPPLEMENTAL MATERIAL
Induction of the XfDSF-biosensor strain by either 2 or 10 µl of a purified fraction of a DSF-containing extract of P. agglomerans 299R expressing X. fastidiosa RpfF (fractions named based on their retention times in minutes in HPLC [HPLC RT]). White bars represent induction of the sensor by 10 µM XfDSF2 or MeOH only. Download
GC-MS chromatograms for esterified natural extracts versus the 2-cis-hexadecenoic acid synthetic standard. (a) 2-cis-hexadecenoic acid; (b) RpfF extract; (c) WT extract; (d) RpfC extract. Download
Sequence identity (shown on black background) and similarity (shown on gray background) of X. fastidiosa (Xf) Tem1 RpfF and P. agglomerans (Pa) 299R F385_3254. The F385_3254 gene encodes a protein with 37% identity to that of X. fastidiosa RpfF. Download
ACKNOWLEDGMENTS
This work was supported by a grant from the California Department of Food and Agriculture Pierce’s Disease and Glassy-winged Sharpshooter Board.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
This work was funded by California Department of Food and Agriculture (CDFA).
Footnotes
Citation Ionescu M, Yokota K, Antonova E, Garcia A, Beaulieu E, Hayes T, Iavarone AT, Lindow SE. 2016. Promiscuous diffusible signal factor production and responsiveness of the Xylella fastidiosa Rpf system. mBio 7(4):e01054-16. doi:10.1128/mBio.01054-16.
REFERENCES
- 1.Purcell AH, Hopkins DL. 1996. Fastidious xylem-limited bacterial plant pathogens. Annu Rev Phytopathol 34:131–151. doi: 10.1146/annurev.phyto.34.1.131. [DOI] [PubMed] [Google Scholar]
- 2.Hopkins DL, Purcell AH. 2002. Xylella fastidiosa: cause of Pierce’s disease of grapevine and other emergent diseases. Plant Dis 86:1056–1066. doi: 10.1094/PDIS.2002.86.10.1056. [DOI] [PubMed] [Google Scholar]
- 3.Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJG, Slater H, Dow JM, Williams P, Daniels MJ. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol 24:555–566. doi: 10.1046/j.1365-2958.1997.3721736.x. [DOI] [PubMed] [Google Scholar]
- 4.Newman KL, Almeida RPP, Purcell AH, Lindow SE. 2004. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc Natl Acad Sci U S A 101:1737–1742. doi: 10.1073/pnas.0308399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee S, Almeida RP, Lindow S. 2008. Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu Rev Phytopathol 46:243–271. doi: 10.1146/annurev.phyto.45.062806.094342. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee S, Wistrom C, Lindow SE. 2008. A cell-cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa. Proc Natl Acad Sci U S A 105:2670–2675. doi: 10.1073/pnas.0712236105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang N, Li JL, Lindow SE. 2012. RpfF-dependent regulon of Xylella fastidiosa. Phytopathology 102:1045–1053. doi: 10.1094/PHYTO-07-12-0146-R. [DOI] [PubMed] [Google Scholar]
- 8.Guilhabert MR, Kirkpatrick BC. 2005. Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute to biofilm maturation to X. fastidiosa and colonization and attenuate virulence. Mol Plant Microbe Interact 18:856–868. doi: 10.1094/MPMI-18-0856. [DOI] [PubMed] [Google Scholar]
- 9.Lindow S, Newman K, Chatterjee S, Baccari C, Lavarone AT, Ionescu M. 2014. Production of Xylella fastidiosa diffusible signal factor in transgenic grape causes pathogen confusion and reduction in severity of Pierce’s disease. Mol Plant Microbe Interact 27:244–254. doi: 10.1094/MPMI-07-13-0197-FI. [DOI] [PubMed] [Google Scholar]
- 10.Deng Y, Wu J, Tao F, Zhang LH. 2011. Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem Rev 111:160–173. doi: 10.1021/cr100354f. [DOI] [PubMed] [Google Scholar]
- 11.Wang LH, He Y, Gao Y, Wu JE, Dong YH, He C, Wang SX, Weng LX, Xu JL, Tay L, Fang RX, Zhang LH. 2004. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol Microbiol 51:903–912. doi: 10.1046/j.1365-2958.2003.03883.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhou L, Yu Y, Chen X, Diab AA, Ruan L, He J, Wang H, He YW. 2015. The multiple DSF-family QS signals are synthesized from carbohydrate and branched-chain amino acids via the FAS elongation cycle. Sci Rep 5:13294. doi: 10.1038/srep13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng Y, Wu J, Yin W, Li P, Zhou J, Chen S, He F, Cai J, Zhang LH. 2016. Diffusible signal factor family signals provide a fitness advantage to Xanthomonas campestris pv. campestris in interspecies competition. Environ Microbiol 18:1534–1545. doi: 10.1111/1462-2920.13244. [DOI] [PubMed] [Google Scholar]
- 14.He YW, Wu J, Cha JS, Zhang LH. 2010. Rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae produces multiple DSF-family signals in regulation of virulence factor production. BMC Microbiol 10:187. doi: 10.1186/1471-2180-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, Pan SQ, Zhang LH. 2008. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J 2:27–36. doi: 10.1038/ismej.2007.76. [DOI] [PubMed] [Google Scholar]
- 16.Deng Y, Wu J, Eberl L, Zhang LH. 2010. Structural and functional characterization of diffusible signal factor family quorum-sensing signals produced by members of the Burkholderia cepacia complex. Appl Environ Microbiol 76:4675–4683. doi: 10.1128/AEM.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies DG, Marques CN. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beaulieu ED, Ionescu M, Chatterjee S, Yokota K, Trauner D, Lindow S. 2013. Characterization of a diffusible signaling factor from Xylella fastidiosa. mBio 4:e00539-12. doi: 10.1128/mBio.00539-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colnaghi Simionato AV, da Silva DS, Lambais MR, Carrilho E. 2007. Characterization of a putative Xylella fastidiosa diffusible signal factor by HRGC-EI-MS. J Mass Spectrom 42:1375–1381. doi: 10.1002/jms.1325. [DOI] [PubMed] [Google Scholar]
- 20.Bi H, Christensen QH, Feng Y, Wang H, Cronan JE. 2012. The Burkholderia cenocepacia BDSF quorum sensing fatty acid is synthesized by a bifunctional crotonase homologue having both dehydratase and thioesterase activities. Mol Microbiol 83:840–855. doi: 10.1111/j.1365-2958.2012.07968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng Y, Liu X, Wu J, Lee J, Chen S, Cheng Y, Zhang C, Zhang LH. 2015. The host plant metabolite glucose is the precursor of diffusible signal factor (DSF) family signals in Xanthomonas campestris. Appl Environ Microbiol 81:2861–2868. doi: 10.1128/AEM.03813-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Cámara M, Williams P, Dow JM. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A 103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.He YW, Xu M, Lin K, Ng YJ, Wen CM, Wang LH, Liu ZD, Zhang HB, Dong YH, Dow JM, Zhang LH. 2006. Genome scale analysis of diffusible signal factor regulon in Xanthomonas campestris pv. campestris: identification of novel cell-cell communication-dependent genes and functions. Mol Microbiol 59:610–622. doi: 10.1111/j.1365-2958.2005.04961.x. [DOI] [PubMed] [Google Scholar]
- 24.Ionescu M, Baccari C, Da Silva AM, Garcia A, Yokota K, Lindow SE. 2013. Diffusible signal factor (DSF) synthase RpfF of Xylella fastidiosa is a multifunction protein also required for response to DSF. J Bacteriol 195:5273–5284. doi: 10.1128/JB.00713-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YM, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 26.Remus-Emsermann MN, Kim EB, Marco ML, Tecon R, Leveau JH. 2013. Draft genome sequence of the phyllosphere model bacterium Pantoea agglomerans 299R. Genome Announc 1:e00036-13. doi: 10.1128/genomeA.00036-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ionescu M, Zaini PA, Baccari C, Tran S, da Silva AM, Lindow SE. 2014. Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc Natl Acad Sci U S A 111:E3910–E3918. doi: 10.1073/pnas.1414944111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, Brandenburg K, Whiteley M. 2008. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol 69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang TP, Lee Wong AC. 2007. Extracellular fatty acids facilitate flagella-independent translocation by Stenotrophomonas maltophilia. Res Microbiol 158:702–711. doi: 10.1016/j.resmic.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Hamed RB, Batchelar ET, Clifton IJ, Schofield CJ. 2008. Mechanisms and structures of crotonase superfamily enzymes-how nature controls enolate and oxyanion reactivity. Cell Mol Life Sci 65:2507–2527. doi: 10.1007/s00018-008-8082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi H, Yu Y, Dong H, Wang H, Cronan JE. 2014. Xanthomonas campestris RpfB is a fatty acyl-CoA ligase required to counteract the thioesterase activity of the RpfF diffusible signal factor (DSF) synthase. Mol Microbiol 93:262–275. doi: 10.1111/mmi.12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou L, Wang XY, Sun S, Yang LC, Jiang BL, He YW. 2015. Identification and characterization of naturally occurring DSF-family quorum sensing signal turnover system in the phytopathogen Xanthomonas. Environ Microbiol 17:4646–4658. doi: 10.1111/1462-2920.12999. [DOI] [PubMed] [Google Scholar]
- 33.Wang XY, Zhou L, Yang J, Ji GH, He YW. 2016. The RpfB-dependent quorum sensing signal turnover system is required for adaptation and virulence in rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact 29:220–230. doi: 10.1094/MPMI-09-15-0206-R. [DOI] [PubMed] [Google Scholar]
- 34.Almeida RP, Killiny N, Newman KL, Chatterjee S, Ionescu M, Lindow SE. 2012. Contribution of rpfB to cell-to-cell signal synthesis, virulence, and vector transmission of Xylella fastidiosa. Mol Plant Microbe Interact 25:453–462. doi: 10.1094/MPMI-03-11-0074. [DOI] [PubMed] [Google Scholar]
- 35.Chatterjee S, Newman KL, Lindow SE. 2008. Cell-to-cell signaling in Xylella fastidiosa suppresses movement and xylem vessel colonization in grape. Mol Plant Microbe Interact 21:1309–1315. doi: 10.1094/MPMI-21-10-1309. [DOI] [PubMed] [Google Scholar]
- 36.Ryan RP, An SQ, Allan JH, McCarthy Y, Dow JM. 2015. The DSF family of cell-cell signals: an expanding class of bacterial virulence regulators. PLoS Pathog 11:e1004986. doi: 10.1371/journal.ppat.1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, Brandenburg K, Whiteley M. 2008. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol 69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suppiger A, Aguilar C, Eberl L. 2016. Evidence for the widespread production of DSF family signal molecules by members of the genus Burkholderia by the aid of novel biosensors. Environ Microbiol Rep 8:38–44. doi: 10.1111/1758-2229.12348. [DOI] [PubMed] [Google Scholar]
- 39.King E, Ward M, Raney D. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44:301–307. [PubMed] [Google Scholar]
- 40.Davis MJ, Whitcomb RF, Gillaspie AG. 1981. Fastidious bacteria of plant vascular tissue and invertebrates (including so called rickettsia-like bacteria), p 2172–2188. In Starr MP, Stolp H, Truper HG, Balows A, Schlegel HG (ed), The prokaryotes: a handbook on habitats, isolation, and identification of bacteria. Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- 41.Brandi M, Clark EM, Lindow SE. 1996. Characterization of the indole-3-acetic acid (IAA) biosynthetic pathway in an epiphytic strain of Erwinia herbicola and IAA production in vitro. Can J Microbiol 42:586–592. doi: 10.1139/m96-079. [DOI] [Google Scholar]
- 42.Loper JE, Lindow SE. 1994. A biological sensor for iron available to bacteria in their habitats on plant surfaces. Appl Environ Microbiol 60:1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torriani A. 1968. Alkaline phosphatase of Escherichia coli, p 212–218. In Grossman L, Moldave K (ed), Methods in enzymology, vol. 12B Academic Press, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Induction of the XfDSF-biosensor strain by either 2 or 10 µl of a purified fraction of a DSF-containing extract of P. agglomerans 299R expressing X. fastidiosa RpfF (fractions named based on their retention times in minutes in HPLC [HPLC RT]). White bars represent induction of the sensor by 10 µM XfDSF2 or MeOH only. Download
GC-MS chromatograms for esterified natural extracts versus the 2-cis-hexadecenoic acid synthetic standard. (a) 2-cis-hexadecenoic acid; (b) RpfF extract; (c) WT extract; (d) RpfC extract. Download
Sequence identity (shown on black background) and similarity (shown on gray background) of X. fastidiosa (Xf) Tem1 RpfF and P. agglomerans (Pa) 299R F385_3254. The F385_3254 gene encodes a protein with 37% identity to that of X. fastidiosa RpfF. Download