ABSTRACT
The ability of cancer cells to degrade the extracellular matrix and invade interstitial tissues contributes to their metastatic potential. We recently showed that overexpression of sorting nexin 9 (SNX9) leads to increased cell invasion and metastasis in animal models, which correlates with increased SNX9 protein expression in metastases from human mammary cancers. Here, we report that SNX9 expression is reduced relative to neighboring normal tissues in primary breast tumors, and progressively reduced in more aggressive stages of non-small-cell lung cancers. We show that SNX9 is localized at invadopodia where it directly binds the invadopodia marker TKS5 and negatively regulates invadopodia formation and function. SNX9 depletion increases invadopodia number and the local recruitment of MT1-MMP by decreasing its internalization. Together, these effects result in increased localized matrix degradation. We further identify SNX9 as a Src kinase substrate and show that this phosphorylation is important for SNX9 activity in regulating cell invasion, but is dispensable for its function in regulating invadopodia. The diversified changes associated with SNX9 expression in cancer highlight its importance as a central regulator of cancer cell behavior.
KEY WORDS: Invadopodia, Src kinase, Extracellular matrix, TKS5, MT1-MMP, SNX9
Summary: SNX9, differentially regulated by Src, alters the ability of cancer cells to degrade the matrix through regulation of MT1-MMP internalization and/or invadopodia formation and function.
INTRODUCTION
In the last decades, considerable effort has been directed towards understanding the complexity of tumorigenesis and metastasis. The non-receptor tyrosine kinase Src, the first identified oncogene (see Guarino, 2010; Irby and Yeatman, 2000 for review), induces the formation of invadopodia (see Murphy and Courtneidge, 2011 for review), which are dynamic and actin-rich protrusions detected in many highly invasive cancer cell lines derived from solid tumors such as breast, melanoma and fibrosarcoma (Linder et al., 2011; Murphy and Courtneidge, 2011). Invadopodia are enriched in matrix-metalloproteases (MMPs) and are necessary at many stages of the metastatic process such as breaking through the basement membrane (Kinjo, 1978) and extravasating through blood vessels (Leong et al., 2014; see Lohmer et al., 2014; Paz et al., 2014 for review). Invadopodia are regulated downstream of many RhoGTPases (Beaty and Condeelis, 2014 for review), formed downstream of Src activation (Chen et al., 1985) and induced in response to a variety of external stimuli (see Linder et al., 2011; Murphy and Courtneidge, 2011 for review). The stabilization and maturation of these highly specialized plasma membrane domains requires the recruitment of actin-regulators (e.g. N-WASP, WAVE, Arp2/3, cortactin) and adaptor proteins [e.g. TKS5 (also known as FISH, KIAA0418 or SH3MD1), TKS4, NCK1] (Buccione et al., 2004; Murphy and Courtneidge, 2011).
Invadopodia function relies on vesicular traffic of key molecules to and from the plasma membrane, including MMPs e.g. MT1-MMP (also known as MMP14), and disintegrin and metalloproteinase domain-containing protein enzymes (e.g. ADAM12 or ADAM15) (Linder, 2007). Not surprisingly, key endocytic regulators such as the large GTPase dynamin localize to invadopodia (Baldassarre et al., 2003; Bruzzaniti et al., 2005; Destaing et al., 2013; Kruchten and McNiven, 2006). Dynamin is a multifunctional protein that, in addition to its role in endocytosis (Doherty and McMahon, 2009; Schmid and Frolov, 2011), also controls actin remodeling (Gu et al., 2010; Menon and Schafer, 2013). Dynamin depletion impairs local matrix degradation at invadopodia (Baldassarre et al., 2003; Destaing et al., 2013; Jiang et al., 2001; McNiven et al., 2004). In addition to dynamin, the membrane-sculpting BAR (Bin–amphiphysin–Rvs) domain proteins, FBP17 (formin-binding protein 17), CIP4 (Cdc42-interacting protein 4) and ASAP1 (ArfGAP with SH3 domain, ankyrin repeat and PH domain 1), have been localized to invadopodia (Bharti et al., 2007; Hu et al., 2011; Kovacs et al., 2006; Yamamoto et al., 2011).
Sorting nexin 9 (SNX9) is a ubiquitously expressed BAR domain protein and a major binding partner of dynamin (Lundmark and Carlsson, 2004, 2005, 2009; Soulet et al., 2005). SNX9 functions in both actin- and clathrin-mediated endocytosis (CME) (Lundmark and Carlsson, 2004, 2009; Soulet et al., 2005; Yarar et al., 2008) and also in clathrin-independent endocytosis (CIE) (Bendris et al., 2016; Yarar et al., 2007). Other studies have implicated SNX9 in actin-rich dorsal ruffle formation (Yarar et al., 2007) and the regulation of actin polymerization by directly binding to Arp2/3 and activating N-WASP (Shin et al., 2008; Yarar et al., 2008, 2007). We have previously shown that SNX9 regulates upstream actin regulators, namely the RhoGTPases RhoA and Cdc42 (Bendris et al., 2016). Consequently, SNX9 expression levels control the motility of cancer cells. SNX9 has also been identified as a direct partner for the invadopodial protein DOCK1 (dedicator of cytokenesis 1), the Drosophila homolog of NCK1 (non-catalytic region of tyrosine kinase adaptor protein 1) (Worby et al., 2002). Finally, SNX9 binds to ADAM9 and ADAM15 and potentially contributes to their trafficking (Howard et al., 1999). Interestingly, SNX9 expression is modified in numerous tumors including invadopodia-expressing cancer cells (Bendris et al., 2016; Mao et al., 2011) (www.nextbio.com, www.oncomine.org). Given these properties, we explored a potential role for SNX9 in invadopodia structure and function, hence in cancer metastasis.
RESULTS
SNX9 expression is lowered in primary tumors
We recently showed that SNX9 expression levels are higher in metastases compared with their respective primary mammary tumors. Consistent with this, we discovered that SNX9 overexpression enhances invasiveness of breast and lung cell lines in vitro and metastasis of breast cancer cells in a chick embryo model (Bendris et al., 2016). Based on these observations, we tested whether SNX9 protein expression varies during tumor progression, expecting to observe an increase in SNX9 levels in more aggressive stages of the disease. Surprisingly, using an immunohistochemical approach on a lung cancer tissue microarray (TMA) containing non-small cell lung cancer (NSCLC) samples from early (stage I) to advanced stage (stage III) disease (Table S1), we observed that SNX9 protein staining was significantly decreased in later, more aggressive stages (Fig. 1A). Similarly, we found that SNX9 expression levels in mammary invasive ductal carcinoma (IDC) were significantly lower in the malignancies compared with normal adjacent tissue (Fig. 1B). Thus, we hypothesized that in primary tumors, as opposed to metastases, SNX9 might fulfill specific functions unrelated to its role in the regulation of cell invasiveness.
Fig. 1.
SNX9 expression in lung and breast cancers. (A) Example of immunohistochemical (IHC) staining of SNX9 in two human NSCLC tumors. Bar chart represents quantification (H-score, see Materials and Methods) of SNX9 staining in stage I (n=97), stage II (n=36) and stage III (n=59) NSCLC. (B) Representative image of IHC staining of SNX9 in human normal breast tissue versus in situ ductal breast carcinoma. Bar chart represents quantification of SNX9 staining in normal versus patient tumor tissues. n=10 for each condition. Scale bars: 200 µm. Results are presented as mean±s.e.m. Statistical significance was evaluated using one-way ANOVA. ****P<0.0001.
Later, more aggressive, stages of cancers exhibit greater invasive activities (Tsim et al., 2010). Given that local invasion by breaking through the basement membrane is a crucial step for cancer cell dissemination from the primary tumors, we tested the involvement of SNX9 in invadopodia-mediated matrix degradation.
SNX9 localizes to invadopodia
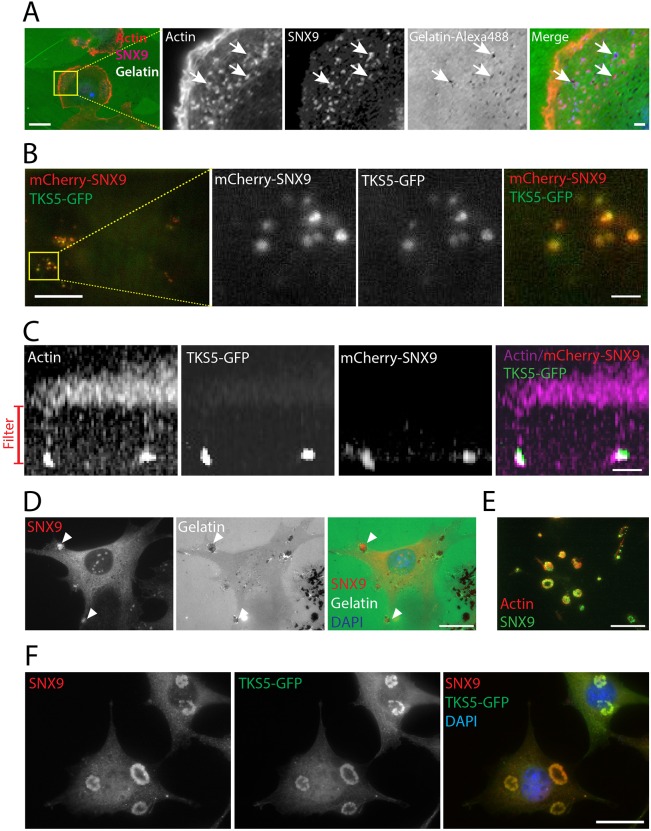
The breast-cancer-derived cell line, MDA-MB-231, has been extensively used as a model to study invadopodia (Artym et al., 2006; Beaty et al., 2014). We first tested for SNX9 localization at functional invadopodia by plating MDA-MB-231 cells on green fluorescent gelatin; dark areas correspond to degraded matrix, reflective of invadopodia activity. Epifluorescence microscopy showed that SNX9 colocalized with actin puncta, some of which corresponded to active areas of matrix degradation (Fig. 2A, arrows). The molecular composition of invadopodia is highly similar to other actin-related structures. Therefore, to unambiguously identify these structures we used the specific invadopodial marker, TKS5 (tyrosine kinase substrate with five SH3 domains), an adaptor protein that is required for invadopodia function (Sharma et al., 2013). Using total internal reflection fluorescence (TIRF) microscopy, which selectively illuminates molecules localized within ∼100 nm of the ventral cell surface, we found that transiently expressed mCherry–SNX9 colocalized with TKS5–GFP (Fig. 2B), further supporting the localization of SNX9 to invadopodia. Time-lapse, dual-channel TIRF microscopy showed that SNX9 appears and disappears with either TKS5 or actin in a dynamic manner at invadopodia (Movies 1–2); which can be distinguished from membrane-bound endocytic structures by their large size (0.5–1 μm).
Fig. 2.
SNX9 is localized to invadopodia and podosomes. (A) Staining of F-actin (phalloidin, in red) and endogenous SNX9 (purple) in MDA-MB-231 cells plated on green fluorescent gelatin. Arrows indicate colocalization of SNX9 with actin at dark areas corresponding to matrix degradation at invadopodia. (B) TIRF microscopy image of invadopodia of MDA-MB-231 cells transiently co-expressing mCherry–SNX9 and TKS5–GFP. Insets in A and B show enlargement of yellow boxes. Scale bars: 10 μm, insets 2 μm. (C) Cells expressing mCherry–SNX9 and TKS5–GFP, plated on a filter containing 1-μm-diameter pores, were stained with phalloidin (purple). Image shows a z-stack of confocal sections of invadopodia extending in through the filter. (D) Staining of endogenous SNX9 (red) in NIH-Src cells plated on green fluorescent gelatin. Arrowheads indicate colocalization of SNX9 where matrix has been degraded at podosomes. (E) TIRF microscopy image showing the colocalization of endogenous SNX9 (green) with actin (red) at podosomes on the ventral plasma membrane. (F) Staining of endogenous SNX9 (red) in NIH-Src cells transiently expressing TKS5–GFP. Scale bars: 10 μm, insets 2 μm in A; 10 μm in C–F.
In cells grown in 3D cultures, invadopodia extend into the surrounding matrix and invadopodial components are specifically distributed along these structures (see Murphy and Courtneidge, 2011 for review). Therefore, to further characterize their relative localization within invadopodia, cells expressing mCherry–SNX9 and TKS5–GFP were cultured in Boyden chambers with 1 μm pores to allow invadopodia extension. Whereas actin staining extended along their entire length, fluorescently tagged SNX9 colocalized with TKS5 mainly at the tips of invadopodia (Fig. 2C).
Podosomes are invadopodial-like structures present in non-cancerous cell types including osteoclasts; they are also induced in Src-transformed fibroblasts (Chen, 1989; see Linder et al., 2011; Murphy and Courtneidge, 2011 for review). Podosomes are highly similar to invadopodia with some notable differences, for example, the adaptor proteins NCK1 and GRB2 are differentially localized to invadopodia or podosomes, respectively (Oser et al., 2011). Given this difference, we investigated if SNX9 was also part of podosomes. As for invadopodia, endogenous SNX9 colocalized with areas of degraded gelatin (Fig. 2D). As previous studies have shown that SNX9 is localized to dorsal ruffles (Yarar et al., 2007), we confirmed that endogenous SNX9 is localized with actin at podosomes found at the ventral plasma membrane by TIRF microscopy (Fig. 2E). Finally, similar to invadopodia, endogenous SNX9 localized with TKS5–GFP in Src-induced podosomes (Fig. 2F), indicating that SNX9 might play similar roles in both structures.
SNX9 is a direct binding partner of the invadopodial marker TKS5
Given that SNX9 and TKS5 are colocalized at invadopodia and that TKS5 directly binds several SNX9 partners including dynamin, N-WASP and NCK1 (Murphy and Courtneidge, 2011), we hypothesized that SNX9 and TKS5 could be direct or indirect binding partners. We first assayed for SNX9 interactions with TKS5 in cell extracts. MDA-MB-231 cells were transfected with either GFP–SNX9 or TKS5–GFP. Cell extracts were subjected to GFP pulldowns followed by western blotting using anti-SNX9 or anti-TKS5 antibodies. SNX9 and TKS5 were detected in TKS5–GFP and GFP–SNX9 pulldowns, respectively (Fig. 3A,B). Interestingly, these experiments also illustrate that although a large fraction of TKS5 is associated with SNX9 (Fig. 3B), only a small subfraction of SNX9 binds TKS5 (Fig. 3A). These data suggest that SNX9 fulfills other functions independent of TKS5.
Fig. 3.
SNX9 directly binds TKS5. (A,B) GFP pulldown assays of TKS5–GFP with endogenous SNX9 (A) or GFP–SNX9 with endogenous TKS5 (B), using MDA-MB-231 cell lysates. (C) Scheme of WT and deletion mutants of SNX9 used in E. (D) Scheme of WT and deletion mutants of SNX9 used in F. (E) HA pulldown of HA-tagged WT or mutant SNX9 using lysates from MDA-MB-231 cells transiently co-expressing exogenous SNX9 with TKS5–GFP. (F) GST pulldown assay of bacterially expressed and purified His–TKS5 using GST–WT-SNX9 or its deletion mutants as bait. Asterisks indicate GST–SNX9 proteins.
Functional domains of SNX9 include an N-terminal SH3 domain followed by an unstructured low complexity region, the protein–lipid interacting PX (phox homology) domain and the BAR domain (Fig. 3C). To further dissect these interactions we co-transfected TKS5–GFP with hemagglutinin (HA)-tagged SNX9 deletion mutants in MDA-MB-231 cells. The deletion mutants used for this experiment consisted of HA–ΔC197 that lacks the BAR domain, HA–ΔC340 that lacks both the BAR domain and a fraction of the PX domain, and HA–ΔC395 consisting of only the SH3 and low complexity domains (Fig. 3C). Using HA–agarose beads, we were able to pull down TKS5–GFP along with all the deletion mutants including the HA–ΔC395 (Fig. 3E), indicating that the region of interaction spans the SH3 and/or low complexity domains. Moreover, although HA–ΔC197 and HA–ΔC340 mutants were poorly expressed compared with wild type (WT) and HA–ΔC395, they pulled down similar amounts of TKS5, suggesting that the BAR domain is inhibitory to SNX9–TKS5 interaction.
Given that TKS5 and SNX9 share several binding partners, we next tested whether the interactions above were direct by using recombinant TKS5 and SNX9 proteins. GST-tagged WT or deletion mutants of SNX9 (Fig. 3D) were tested for direct binding to full length histidine-tagged TKS5 in vitro. WT-SNX9 was able to pull down TKS5 protein, demonstrating a direct interaction. More TKS5 was pulled down with ΔPX-SNX9 as compared with WT, suggesting that the PX domain, similar to the BAR domain, has an inhibitory effect on TKS5–SNX9 interactions. In this experiment, SNX9–TKS5 interactions were not dependent on the SH3 domain of SNX9 (Fig. 3F). This result, combined with the pulldown experiments using recombinant proteins, demonstrates that SNX9 interacts directly with TKS5 through its low complexity region. Further studies are needed to identify the specific sites required for TKS5–SNX9 interactions.
SNX9 negatively regulates invadopodia formation
To explore the role of SNX9 at invadopodia, SNX9 expression was suppressed by siRNA-mediated knockdown in MDA-MB-231. We plated control and SNX9-depleted cells (231-siCTR and 231-siSNX9, respectively) on gelatin-coated coverslips and co-stained for actin and cortactin to visualize invadopodia (Fig. 4A). SNX9 depletion not only increased the number of cells expressing invadopodia (Fig. 4B; see Materials and Methods) but also increased the number of invadopodia per cell (Fig. 4C). To ensure that the effect of SNX9 on invadopodia is not cell-type-specific, we depleted SNX9 in other cancer cell lines, namely SCC61 (oral squamous carcinoma) and HT1080 (fibrosarcoma). Consistent with the results obtained in MDA-MB231 cells, SNX9 knockdown increased the number of invadopodia in both cell lines (Fig. S1). Taken together, these results show that SNX9 is a negative regulator of invadopodia formation in cancer cells.
Fig. 4.
SNX9 negatively regulates invadopodia formation. (A) 231-CTR or 231-siSNX9 cells were plated on gelatin and stained for cortactin (green) and F-actin (phalloidin, in red) to visualize invadopodia. Insets represent enlargement of yellow boxes. Scale bars: 20 μm, insets 4 μm. (B,C) Quantification of percentage of cells expressing invadopodia (B) or of the relative number of invadopodia (C) in conditions used in A. ∼200 cells were counted for each condition, n=3; *P≤0.05, ****P<0.0001. Results are presented as mean±s.e.m. Statistical significance was evaluated using one-tailed Mann–Whitney test. (D) 231-CTR or 231-siSNX9 cells plated on a filter containing 1-μm-diameter pores. Images represent a z-stack of confocal sections of invadopodia extending through the filter. F-actin is visualized in red and cortactin in green. Two independent siRNAs against SNX9 were used. (E) Western blot analysis of SNX9 expression under conditions used in D showing knockdown of SNX9 expression in MDA-MB-231 cells transfected with SNX9 siRNA #1 and #2. GAPDH was used as loading control.
We have previously shown that SNX9 regulates RhoA and Cdc42 (Bendris et al., 2016). As RhoGTPases are master regulators of the actin cytoskeleton at invadopodia (Beaty and Condeelis, 2014), and actin is one of the core components found throughout the structure, we asked whether invadopodia were properly protruding from SNX9-depleted cells. To address this question, 231-siCTR or 231-siSNX9 cells were plated on collagen-coated filters, allowing the cells to extend invadopodia through the 1-μm-diameter pores. The structures were visualized using actin (red) and cortactin (green) staining. We used confocal microscopy to reconstitute a z-stack image of extending invadopodia. Although difficult to quantify, the number of invadopodia extending through the pores was consistently higher in 231-siSNX9 cells as compared with control. This effect was also reproduced using two independent siRNAs against SNX9 (Fig. 4D,E). Given that the two independent siRNA against SNX9 gave similar results, we used siRNA #1 (Yarar et al., 2007; Bendris et al., 2016), also used in Fig. 4A and in Fig. S1, throughout this work. These results demonstrate that SNX9 depletion favors the formation of invadopodia in both 2D and 3D cultures.
SNX9 depletion enhances invadopodia function by impairing MT1-MMP endocytosis
To investigate whether the increased invadopodia formation in the absence of SNX9 corresponds to enhanced function, we next measured substrate degradation. 231-siCTR or 231-siSNX9 cells were plated on green fluorescent gelatin matrix for 3 h and invadopodial-associated degradation areas were quantified (see Materials and Methods). 231-siSNX9 cells degraded the matrix more efficiently compared with the controls (Fig. 5A,B). Similarly, SNX9 knockdown increased the matrix degradation abilities of NIH-Src cells (Fig. S2A,B).
Fig. 5.
SNX9 regulates MT1-MMP internalization and invadopodia activity. (A) 231-CTR or 231-siSNX9 cells were plated on green fluorescent gelatin and stained for F-actin (phalloidin, in red). Dark areas on the green channel correspond to matrix degradation. (B) Corresponding quantification of invadopodia-dependent matrix degradation (see Materials and Methods). n=3, ***P<0.005 (au=arbitrary unit). (C) 231-CTR or 231-siSNX9 cells were incubated with anti-MT1-MMP–Alexa-Fluor-448 antibody at 37°C for 20 or 60 min to allow the internalization of the antibody. The contours of cells are outlined in yellow. Arrows point to vacuoles containing MT1-MMP. 231-siSNX9 contain fewer vacuoles compared with 231-siCTR cells. (D) Corresponding quantification of anti-MT1-MMP antibody internalization. n=3; *P≤0.05. (E) TIRF microscopy image of 231-CTR or 231-siSNX9 stained using red phalloidin and with anti-MT1-MMP–Alexa-Fluor-448 antibody. Insets show enlargement of yellow boxes. Results on bar charts are presented as mean±s.e.m. Statistical significance was evaluated using one-tailed Mann–Whitney test. Scale bars: 20 μm in A; 10 μm in C; 10 μm, insets 2 μm in E. Arrows in E indicate colocalization of actin with MT1-MMP at invadopodia.
Previous studies have indicated that SNX18, which is closely related to SNX9, can be functionally redundant with respect to its role in CME (Park et al., 2010). Therefore, we tested whether SNX18 is also involved in the regulation of matrix degradation. Surprisingly, siRNA knockdown of SNX18 decreased matrix degradation, suggesting that SNX18 and SNX9 are not functionally redundant in regulating matrix degradation in this regard (Fig. S2C–E). Further work is needed to investigate this functional difference.
As only mature invadopodia are able to degrade the matrix and TKS5 is a marker for mature invadopodia (Sharma et al., 2013), we also stained for endogenous TKS5 in control and SNX9-depleted MDA-MB-231 cells. In accordance with the previous results, the number of TKS5- and actin-positive structures, indicative of mature invadopodia (Fig. S2F, arrows), was increased in SNX9-deficient cells. This observation suggests that in absence of SNX9 not only the number of invadopodia is increased but also their maturation, which is in accordance with the previous result showing increased matrix degradation of 231-siSNX9 (Fig. 5A,B).
Degradation-active invadopodia are also characterized by the local enrichment in matrix proteases, mainly MT1-MMP (Poincloux et al., 2009), whose localization is controlled by membrane trafficking. SNX9 is involved in multiple endocytic pathways including CME, CIE and actin-mediated bulk uptake (Bendris et al., 2016; Ma and Chircop, 2012; Yarar et al., 2007). MT1-MMP is internalized via clathrin- and caveolin-dependent pathways (Jiang et al., 2001; Remacle et al., 2003; see Poincloux et al., 2009 for review). Given that matrix degradation was enhanced following SNX9 depletion, we hypothesized that SNX9 might be involved in MT1-MMP internalization. We first approached the question by measuring MT1-MMP uptake using a surface biotinylation assay. Although we detected and measured an increase in surface MT1-MMP in 231-MMP as expected (Fig. S3A,B), we could not follow its internalization using this method because of low signal:noise. We then developed a microscopy-based approach using anti-MT1-MMP–Alexa-Fluor-488 antibody to quantify its internalization. For this, cells were incubated in the continuous presence of anti-MT1-MMP antibody at 37°C for either 20 or 60 min (Fig. 5C). Control cells showed accumulation of the antibody in perinuclear vesicles whose size and intensity increased over time. In contrast, SNX9-depleted cells showed weaker signals and smaller vesicles at both time points (Fig. 5C). Quantification of fluorescence intensities after 60 min of uptake revealed that SNX9 depletion resulted in a significant decrease in bulk MT1-MMP internalization (Fig. 5C,D). Correspondingly, we observed an increase in cell surface MT1-MMP detected by incubating 231-siCTR and 231-siSNX9 cells with anti-MT1-MMP antibodies at 4°C (Fig. S3C,D). Taken together, these observations indicate that in the absence of SNX9, MT1-MMP internalization is impaired, which allows its accumulation at the plasma membrane. Finally, by immunofluorescence labeling and TIRF microscopy, we found a striking increase in localized MT1-MMP staining (green) at invadopodia (actin labeled in red) in 231-siSNX9 cells compared with 231-siCTR cells (Fig. 5E, arrows). Taken together, these results demonstrate that SNX9 depletion enhances the recruitment of the maturation marker TKS5 to invadopodia and also directly impairs MT1-MMP internalization, resulting in increased matrix degradation.
SNX9 is a Src substrate
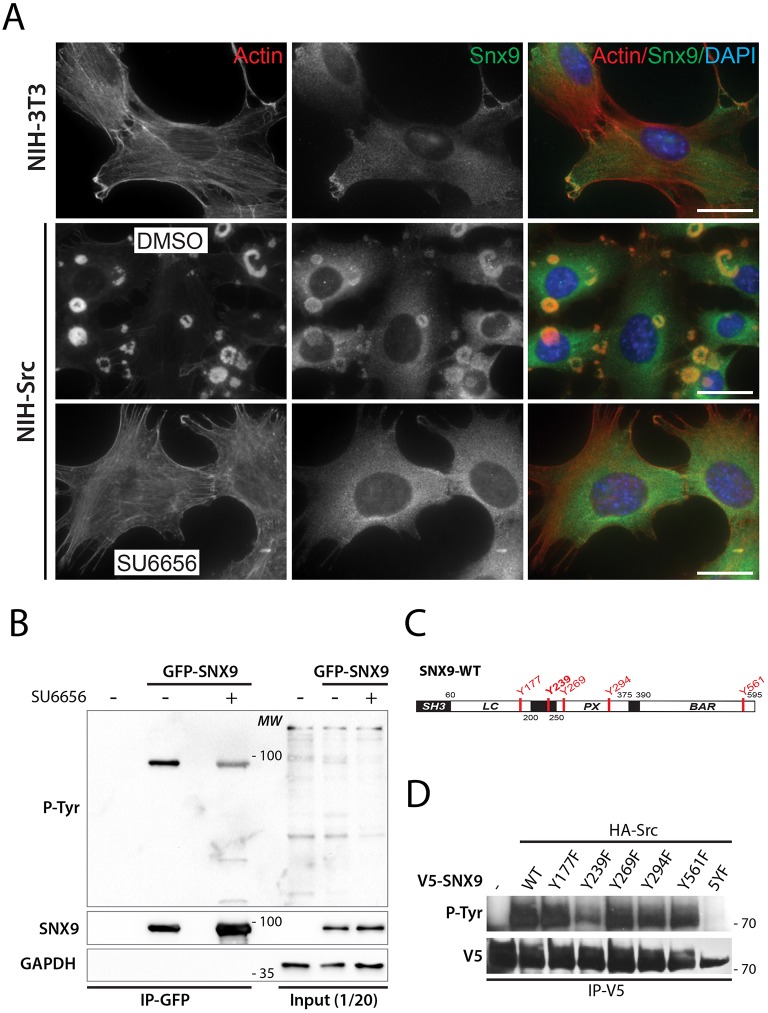
SNX9 is subject to post-translational modifications such as tyrosine phosphorylation by the protein kinase ACK (Lin et al., 2002). ACK is activated by Cdc42 GTPase and by its phosphorylation downstream of Src (Yang et al., 2001a). Src kinase is instrumental in invadopodia biology as Src phosphorylates many invadopodia and podosome components, including TKS5 and cortactin (Ayala et al., 2008; Lock et al., 1998). Moreover, the oncogenic transformation of NIH3T3 cells by Src overexpression induces podosome formation and recruitment of TKS5 (Oikawa et al., 2008). Based on this, we asked whether SNX9 might be phosphorylated downstream of Src. We first assessed the SNX9 localization pattern in NIH-Src compared with NIH-3T3 cells. Similarly to TKS5, we found that Src transformation also induced an enrichment of SNX9 at podosomes (Fig. 6A, upper and middle panels). As expected, the localization of SNX9 to podosomes was disrupted following the treatment with the specific Src inhibitor SU6656 (Fig. 6A, lower panels).
Fig. 6.
SNX9 is phosphorylated by Src. (A) NIH-3T3 or NIH-Src were treated with Src inhibitor SU6656 or DMSO (control). Cells were then stained for F-actin and for endogenous SNX9. Scale bars: 10 μm. (B) Western blot comparing exogenous SNX9 phosphorylation in NIH-Src cells transiently expressing GFP–SNX9, mock-treated or treated with SU6656. Tyrosine phosphorylation was evaluated on immunoprecipitated GFP–SNX9 using an anti-phospho-tyrosine antibody. GAPDH was used as loading control. (C) Schematic presentation of SNX9 protein domains. Tyrosines, identified by mass spectrometry (see Fig. S4B), phosphorylated by Src are marked in red. (D) HA–Src and V5–WT-SNX9 or the indicated mutants were transiently co-expressed in HEK293 cells. V5–SNX9 was then immunoprecipitated and tyrosine phosphorylation was assessed using an anti-phospho-tyrosine antibody. 5YF represents the quintuple mutant, where all phosphorylated tyrosines were mutated (Y177F, Y239F, Y269F, Y294F and Y561F).
It has previously been suggested that Src kinase phosphorylates SNX9 in Drosophila cells either directly or indirectly via ACK (Worby et al., 2002). To determine whether SNX9 is a direct substrate for Src, serum-starved NIH-Src cells transiently expressing GFP–SNX9 were stimulated with serum-containing medium in the presence or absence of 10 μM SU6656, followed by a GFP pulldown. Using an anti-phospho-tyrosine antibody, we found that SNX9 is indeed phosphorylated on tyrosine residue(s) and that this phosphorylation is reduced upon inhibition of Src (Fig. 6B). We next investigated whether Src directly phosphorylates SNX9 by incubating recombinant SNX9 with purified Src. We observed a time-dependent increase in SNX9 tyrosine phosphorylation, confirming that SNX9 is a substrate for Src (Fig. S4A). To identify Src phosphorylation sites on SNX9, HEK-293 cells were transiently transfected with HA–Src and V5–SNX9 expression plasmids. Immunoprecipitated V5–SNX9 was analyzed by liquid chromatography mass spectroscopy (LC-MS-MS) (Fig. S4B). Five tyrosine residues, Y177, Y239, Y269, Y294 and Y561, distributed in multiple domains (Fig. 6C), were identified, which were not detected when V5–SNX9 was expressed alone (not shown).
Finally, we generated SNX9 mutants in which all five tyrosines were mutated together (5YF-SNX9) or individually. SNX9 phosphorylation mutants were then co-expressed with HA–Src, immunoprecipitated and probed for tyrosine phosphorylation. Src no longer phosphorylated 5YF-SNX9, confirming our identification of the sites. Y177F, Y269F, Y294F and Y561F mutants showed similar phosphorylation compared with WT-SNX9. However, the SNX9-Y239F mutant showed a dramatic decrease in phosphorylation, indicating that Y239 is the major Src phosphorylation site on SNX9 (Fig. 6D).
Src phosphorylation differentially regulates SNX9 function
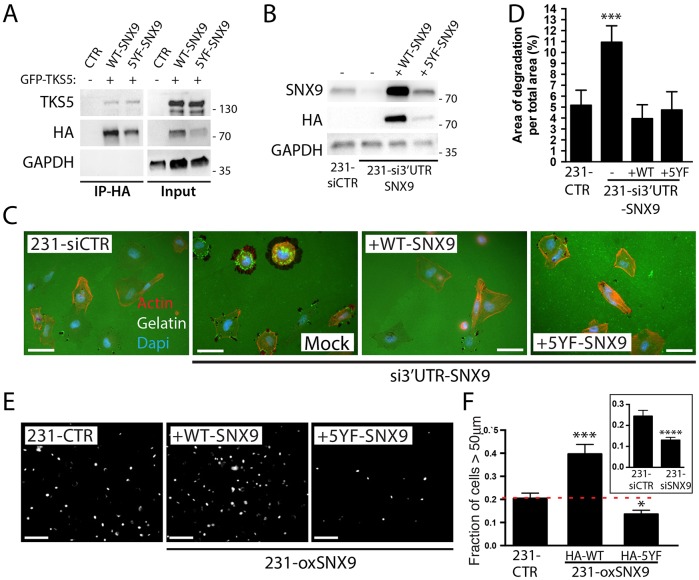
We have shown that SNX9 depletion increases matrix degradation. Given that Src is essential for invadopodia formation, we hypothesized that Src-induced phosphorylation of SNX9 might be important for its function at invadopodia. We first evaluated if the non-phosphorylatable SNX9 mutant is still able to bind to TKS5. MDA-MB-231 cells were co-transfected with TKS5–GFP and HA–WT-SNX9 or HA–5YF-SNX9. Cell extracts were used to immunoprecipitate HA–SNX9. Both WT-SNX9 and 5YF-SNX9 efficiently co-immunoprecipitated TKS5, indicating that Src phosphorylation of SNX9 was not important for SNX9–TKS5 binding (Fig. 7A), confirming in vitro binding results (Fig. 3E,F) using non-phosphorylated recombinant proteins. We examined the effect of 5YF-SNX9 on matrix degradation and on MT1-MMP endocytosis. We used an siRNA directed against the 3′UTR of SNX9 to specifically knockdown the endogenous without affecting the expression of transfected SNX9 (Fig. 7B), and first confirmed that WT-SNX9 was able to restore matrix degradation to control levels after SNX9 depletion (Fig. 7C,D). Surprisingly, 5YF-SNX9 was as efficient as WT-SNX9 in reducing matrix degradation after SNX9 depletion (Fig. 7C,D), suggesting that Src phosphorylation is not crucial for the contribution of SNX9 to matrix degradation activity regulation.
Fig. 7.
Src phosphorylation differentially regulates SNX9 functions. (A) HA pulldown assay of HA–WT-SNX9 or HA–5YF-SNX9 transiently expressed with TKS5–GFP in MDA-MB-231 cells. (B) Western blot analysis of SNX9 expression after SNX9 depletion and rescue with HA–WT-SNX9 or HA–5YF-SNX9. GAPDH was used as loading control in A,B. (C,D) Cells depleted of endogenous SNX9 using a siRNA against its 3′UTR were either transfected with HA–WT-SNX9 or HA–5YF-SNX9, and plated on green fluorescent gelatin. F-actin was subsequently stained using red phalloidin (C) and matrix degradation areas quantified (D). (E) 231-CTR or 231-oxSNX9 cells expressing either WT-SNX9 or 5YF-SNX9 were used in an inverted 3D cell invasion assay through a collagen I matrix. Cell nuclei were stained and images were taken every 25 μm. Figure shows Hoechst staining, showing nuclei distribution at 50 μm, from a representative experiment. (F) Quantification of nuclei distribution in the inverted invasion assay described in E. Inset shows invasion ratio of 231-siSNX9 versus 231-siCTR, confirming previous results (Bendris et al., 2016). n=3; *P≤0.05, ***P<0.005, ****P<0.0001. Results on bar charts are presented as mean±s.e.m. Statistical significance was evaluated using one-tailed Mann–Whitney test. Scale bars: 20 μm in C, 25 μm in F.
We recently discovered that SNX9 is a regulator of cancer cell invasion. Indeed, SNX9 depletion impairs (also see inset in Fig. 7F), whereas the overexpression of low amounts of SNX9 increases cell invasiveness (Bendris et al., 2016). Therefore, we assessed the effect of Src phosphorylation on the ability of SNX9 to increase the invasive potential of MDA-MB-231. We used an inverted invasion assay in which cells invade upwards through a 3D collagen I matrix attracted by a gradient of serum present in the overlying culture media (Arsic et al., 2012; Sanz-Moreno et al., 2008; Bendris et al., 2016). The distribution of nuclei stained with Hoechst was quantified on microscope z-sections spanning the collagen plug. In this case, whereas the expression of WT-SNX9 increased cell invasion, the expression of 5YF-SNX9 had the opposite effect (Fig. 7E,F, compare panel F with inset on same panel), suggesting that Src phosphorylation of SNX9 is essential for its function in cell invasion and that 5YF-SNX9 exerts a dominant-negative effect on endogenous SNX9 function in cell invasion (Fig. 7E,F). Taken together, these results establish that SNX9 has diverse activities in invadopodia function and in cell invasion, and that Src differentially regulates these functions.
DISCUSSION
SNX9 is a multifaceted scaffold regulating diverse cellular processes that modulate cancer cell behavior. In this work, we report that SNX9 is localized at invadopodia and negatively regulates their formation and function. This activity likely reflects both the direct interaction between SNX9 and the invadopodial maturation marker TKS5 and the role of SNX9 in MT1-MMP internalization. We also show that SNX9 is directly phosphorylated by Src kinase and that this phosphorylation alters its ability to regulate cancer cell invasiveness without interfering with SNX9 regulation of invadopodia. We previously showed by immunohistochemistry that SNX9 levels increase in metastases relative to their parent breast cancer tumors (Bendris et al., 2016) and here show that SNX9 levels decrease in more invasive primary breast and NSCLC tumors. Together, these observations reveal the complexity of SNX9 function in cancer cells that can be regulated by differential, stage-specific changes in SNX9 expression, and also, potentially, by post-translational modification.
SNX9 is both a direct, via N-WASP (Yarar et al., 2008, 2007), and indirect, via RhoGTPases (Bendris et al., 2016), regulator of the actin cytoskeleton. We have previously shown a role for SNX9 in the regulation of cell invasion that is likely independent of its role in endocytosis and more reflective of its role in regulating the actin cytoskeleton (Bendris et al., 2016). Here, we propose that the role of SNX9 in regulating invadopodia results from at least three SNX9 activities that are not mutually exclusive. First, SNX9 can negatively regulate invadopodia through inactivation of RhoA (Bendris et al., 2016), as it is well-documented that formation of invadopodia is triggered by the stimulation of RhoA in cancer cells (Roh-Johnson et al., 2014; Bravo-Cordero et al., 2011). Second, our results suggest that SNX9 might regulate invadopodia maturation, function and/or turnover through its interactions with TKS5. For example, by reducing TKS5 recruitment to these structures (i.e. maintaining TKS5 in the cytosol) SNX9 might impair invadopodia maturation. Alternatively, given that a larger fraction of TKS5 appears to interact with SNX9 than vice versa, SNX9 binding might compete for the interaction of other proteins that either recruit and/or are recruited by TKS5 for invadopodia maturation and function. Third, SNX9 functions in MT1-MMP endocytosis and hence can regulate invadopodia function. As TKS5 and SNX9 are preferentially localized at the tip of invadopodia where MT1-MMP is also concentrated (Yu et al., 2012), TKS5 might locally interfere with SNX9 function in CME to stabilize MT1-MMP at these structures.
SNX9 appears to play distinct roles in the regulation of invadopodia versus cell invasion through collagen, despite the fact that the latter should increase local matrix degradation and hence facilitate cell invasion. We speculate that the imbalance between RhoA and Cdc42 activation resulting from SNX9 knockdown (Bendris et al., 2016) leads to changes in the actin cytoskeleton that consequently impede cell invasion in spite of the increased matrix degradation, revealing the complex nature of these processes. Similarly, it was reported that focal adhesion kinase (FAK) depletion increases invadopodia number and function but failed to increase cell invasion of breast cancer cells (Chan et al., 2009). Likewise, a non-phosphorylatable mutant of MT1-MMP is retained at the plasma membrane and increases matrix degradation downstream of MT1-MMP, yet negatively influences cell invasion (Williams and Coppolino, 2011).
SNX9 expression levels vary in cancers and in this work we discover that SNX9 is directly phosphorylated by Src. Interestingly, mass spectrometry studies using various tumor samples confirmed that the tyrosines Y177 and Y239 on SNX9 are phosphorylated (www.phosphosite.org) and that Y239 phosphorylation is particularly enriched in NIH-Src versus NIH-3T3 cells (Luo et al., 2008; Rush et al., 2005), correlating with our result that Y239 is the main phosphorylation site. Cells depleted of endogenous SNX9 showed increased matrix degradation. We found that phosphorylation of SNX9 by Src does not control its activity in regulating invadopodia function but does control SNX9 function in the regulation of cell invasion. More experimental work is needed to investigate this axis. Taken together, these results again point to the diversity of the cellular functions of SNX9.
Invadopodia formation is triggered by external cues such as EGF. Interestingly, SNX9 is degraded downstream of EGFR internalization (Childress et al., 2006) and reciprocally, SNX9 also contributes to EGFR degradation, consistent with our finding that SNX9 depletion enhances invadopodia formation in vitro. In parallel, EGF also activates Src (Murphy and Courtneidge, 2011; Osherov and Levitzki, 1994; Parsons and Parsons, 2004), which in turn triggers a cascade of signaling pathways including the activation of other kinases such as ACK2 (Yang et al., 2001a). In EGF-treated cells, SNX9 is directly phosphorylated by ACK (Lin et al., 2002), suggesting that Src induces SNX9 phosphorylation both directly and indirectly. Consequently, although we report that Src does not directly regulate SNX9 function at invadopodia, we cannot rule out an indirect effect via ACK. Interestingly, ACK is also activated downstream of Cdc42 (Yang et al., 2001b). Whether ACK is localized at invadopodia and/or podosomes, as well as the role of ACK-dependent phosphorylation of SNX9 on invadopodia function and on cell invasion, has yet to be explored. Taken together, these observations reveal an intricate regulation of SNX9 functions by means of phosphorylation.
The potential contribution of SNX9 to cancer cell metastasis is complex as the SNX9 expression level seems to be bimodal, where it is lowered in primary tumors but then increased in metastases. We showed experimentally that lowering SNX9 expression increases invadopodia number, but, at the same time, maintaining a substantial amount of SNX9 is crucial for cell invasiveness. Correspondingly, SNX9 expression is not completely repressed in primary tumors but rather overall diminished while also heterogeneous between cells. We speculate that, to sustain SNX9-mediated cell invasion, SNX9 levels might be locally down-regulated in invadopodia. Another possibility would be that cells with low levels of SNX9 do not disseminate, but rather facilitate, by locally degrading the ECM, the dissemination of cancer cells with higher amounts of SNX9, which correlates with the increase in SNX9 expression in metastases (Bendris et al., 2016). Finally, low amounts of SNX9 in the primary tumor might not be instrumental for cell metastasis but could be important to sustain growth factor signaling (e.g. by EGF), resulting from the impairment of endocytosis, thereby enhancing other aggressive characteristics of malignant cells.
Taken together, the results presented here and elsewhere (Bendris et al., 2016) point to complex and diverse functions for SNX9 in both primary tumors and metastases, where, as a scaffold, it can coordinate the activation of RhoGTPases, changes in actin cytoskeleton and cell morphology, as well as endocytic membrane trafficking.
MATERIAL AND METHODS
Cell culture
Cell lines
MDA-MB-231 cells were a gift from R. Brekken (UT Southwestern Medical Center, Dallas). HT1080 and SCC61 cells were a gift from S. Courtneidge (Sanford Burnham, San Diego). MDA-MB-231, HT1080 and HEK293 cells were cultivated in DMEM (Invitrogen) supplemented with 10% FBS (Sigma Aldrich). SCC61 cells were maintained in DMEM supplemented with 20% FBS+0.4 µg ml−1 hydrocortisone (Sigma Aldrich). H1299 cells were cultivated in RPMI (Invitrogen) supplemented with 5% FBS. Cells were regularly tested for mycoplasma contaminations.
Transfections
Lipofectamine-RNAi-Max or Lipofectamine and Lipofectamine-2000 (Invitrogen) were used for siRNA or plasmid delivery into cells, respectively.
siRNA
SNX9 siRNA #1 sense: UAAGCACUUUGACUGGUUAUU; SNX9 siRNA #2 sense: AACAGUCGUGCUAGUUCCUCA; 3UTR-SNX9 siRNA sense: GGGACUUUGUAGAGAAUUU. ON-TARGETplus SMART pool SNX18 siRNA (112574, Dharmacon). We used AllStars® negative control siRNA in all knockdown experiments (Qiagen). siRNA was delivered to cells using Lipofectamine-RNAi-Max. Two rounds of siRNA treatment were performed on day 1 and 2. Cells were then used on day 3.
Generation of stable cell lines
Cells were transfected with the appropriate plasmid, followed by antibiotic selection using 1 mg ml−1 G418 (Life Technologies). We generated and used two independent non-clonal cell lines stably expressing WT or mutant SNX9.
Cell culture in Boyden chamber
We used 12-well plates containing inserts with 1-μm-diameter pores. 5×104 cells from each condition were seeded on the upper part of the Boyden chamber in medium containing 1% FBS. Inserts were immersed in medium supplemented with 10% FBS+20 ng ml−1 EGF. Cells used in Fig. 2C were transfected 24 h prior the plating. Invadopodia were allowed to extend for ∼16 h, cells were then fixed, permeabilized and stained with phalloidin only (Fig. 2C) or with phalloidin and anti-cortactin antibody (Fig. 4D).
Plasmids
SNX9 and N-WASP constructs were previously described (Yarar et al., 2007). pcDNA3-TKS5-GFP plasmid was a gift from S. Courtneidge. We transferred TKS5 cDNA insert into a pGEX vector to allow bacterial protein expression.
SNX9 deletion mutants
pcDNA3-SNX9 wild-type plasmid was used as a template to construct the carboxyl-terminal deletion mutants. Briefly, DNA primers to generate each deletion construct (SH3PX1-ΔC197, -ΔC339, -ΔC395) were engineered with BamH1 and EcoR1 restriction sites. The PCR products were ligated into the pCR2-TOPO vector, followed by restriction digests with BamH1 and EcoR1. DNA products were resolved on a gel, and then the DNA inserts were excised from the gel and purified with the Qiaquick Gel Extraction kit. The resulting DNA inserts were ligated into the HA-tagged pcDNA3 expression vector using T4 DNA ligase.
SNX9 point mutants
Y177F, Y239F, Y269F, Y294F and Y561F mutants of SNX9, as well as the quintuple mutant (5YF mutant), Y177F/Y239F/Y269F/Y294F/Y561F, were generated by PCR with the QuikChange Site-Directed Mutagenesis kit using pcDNA3-SNX9 wild-type construct as a template.
Antibodies and reagents
For western blot experiments, the following antibodies were used: anti-SNX9 (Yarar et al., 2008, 2007; 1/2000), anti-SNX9 (HPA031410, Sigma Aldrich; 1/1000); anti-TKS5 and anti-cortactin (#09-403 and #4F11, Millipore; 1/1000); anti-SNX18 and anti-MT1-MMP (from Abcam, ab111706 and ab51074, respectively; 1/2000) and anti-MT1-MMP–Alexa-Fluor-448 (FAB9181G, R&D Systems; 1/400); anti-HA and anti-c-Myc (MMS-120P and MMS-150P, respectively; 1/1000) from Covance; anti-phosphotyrosine (94G10, Upstate Biotechnology; 1/500); anti-His (#2365, Cell Signaling; 1/5000 and #27471001, GE Healthcare; 1/5000); anti-GAPDH (G9545, Sigma Aldrich; 1/10000); and anti-V5 (R960-25; 1/1000) and HRP-conjugated antibodies were purchased from Invitrogen [31460 (goat-anti rabbit) and 32430 (goat-anti mouse); 1/5000]. Oregon-Green-488-conjugated gelatin (G13186; prepared according to the manufacturer's instructions), Alexa-Fluor-561-conjugated phalloidin (A12380; 1/10,000) and Alexa-Fluor-conjugated secondary antibodies (A-A11001, A-11008 and A-11011; 1/800) were obtained from Life Technologies; and gelatin from bovine skin (G-6650), SU6656 (S9692) were purchased from Sigma Aldrich. Protein-G agarose beads and T4 DNA ligase were purchased from Invitrogen. QuikChange Site-Directed Mutagenesis and Qiaquick Gel Extraction kits were purchased from Stratagene and Qiagen, respectively.
Immunohistochemistry of cancer tissues
Non-small cell lung carcinoma (NSCLC) tissue microarray (see Table S1) and breast patient samples were obtained and processed by the Department of Translational Molecular Pathology at the University of Texas – MD Anderson Cancer Center and the UT Southwestern Tissue Resource facility, respectively. SNX9 staining was performed using a rabbit polyclonal antibody (HPA031410, Sigma Aldrich; 1/700). TMA slides were stained using an automated stainer (Dako Autostainer Plus), using a 1:850 dilution, following the Human Protein Atlas protocol (http://www.proteinatlas.org). Human colon and kidney were used as positive control; a non-primary antibody tissue section was used as negative control. The TMA slides were read by two pathologists (H.L. and J.R.), who agreed on the optimization, staining pattern and evaluation of the cases. SNX9 IHC expression on the NSCLC TMA was analyzed using the H-score method, which includes the evaluation of percentage of tumor cells positive (0 to 100) and intensity of the immunoreaction (0 to 3+) providing a final score ranging from 0 to 300.
Breast tissues were processed using an identical protocol as above. All breast tumors represented invasive ductal carcinoma, and for each patient, we evaluated SNX9 expression in the tumor and in the normal breast tissue.
Cell imaging
Fixed cells were imaged using a microscope with 40×, 60× or 100×1.49 NA objective (Nikon) mounted on a Ti-Eclipse inverted microscope. Images were collected using a charge-coupled device camera (CoolSNAP HD2; Roper Scientific) driven by MicroManager software (Vale Lab, UCSF).
Extending invadopodia through Boyden chamber filters were imaged using a Zeiss LSM 780 inverted confocal microscope with z-intervals of 0.5 μm, using a 63×/1.40 NA oil immersion objective. Image deconvolution and z-stack reconstructions were performed using Imaris 7.2.2 sofware (Bitplane).
Fluorescent matrix degradation and quantification
Fluorescent gelatin solutions were prepared as recommended by the manufacturers. Gelatin-coated coverslips were prepared as in Diaz et al. (2013). Briefly, 0.2 mg ml−1 gelatin solution was prepared in 2% sucrose. Glass coverslips were coated with warm gelatin solution, air-dried, then cross linked with 0.5% cold glutaraldehyde solution for 15 min followed by extensive washing. Sodium borohydryde solution at 5 mg ml−1 was used to quench gelatin auto-fluorescence, for 3 min. Before use, coverslips were equilibrated with complete medium for at least 1 h. Cells were plated for 4–7 h for MDA-MB-231 or 7 h for HT1080 and SCC61.
Quantification of normalized matrix degradation relative to the total cell area was done using the procedure as described in (Martin et al., 2012). In brief, cells were first stained with phalloidin. Cell area on microscopy images was quantified by manually segmenting cell area using phalloidin intensity, and degraded area was measured by manually segmenting the inverted FITC-gelatin intensity. Total cell area was estimated for ∼150 cells per condition from three independent experiments. Data are presented as mean±s.e.m.
Anti-MT1-MMP antibody internalization and quantification
231-siCTR or 231-siSNX9 cells were deposited at 3×104 cells/well in 96-well plates (black, clear bottom), in five replicates. We used the anti-MT1-MMP14 antibody for 20 min or 1 h at 37°C, or for 1 h at 4°C. Cells were then fixed, permeabilized and stained with phalloidin. Corrected total cell fluorescence intensities of one microscopy image for each well were calculated relative to the cell area (using phalloidin staining on the same cells) and mean fluorescence of background readings (100 cells/condition/experiment) using ImageJ (NIH). Data are presented as mean±s.e.m. (normalized to control cells).
Biotinylation of surface-bound MT1-MMP and quantification
Cells were washed with cold PBS and maintained on ice. After 30 min, the PBS was replaced by cold PBS containing 0.2 mg ml−1 of Sulfo-NHS-SS-Biotin (21331, Thermo Fisher) and incubated for 1 h at 4°C. After extensive washes to remove unbound biotin, cells were lysed (10 mM Tris HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40) and biotinylated proteins were pulled down using 10 μl of streptavidin–agarose-bead slurry (69203-3, Millipore) under agitation, for 1 h at 4°C. Surface-labeled MT1-MMP was detected after SDS-PAGE and western blotting using anti-MT1-MMP antibody. ‘Total’ fraction represents 1/50 of total lysate. We used ImageJ software to measure band intensities. Results are expresses as the fraction of surface-bound biotinylated MT1-MMP/total MT1-MMP normalized to GAPDH intensity.
Immunoprecipitations, pulldowns and recombinant protein interactions
Cells were lysed (10 mM Tris pH 7.4, 5 mM MgCl2, 150 mM NaCl, 1% Triton X-100, 1 mM sodium orthovanadate, 5 mM beta-glycerol phosphate, 10 μg ml−1 aprotinin, 10 μg ml−1 leupeptin). Cell lysates were incubated with anti-V5 antibody for 2–20 h, with rotation at 4°C. Protein-G agarose beads were added, rotating for an additional hour. The beads were then precipitated in a microfuge and washed 3–4 times with cold cell lysis buffer.
Pulldown assays were performed using GFP-trap beads®(Chromo-Tek) according to manufacturer instructions as reported by Arsic et al. (2012). After washes, beads were resuspended in Laemmli buffer for western blot analysis. Input fraction represents 1/20 of total lysate. In vitro binding assays were performed with bacterially produced GST-fused SNX9 or His-fused TKS5 constructs. SNX9 proteins were purified on glutathione-conjugated agarose beads (Agarose Beads Technology) according to standard methods (Yarar et al., 2008). SNX9 beads were incubated in NP40 buffer (10 mM Tris HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.05% NP40) with His–TKS5 for 4 h at 4°C, extensively washed with the same buffer and resuspended in Laemmli buffer. Input fraction represents 1/20 of total recombinant TKS5. Acrylamide gels were either loaded for western blot analysis and further blotted with TKS5 antibody or stained with Coomassie Blue to visualize SNX9 recombinant proteins.
Mass spectrometry analysis
HEK293 cells were transfected with HA–Src and V5–SNX9 plasmids. After 24 h, cells were lysed in the lysis buffer and V5–SNX9 was immunoprecipitated as previously described using ∼10 mg total protein lysates. After boiling, samples were run on an SDS-PAGE gel, stained with Coomassie Blue, and V5–SNX9 band was excised. Aliquots from the samples were set aside and tyrosine phosphorylation was confirmed by western blotting. The samples were then submitted to the Cornell Biotechnology Resource Center Proteomics and Mass Spectrometry Core Facility.
In vitro tyrosine kinase assay
In vitro tyrosine phosphorylation of SNX9 was determined by incubating 50 ng recombinant His–Src or His–FAK with 500 ng bacterially purified His–SH3PX1 in kinase reaction buffer (50 mM Tris HCl pH 7.4, 2 mM MgCl2, 10 mM MnCl2, 1 μM ATP, 10 μCi [32P]-ATP). Reactions were carried out at room temperature for various lengths of time (5–120 min), and then quenched with the addition of 5× SDS loading buffer.
Inverted invasion assay
Experiments were performed according to Arsic et al. (2012) and Bendris et al. (2016). Invasion assays were performed using 96-well plates (PerkinElmer). Briefly, cells were suspended in 2.3 mg ml−1 serum-free liquid bovine collagen I (Advanced Biomatrix) at 5×104 cells ml−1. Aliquots of 100 µl were dispensed into the wells followed by a centrifugation at 1000 rpm of the plate for 5 min at 4°C. The plate was then transferred at 37°C to allow collagen polymerization, then 30 µl of medium containing 5% FBS was added on top. After 36 h, cells were fixed with paraformaldehyde and stained with Hoechst-33342 (Invitrogen). Quantification of invaded cells was performed according to Bendris et al. (2016). Invasion ratio represents the sum of cell counts at 50, 100 and 150 μm over cell counts at 0 μm. Results were obtained from at least three independent experiments including five replicates, on each day. Bar charts are plotted as mean±s.e.m. of all experiments.
Statistical analysis
All experiments have been repeated at least three times. Data are reported as arithmetic means±s.e.m. Statistical analyses were performed using either nonparametric Mann–Whitney or column statistics, using GraphPad Prism software. Statistical significance was defined as P≤0.05.
Acknowledgements
We thank R. Cerione for providing reagents and communicating unpublished data regarding SNX9 phosphorylation sites and S. Courtneidge for providing cell lines, reagents and for helpful discussions at the outset of these studies. L. Machesky provided the ImageJ plug-in for the quantification of invadopodia-related matrix degradation. We are grateful to B. Lemmers for reading and commenting on the manuscript. We thank D. Reed and A. Mohanakrishnan for help in recombinant protein production and the UT Southwestern Microscopy Core Facility for confocal microscopy support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conception, design and interpretation of data: N.B., S.L.S. Writing, review, and/or revision of the manuscript: N.B., S.L.S. Experiments for the identification of Src phosphorylation sites (Fig. 6D and Fig. 3S) and generation of SNX9 mutants (Fig. 3C): C.J.S.S. IHC staining, observation and quantification (Fig. 1): J.R.-C., H.L., A.W.W. Remaining experiments: N.B., Microscopy: N.B. and C.R.R. for confocal microscopy. Quantification of matrix degradation and of MT1-MMP internalization: C.R.R. All authors read and commented on the manuscript.
Funding
This work was supported by the National Institutes of Health [grants GM42455 and MH61345 to S.L.S.] N.B. was partially supported by a Cancer Prevention and Research Institute of Texas training grant [RP140110]. J.R.-C. and H.L. are supported by National Cancer Institute Lung Cancer SPORE [grant P50CA70907] and Cancer Center Support National Institutes of Health [grant CA-16672]. A.W.W. is supported by the National Institutes of Health [R01CA163863]. UT Southwestern Medical Center Tissue Resource (UTSTR) facility (National Institutes of Health) is supported by [5P30CA142543-04]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.188045.supplemental
References
- Arsic N., Bendris N., Peter M., Begon-Pescia C., Rebouissou C., Gadéa G., Bouquier N., Bibeau F., Lemmers B. and Blanchard J. M. (2012). A novel function for Cyclin A2: control of cell invasion via RhoA signaling. J. Cell Biol. 196, 147-162. 10.1083/jcb.201102085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym V. V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K. M. and Mueller S. C. (2006). Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66, 3034-3043. 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- Ayala I., Baldassarre M., Giacchetti G., Caldieri G., Tete S., Luini A. and Buccione R. (2008). Multiple regulatory inputs converge on cortactin to control invadopodia biogenesis and extracellular matrix degradation. J. Cell Sci. 121, 369-378. 10.1242/jcs.008037 [DOI] [PubMed] [Google Scholar]
- Baldassarre M., Pompeo A., Beznoussenko G., Castaldi C., Cortellino S., McNiven M. A., Luini A. and Buccione R. (2003). Dynamin participates in focal extracellular matrix degradation by invasive cells. Mol. Biol. Cell 14, 1074-1084. 10.1091/mbc.E02-05-0308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty B. T. and Condeelis J. (2014). Digging a little deeper: the stages of invadopodium formation and maturation. Eur. J. Cell Biol. 93, 438-444. 10.1016/j.ejcb.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty B. T., Wang Y., Bravo-Cordero J. J., Sharma V. P., Miskolci V., Hodgson L. and Condeelis J. (2014). Talin regulates moesin-NHE-1 recruitment to invadopodia and promotes mammary tumor metastasis. J. Cell Biol. 205, 737-751. 10.1083/jcb.201312046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendris N., Williams K. C., Reis C. R., Welf E. S., Chen P.-H., Lemmers B., Hahne M., Leong H. S. and Schmid S. L. (2016). SNX9 promotes metastasis by enhancing cancer cell invasion via differential regulation of RhoGTPases. Mol. Biol. Cell 27, 1409-1419. 10.1091/mbc.e16-02-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti S., Inoue H., Bharti K., Hirsch D. S., Nie Z., Yoon H.-Y., Artym V., Yamada K. M., Mueller S. C., Barr V. A. et al. (2007). Src-dependent phosphorylation of ASAP1 regulates podosomes. Mol. Cell. Biol. 27, 8271-8283. 10.1128/MCB.01781-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero J. J., Oser M., Chen X., Eddy R., Hodgson L. and Condeelis J. (2011). A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr. Biol. 21, 635-644. 10.1016/j.cub.2011.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzaniti A., Neff L., Sanjay A., Horne W. C., De Camilli P. and Baron R. (2005). Dynamin forms a Src kinase-sensitive complex with Cbl and regulates podosomes and osteoclast activity. Mol. Biol. Cell 16, 3301-3313. 10.1091/mbc.E04-12-1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccione R., Orth J. D. and McNiven M. A. (2004). Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat. Rev. Mol. Cell Biol. 5, 647-657. 10.1038/nrm1436 [DOI] [PubMed] [Google Scholar]
- Chan K. T., Cortesio C. L. and Huttenlocher A. (2009). FAK alters invadopodia and focal adhesion composition and dynamics to regulate breast cancer invasion. J. Cell Biol. 185, 357-370. 10.1083/jcb.200809110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-T. (1989). Proteolytic activity of specialized surface protrusions formed at rosette contact sites of transformed cells. J. Exp. Zool. 251, 167-185. 10.1002/jez.1402510206 [DOI] [PubMed] [Google Scholar]
- Chen W.-T., Chen J.-M., Parsons S. J. and Parsons J. T. (1985). Local degradation of fibronectin at sites of expression of the transforming gene product pp60src. Nature 316, 156-158. 10.1038/316156a0 [DOI] [PubMed] [Google Scholar]
- Childress C., Lin Q. and Yang W. (2006). Dimerization is required for SH3PX1 tyrosine phosphorylation in response to epidermal growth factor signalling and interaction with ACK2. Biochem. J. 394, 693-698. 10.1042/BJ20050576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O., Ferguson S. M., Grichine A., Oddou C., De Camilli P., Albiges-Rizo C. and Baron R. (2013). Essential function of dynamin in the invasive properties and actin architecture of v-Src induced podosomes/invadosomes. PLoS ONE 8, e77956 10.1371/journal.pone.0077956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz B., Yuen A., Iizuka S., Higashiyama S. and Courtneidge S. A. (2013). Notch increases the shedding of HB-EGF by ADAM12 to potentiate invadopodia formation in hypoxia. J. Cell Biol. 201, 279-292. 10.1083/jcb.201209151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty G. J. and McMahon H. T. (2009). Mechanisms of endocytosis. Annu. Rev. Biochem. 78, 857-902. 10.1146/annurev.biochem.78.081307.110540 [DOI] [PubMed] [Google Scholar]
- Gu C., Yaddanapudi S., Weins A., Osborn T., Reiser J., Pollak M., Hartwig J. and Sever S. (2010). Direct dynamin-actin interactions regulate the actin cytoskeleton. EMBO J. 29, 3593-3606. 10.1038/emboj.2010.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino M. (2010). Src signaling in cancer invasion. J. Cell. Physiol. 223, 14-26. 10.1002/jcp.22011 [DOI] [PubMed] [Google Scholar]
- Howard L., Nelson K. K., Maciewicz R. A. and Blobel C. P. (1999). Interaction of the metalloprotease disintegrins MDC9 and MDC15 with two SH3 domain-containing proteins, endophilin I and SH3PX1. J. Biol. Chem. 274, 31693-31699. 10.1074/jbc.274.44.31693 [DOI] [PubMed] [Google Scholar]
- Hu J., Mukhopadhyay A., Truesdell P., Chander H., Mukhopadhyay U. K., Mak A. S. and Craig A. W. (2011). Cdc42-interacting protein 4 is a Src substrate that regulates invadopodia and invasiveness of breast tumors by promoting MT1-MMP endocytosis. J. Cell Sci. 124, 1739-1751. 10.1242/jcs.078014 [DOI] [PubMed] [Google Scholar]
- Irby R. B. and Yeatman T. J. (2000). Role of Src expression and activation in human cancer. Oncogene 19, 5636-5642. 10.1038/sj.onc.1203912 [DOI] [PubMed] [Google Scholar]
- Jiang A., Lehti K., Wang X., Weiss S. J., Keski-Oja J. and Pei D. (2001). Regulation of membrane-type matrix metalloproteinase 1 activity by dynamin-mediated endocytosis. Proc. Natl. Acad. Sci. USA 98, 13693-13698. 10.1073/pnas.241293698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo M. (1978). Lodgement and extravasation of tumour cells in blood-borne metastasis: an electron microscope study. Br. J. Cancer 38, 293-301. 10.1038/bjc.1978.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs E. M., Makar R. S. and Gertler F. B. (2006). Tuba stimulates intracellular N-WASP-dependent actin assembly. J. Cell Sci. 119, 2715-2726. 10.1242/jcs.03005 [DOI] [PubMed] [Google Scholar]
- Kruchten A. E. and McNiven M. A. (2006). Dynamin as a mover and pincher during cell migration and invasion. J. Cell Sci. 119, 1683-1690. 10.1242/jcs.02963 [DOI] [PubMed] [Google Scholar]
- Leong H. S., Robertson A. E., Stoletov K., Leith S. J., Chin C. A., Chien A. E., Hague M. N., Ablack A., Carmine-Simmen K., McPherson V. A. et al. (2014). Invadopodia are required for cancer cell extravasation and are a therapeutic target for metastasis. Cell Rep. 8, 1558-1570. 10.1016/j.celrep.2014.07.050 [DOI] [PubMed] [Google Scholar]
- Lin Q., Lo C. G., Cerione R. A. and Yang W. (2002). The Cdc42 target ACK2 interacts with sorting nexin 9 (SH3PX1) to regulate epidermal growth factor receptor degradation. J. Biol. Chem. 277, 10134-10138. 10.1074/jbc.M110329200 [DOI] [PubMed] [Google Scholar]
- Linder S. (2007). The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 17, 107-117. 10.1016/j.tcb.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Linder S., Wiesner C. and Himmel M. (2011). Degrading devices: invadosomes in proteolytic cell invasion. Annu. Rev. Cell Dev. Biol. 27, 185-211. 10.1146/annurev-cellbio-092910-154216 [DOI] [PubMed] [Google Scholar]
- Lock P., Abram C. L., Gibson T. and Courtneidge S. A. (1998). A new method for isolating tyrosine kinase substrates used to identify fish, an SH3 and PX domain-containing protein, and Src substrate. EMBO J. 17, 4346-4357. 10.1093/emboj/17.15.4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmer L. L., Kelley L. C., Hagedorn E. J. and Sherwood D. R. (2014). Invadopodia and basement membrane invasion in vivo. Cell Adh. Migr. 8, 246-255. 10.4161/cam.28406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R. and Carlsson S. R. (2004). Regulated membrane recruitment of dynamin-2 mediated by sorting nexin 9. J. Biol. Chem. 279, 42694-42702. 10.1074/jbc.M407430200 [DOI] [PubMed] [Google Scholar]
- Lundmark R. and Carlsson S. R. (2005). Expression and properties of sorting nexin 9 in dynamin-mediated endocytosis. Methods Enzymol. 404, 545-556. 10.1016/S0076-6879(05)04048-6 [DOI] [PubMed] [Google Scholar]
- Lundmark R. and Carlsson S. R. (2009). SNX9 - a prelude to vesicle release. J. Cell Sci. 122, 5-11. 10.1242/jcs.037135 [DOI] [PubMed] [Google Scholar]
- Luo W., Slebos R. J., Hill S., Li M., Brábek J., Amanchy R., Chaerkady R., Pandey A., Ham A.-J. and Hanks S. K. (2008). Global impact of oncogenic Src on a phosphotyrosine proteome. J. Proteome Res. 7, 3447-3460. 10.1021/pr800187n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M. P. C. and Chircop M. (2012). SNX9, SNX18 and SNX33 are required for progression through and completion of mitosis. J. Cell Sci. 125, 4372-4382. 10.1242/jcs.105981 [DOI] [PubMed] [Google Scholar]
- Mao X., Boyd L. K., Yanez-Munoz R. J., Chaplin T., Xue L., Lin D., Shan L., Berney D. M., Young B. D. and Lu Y. J. (2011). Chromosome rearrangement associated inactivation of tumour suppressor genes in prostate cancer. Am. J. Cancer Res. 1, 604-617. [PMC free article] [PubMed] [Google Scholar]
- Martin K. H., Hayes K. E., Walk E. L., Ammer A. G., Markwell S. M. and Weed S. A. (2012). Quantitative measurement of invadopodia-mediated extracellular matrix proteolysis in single and multicellular contexts. J. Vis. Exp. 66, e4119 10.3791/4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNiven M. A., Baldassarre M. and Buccione R. (2004). The role of dynamin in the assembly and function of podosomes and invadopodia. Front Biosci. 9, 1944-1953. 10.2741/1348 [DOI] [PubMed] [Google Scholar]
- Menon M. and Schafer D. A. (2013). Dynamin: expanding its scope to the cytoskeleton. Int. Rev. Cell Mol. Biol. 302, 187-219. 10.1016/B978-0-12-407699-0.00003-0 [DOI] [PubMed] [Google Scholar]
- Murphy D. A. and Courtneidge S. A. (2011). The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413-426. 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T., Itoh T. and Takenawa T. (2008). Sequential signals toward podosome formation in NIH-src cells. J. Cell Biol. 182, 157-169. 10.1083/jcb.200801042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M., Dovas A., Cox D. and Condeelis J. (2011). Nck1 and Grb2 localization patterns can distinguish invadopodia from podosomes. Eur. J. Cell Biol. 90, 181-188. 10.1016/j.ejcb.2010.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherov N. and Levitzki A. (1994). Epidermal-growth-factor-dependent activation of the src-family kinases. Eur. J. Biochem. 225, 1047-1053. 10.1111/j.1432-1033.1994.1047b.x [DOI] [PubMed] [Google Scholar]
- Park J., Kim Y., Lee S., Park J. J., Park Z. Y., Sun W., Kim H. and Chang S. (2010). SNX18 shares a redundant role with SNX9 and modulates endocytic trafficking at the plasma membrane. J. Cell Sci. 123, 1742-1750. 10.1242/jcs.064170 [DOI] [PubMed] [Google Scholar]
- Parsons S. J. and Parsons J. T. (2004). Src family kinases, key regulators of signal transduction. Oncogene 23, 7906-7909. 10.1038/sj.onc.1208160 [DOI] [PubMed] [Google Scholar]
- Paz H., Pathak N. and Yang J. (2014). Invading one step at a time: the role of invadopodia in tumor metastasis. Oncogene 33, 4193-4202. 10.1038/onc.2013.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poincloux R., Lizarraga F. and Chavrier P. (2009). Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J. Cell Sci. 122, 3015-3024. 10.1242/jcs.034561 [DOI] [PubMed] [Google Scholar]
- Remacle A., Murphy G. and Roghi C. (2003). Membrane type I-matrix metalloproteinase (MT1-MMP) is internalised by two different pathways and is recycled to the cell surface. J. Cell Sci. 116, 3905-3916. 10.1242/jcs.00710 [DOI] [PubMed] [Google Scholar]
- Roh-Johnson M., Bravo-Cordero J. J., Patsialou A., Sharma V. P., Guo P., Liu H., Hodgson L. and Condeelis J. (2014). Macrophage contact induces RhoA GTPase signaling to trigger tumor cell intravasation. Oncogene 33, 4203-4212. 10.1038/onc.2013.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X.-M., Polakiewicz R. D. and Comb M. J. (2005). Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94-101. 10.1038/nbt1046 [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V., Gadea G., Ahn J., Paterson H., Marra P., Pinner S., Sahai E. and Marshall C. J. (2008). Rac activation and inactivation control plasticity of tumor cell movement. Cell 135, 510-523. 10.1016/j.cell.2008.09.043 [DOI] [PubMed] [Google Scholar]
- Schmid S. L. and Frolov V. A. (2011). Dynamin: functional design of a membrane fission catalyst. Annu. Rev. Cell Dev. Biol. 27, 79-105. 10.1146/annurev-cellbio-100109-104016 [DOI] [PubMed] [Google Scholar]
- Sharma V. P., Eddy R., Entenberg D., Kai M., Gertler F. B. and Condeelis J. (2013). Tks5 and SHIP2 regulate invadopodium maturation, but not initiation, in breast carcinoma cells. Curr. Biol. 23, 2079-2089. 10.1016/j.cub.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N., Ahn N., Chang-Ileto B., Park J., Takei K., Ahn S.-G., Kim S.-A., Di Paolo G. and Chang S. (2008). SNX9 regulates tubular invagination of the plasma membrane through interaction with actin cytoskeleton and dynamin 2. J. Cell Sci. 121, 1252-1263. 10.1242/jcs.016709 [DOI] [PubMed] [Google Scholar]
- Soulet F., Yarar D., Leonard M. and Schmid S. L. (2005). SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol. Biol. Cell 16, 2058-2067. 10.1091/mbc.E04-11-1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsim S., O'Dowd C. A., Milroy R. and Davidson S. (2010). Staging of non-small cell lung cancer (NSCLC): a review. Respir. Med. 104, 1767-1774. 10.1016/j.rmed.2010.08.005 [DOI] [PubMed] [Google Scholar]
- Williams K. C. and Coppolino M. G. (2011). Phosphorylation of membrane type 1-matrix metalloproteinase (MT1-MMP) and its vesicle-associated membrane protein 7 (VAMP7)-dependent trafficking facilitate cell invasion and migration. J. Biol. Chem. 286, 43405-43416. 10.1074/jbc.M111.297069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby C. A., Simonson-Leff N., Clemens J. C., Huddler D. Jr, Muda M. and Dixon J. E. (2002). Drosophila Ack targets its substrate, the sorting nexin DSH3PX1, to a protein complex involved in axonal guidance. J. Biol. Chem. 277, 9422-9428. 10.1074/jbc.M110172200 [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Sutoh M., Hatakeyama S., Hashimoto Y., Yoneyama T., Koie T., Saitoh H., Yamaya K., Funyu T., Nakamura T. et al. (2011). Requirement for FBP17 in invadopodia formation by invasive bladder tumor cells. J. Urol. 185, 1930-1938. 10.1016/j.juro.2010.12.027 [DOI] [PubMed] [Google Scholar]
- Yang W., Lin Q., Zhao J., Guan J.-L. and Cerione R. A. (2001a). The nonreceptor tyrosine kinase ACK2, a specific target for Cdc42 and a negative regulator of cell growth and focal adhesion complexes. J. Biol. Chem. 276, 43987-43993. 10.1074/jbc.M104819200 [DOI] [PubMed] [Google Scholar]
- Yang W., Lo C. G., Dispenza T. and Cerione R. A. (2001b). The Cdc42 target ACK2 directly interacts with clathrin and influences clathrin assembly. J. Biol. Chem. 276, 17468-17473. 10.1074/jbc.M010893200 [DOI] [PubMed] [Google Scholar]
- Yarar D., Waterman-Storer C. M. and Schmid S. L. (2007). SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev. Cell 13, 43-56. 10.1016/j.devcel.2007.04.014 [DOI] [PubMed] [Google Scholar]
- Yarar D., Surka M. C., Leonard M. C. and Schmid S. L. (2008). SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic 9, 133-146. 10.1111/j.1600-0854.2007.00675.x [DOI] [PubMed] [Google Scholar]
- Yu X., Zech T., McDonald L., Gonzalez E. G., Li A., Macpherson I., Schwarz J. P., Spence H., Futó K., Timpson P. et al. (2012). N-WASP coordinates the delivery and F-actin-mediated capture of MT1-MMP at invasive pseudopods. J. Cell Biol. 199, 527-544. 10.1083/jcb.201203025 [DOI] [PMC free article] [PubMed] [Google Scholar]