ABSTRACT
The transcription factor EB (TFEB) plays a pivotal role in the regulation of basic cellular processes, such as lysosomal biogenesis and autophagy. The subcellular localization and activity of TFEB are regulated by mechanistic target of rapamycin (mTOR)-mediated phosphorylation, which occurs at the lysosomal surface. Phosphorylated TFEB is retained in the cytoplasm, whereas dephosphorylated TFEB translocates to the nucleus to induce the transcription of target genes. Thus, a lysosome-to-nucleus signaling pathway regulates cellular energy metabolism through TFEB. Recently, in vivo studies have revealed that TFEB is also involved in physiological processes, such as lipid catabolism. TFEB has attracted a lot of attention owing to its ability to induce the intracellular clearance of pathogenic factors in a variety of murine models of disease, such as Parkinson's and Alzheimer's, suggesting that novel therapeutic strategies could be based on the modulation of TFEB activity. In this Cell Science at a Glance article and accompanying poster, we present an overview of the latest research on TFEB function and its implication in human diseases.
KEY WORDS: TFEB, TFE3, MiT family, mTOR, Lysosomes, Autophagy, Lysosomal storage disorders
Summary: The article discusses the roles of TFEB as a regulator of lysosomal biogenesis and intracellular clearance, and its involvement in human diseases.
Introduction
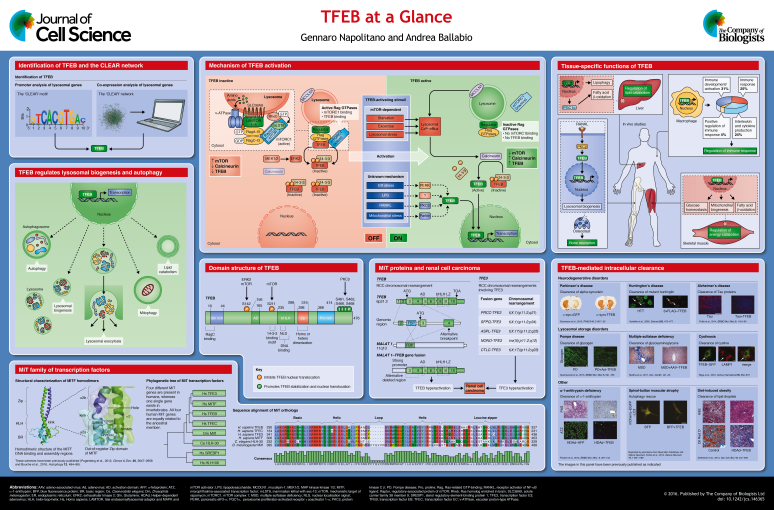
The transcription factor EB (TFEB) is a member of the microphthalmia family of basic helix-loop-helix– leucine-zipper (bHLH-Zip) transcription factors (MiT family) (Steingrímsson et al., 2004). The role of MiT transcription factors in the regulation of basic cellular processes has only recently become more clear. In particular, recent work has shown that TFEB has an important function in organelle biogenesis and metabolic processes. Here, we focus on the mechanism of TFEB activation and on its role in regulating lysosomal function and autophagy. We also describe TFEB tissue-specific functions, as well as the implication of aberrant TFEB activity in human diseases. Finally, we discuss how induction of intracellular clearance by TFEB ameliorates disease phenotypes in mouse models.
MiT family of transcription factors
Four members of the MiT family have been identified: microphthalmia-associated transcription factor (MITF), TFEB, TFE3 and TFEC (Steingrímsson et al., 2004) (see poster). MiT proteins share an identical basic region, which is required for DNA binding, and highly similar HLH and Zip regions that are important for their dimerization; however, outside of these regions they are quite different (Steingrímsson et al., 2004). TFEB, MITF and TFE3 also contain a conserved activation domain that is important for their transcriptional activation (Beckmann et al., 1990; Sato et al., 1997). The activation domain is missing in TFEC, which is the most divergent member of the family and appears to inhibit, rather than activate, transcription (Zhao et al., 1993).
MiT members bind the palindromic CACGTG E-box, a motif also recognized by other bHLH-Zip transcription factors, such as MYC, MAX and MAD proteins (Hemesath et al., 1994). Unlike other bHLH-Zip transcription factors, MiT proteins also bind the asymmetric TCATGTG M-box sequence (Aksan and Goding, 1998). MiT transcription factors bind DNA in the form of both homodimers and heterodimers with any other family member (Hemesath et al., 1994; Pogenberg et al., 2012). However, MiT proteins are not able to heterodimerize with other bHLH-Zip transcription factors (Hemesath et al., 1994). Structural data indicate that a conserved three-residue shift (‘kink’, see poster) within the Zip domain of the MiT members generates an unusual out-of-register leucine zipper that allows for specific heterodimerization among MiT members, while preventing binding to other bHLH-Zip transcription factors (Pogenberg et al., 2012). However, the functional relevance of MiT homodimers compared to heterodimers is still unknown.
All four MiT members are conserved in vertebrates; however, a single MiT ortholog is found in lower organisms, called Mitf in Drosophila melanogaster (Hallsson et al., 2004) and HLH-30 in Caenorhabditis elegans, respectively (Rehli et al., 1999). Invertebrate MiT orthologs all have conserved basic regions and HLH-Zip domains (Bouche et al., 2016), suggesting that they bind DNA in a similar way to mammalian MiT members. Interestingly, Drosophila Mitf is equally related to both human MITF and TFEB (Bouche et al., 2016). Furthermore, the specific functions exerted by the different mammalian MiT members appear to be exploited by a single protein in invertebrate organisms, suggesting that the common ancestor gene underwent multiple rounds of duplication that allowed a functional specialization of the mammalian MiT proteins.
TFEB as a master regulator of lysosomal function and autophagy
Lysosomes are crucial components of the cellular degradation and recycling system, and their correct function is required to maintain proper cell homeostasis (Ballabio, 2016). These organelles are indeed involved in a number of essential cellular processes, including endocytosis, autophagy and lysosomal exocytosis (Settembre et al., 2013b).
Although lysosomes were originally described as static organelles devoted to terminal degradation of waste material, this concept has been challenged by the recent discoveries that lysosomal biogenesis and function are subject to finely tuned transcriptional regulation. Microarray analysis revealed that genes encoding for lysosomal proteins are co-expressed in different cell types and under different conditions (Sardiello et al., 2009). Subsequent promoter analysis of lysosomal genes revealed that they share a common 10-base E-box-like palindromic sequence, the so-called coordinated lysosomal expression and regulation (CLEAR) motif (see poster). TFEB has been shown to directly bind to CLEAR elements, thereby promoting the expression of the entire network of genes that contains the CLEAR regulatory motif in their promoter (namely the CLEAR network) (Palmieri et al., 2011; Sardiello et al., 2009). Accordingly, TFEB overexpression results in an increased number of lysosomes and higher levels of lysosomal enzymes, thus enhancing lysosomal catabolic activity (Sardiello et al., 2009). These findings demonstrate that lysosomal biogenesis and function are globally coordinated by transcriptional regulation.
Subsequent work has shown that TFEB orchestrates the expression of a broader number of genes, which are not only involved in lysosomal biogenesis and function, but also in autophagy and lysosomal exocytosis (Palmieri et al., 2011). In particular, TFEB has been shown to bind to the promoter regions of numerous autophagy genes and to induce autophagosome biogenesis and autophagosome–lysosome fusion (Settembre et al., 2011). Interestingly, TFEB overexpression results in the enhanced degradation of bulk autophagy substrates such as long-lived proteins (Settembre et al., 2011), as well as in the clearance of lipid droplets and damaged mitochondria (Nezich et al., 2015; Settembre et al., 2013a), indicating that this transcription factor also plays a role in modulating organelle-specific autophagy, such as lipophagy and mitophagy. TFEB has also been found to induce lysosomal exocytosis (Medina et al., 2011), a process by which lysosomes fuse to the plasma membrane and secrete their content to the extracellular space.
Therefore, by modulating the processes of lysosomal biogenesis, autophagy and lysosomal exocytosis, TFEB coordinates a transcriptional program able to control the main cellular degradative pathways and to promote intracellular clearance. Importantly, TFEB does not regulate the basal transcription of its targets but rather enhances their transcriptional levels to respond to environmental cues. Following the identification of TFEB, additional regulators of autophagic–lysosomal function have also been identified (reviewed in Feng et al., 2015).
Regulation of TFEB activity
The activity of TFEB is strictly regulated through post-translational modifications, protein–protein interactions and spatial organization (see poster). In resting cells, under nutrient-rich conditions, TFEB is largely cytosolic and inactive (Sardiello et al., 2009; Settembre et al., 2011). Upon starvation or under conditions of lysosomal dysfunction, TFEB rapidly translocates to the nucleus and activates the transcription of its target genes.
The cellular localization and activity of TFEB are mainly controlled by its phosphorylation status. Two particular serine residues in the TFEB protein play a crucial role in determining its subcellular localization, Ser142 (Settembre et al., 2011, 2012) and Ser211 (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012). When both of these two serine residues are phosphorylated, TFEB is kept inactive in the cytosol (see poster). Accordingly, variants of TFEB carrying Ser-to-Ala mutations of either Ser142 or Ser211 are always nuclear and constitutively active (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2011, 2012). Phosphorylation of Ser211, in particular, has been shown to serve as a docking site for the chaperone 14-3-3, which sequesters TFEB in the cytosol and prevents its nuclear translocation, probably by masking its nuclear localization signal (NLS) (Martina et al., 2012; Roczniak-Ferguson et al., 2012).
Mechanistic target of rapamycin complex 1 (mTORC1) and extracellular signal-regulated kinase 2 (ERK2, also known as MAPK1), both master controllers of cellular growth, are the main protein kinases known to phosphorylate TFEB under nutrient-rich conditions in most cell types (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2011, 2012) (see poster). In osteoclasts, TFEB has also been shown to be phosphorylated in its C-terminal region by protein kinase Cβ (PKCβ) upon stimulation with receptor activator of nuclear factor κB ligand (RANKL) (Ferron et al., 2013).
Remarkably, mTORC1 activation occurs at the lysosomal membrane (Sancak et al., 2010). In the presence of nutrients, a mechanism involving the v-ATPase complex promotes the activation of the small Rag (Ras-related GTP-binding) GTPases, which recruit mTORC1 to the lysosomal membrane, thus promoting its activation through the small GTPase Rheb (Sancak et al., 2010, 2008; Zoncu et al., 2011, see poster). Interestingly, active Rag GTPases also bind to TFEB and recruit it to the lysosomal membrane (Martina and Puertollano, 2013), thereby promoting its phosphorylation by mTORC1. This suggests that mTORC1-mediated TFEB phosphorylation can occur at the lysosomal membrane. Multiple lines of evidence have established the lysosome as the main cellular location where mTORC1 activation by amino acids occurs (Sancak et al., 2010, 2008; Zoncu et al., 2011). However, it is still debated whether substrate phosphorylation by mTORC1 also occurs at the lysosome or whether active mTORC1 is released into the cytosol to exert its kinase activity there. Similar to mTORC1, ERK2 also localizes to several subcellular compartments, including the lysosome (Nada et al., 2009). It remains unclear whether ERK2 phosphorylates TFEB at the lysosomal surface or in other subcellular compartments.
Upon starvation or lysosomal stress, mTORC1 is released from the lysosomal membrane and becomes inactive (Sancak et al., 2010). Interestingly, nutrient deprivation concomitantly induces the release of lysosomal Ca2+ through the Ca2+ channel mucolipin 1 (MCOLN1); this activates the phosphatase calcineurin, which in turn dephosphorylates TFEB and promotes its nuclear translocation (Medina et al., 2015) (see poster). Depletion of MCOLN1 inhibits lysosomal Ca2+ release and calcineurin activation, thus preventing TFEB activation and autophagy induction upon nutrient deprivation (Medina et al., 2015).
The signaling cascades described above highlight a central role for the lysosome as a signaling hub able to sense nutrient availability and coordinate the activation of a transcriptional program that allows a finely tuned adaptation of the cell to arising metabolic demands. Intriguingly, many factors that regulate TFEB activity (i.e. most v-ATPase subunits, the Ca2+ channel MCOLN1, the lysosomal ‘platform’ itself) are themselves transcriptionally regulated by TFEB, providing evidence that lysosomal adaptation to environmental changes is a self-sustaining response that is regulated by multiple feedback loops. In addition, TFEB activation also promotes its own transcription (Settembre et al., 2013a), which represents an additional feedback-loop that further sustains lysosomal signaling and function.
The same mechanisms that regulate TFEB activity also appear to control the activity of other MiT members. In the case of TFE3, nutrient availability, mTORC1 activity, as well as its binding to Rag GTPases and to 14-3-3, also play a role in its cytosolic retention and inhibition of its activity (Martina et al., 2014). Furthermore, binding to Rag GTPases and mTORC1 activity also regulate the localization of several MITF isoforms (Martina and Puertollano, 2013), indicating that MiT transcription factors share similar activation mechanisms. In addition, TFE3 overexpression in ARPE19 cells (a cell line derived from retinal pigment epithelium) induces the transcription of a set of genes that is similar to that induced by TFEB (Martina et al., 2014), suggesting a similar function for both factors. However, the strikingly different phenotypes observed upon knockout of the respective genes in mice (see Box 1) indicate that MiT transcription factors have specific functions and limited redundancy.
Box 1. MiT proteins and lysosomal signaling in development and differentiation.
MiT transcription factors regulate important developmental and differentiation processes. Among the different MiT members, the role of MITF in development and differentiation has been extensively characterized. Analysis of several mouse models carrying mutations in the Mitf locus has revealed that MITF is required for the development of melanocytes, retinal pigment epithelial cells, osteoclasts and mast cells (Hodgkinson et al., 1993; Steingrímsson et al., 2004). Accordingly, mice carrying Mitf mutations show coat color defects, smaller eyes and osteopetrosis (Hansdottir et al., 2004; Hodgkinson et al., 1993). In addition, Mitf mutant mice show a reduction in the cell numbers of several other cell types, including NK cells, macrophages and B cells (Roundy et al., 1999; Stechschulte et al., 1987). The role of TFE3 in development is less well characterized. Although TFE3-null mice are viable and do not show any apparent developmental defects, closer characterization of these mice has revealed that TFE3 is important for osteoclast development (Steingrimsson et al., 2002) and for the control of peritoneal mast cell number (Yagil et al., 2012). Furthermore, TFE3 knockdown and overexpression experiments in embryonic stem cells (ESCs) have indicated that this transcription factor sustains ESC self-renewal and restricts their exit from pluripotency (Betschinger et al., 2013). TFEB-null mice show defective placental vascularization and embryonic lethality at E9.5–10.5 (Steingrimsson et al., 1998). Furthermore, TFEB activity has been associated with dendritic cell maturation (Samie and Cresswell, 2015) and osteoclast differentiation (Ferron et al., 2013). TFEB depletion in ESCs has been associated with defective differentiation into the endodermal lineage (Young et al., 2016). These defects appear to be caused by an impaired ability of TFEB-deficient ESCs to induce the canonical Wnt signaling; this is thought to be due to the lack of a fully functional lysosomal compartment (Young et al., 2016), which is required for proper Wnt signaling (Cruciat et al., 2010). A functional lysosomal compartment is also required for Notch activation (Vaccari et al., 2010). Accordingly, depletion of Drosophila MITF results in impaired Notch signaling and developmental defects (Tognon et al., 2016). Thus MiT proteins, by controlling lysosomal homeostasis, ensure proper activation of key signaling pathways that orchestrate developmental and differentiation processes.
Recently, ER stress has been shown to induce the nuclear translocation of TFEB and TFE3 as part of the integrated stress response (Martina et al., 2016). In this case, activation of TFEB and/or TFE3 appears to be mTOR-independent and is achieved through a protein kinase RNA-like endoplasmic reticulum kinase (PERK, also known as EIF2AK3)-dependent mechanism that induces activation of calcineurin and nuclear translocation of TFEB and/or TFE3 (Martina et al., 2016). However, the exact role of PERK in the activation of these transcription factors is still unclear.
Further studies will be needed to fully understand how different pathways communicate with each other to modulate TFEB activity and finely tune the final transcriptional outcome.
Functional in vivo studies of TFEB
The use of animal models, both lower organisms and mammals, has been very helpful to further elucidate TFEB function. TFEB-null mice die at embryonic day (E)9.5–10.5 because of defective placental vascularization (Steingrimsson et al., 1998). Recent evidence has shown that TFEB governs endodermal specification during embryoid body formation (Young et al., 2016) (see also Box 1).
The embryonic lethality of TFEB-null mice has precluded a systematic characterization of TFEB function in different tissues; therefore, the generation of conditional mouse models has been the only way to circumvent this problem. The use of such mice has revealed that TFEB has specialized functions in different tissues (see poster).
Liver-specific deletion and viral-mediated overexpression of TFEB in the liver has revealed a central role for this protein in regulating liver lipid metabolism (Settembre et al., 2013a) (see poster). TFEB has been shown to control lipid catabolism and to directly regulate the transcription of peroxisome proliferator-activated receptor-γ coactivator 1α (Pgc1α, also known as PPARGC1A), a key regulator of energy metabolism (Lin et al., 2005). TFEB depletion in the liver results in impaired liver catabolism and exacerbated metabolic imbalance in obese mice, whereas TFEB overexpression has the opposite effect and rescues obesity and associated metabolic syndrome (Settembre et al., 2013a).
Another tissue-specific function of TFEB has been described in osteoclasts (Ferron et al., 2013). As mentioned above, TFEB activity in osteoclasts is controlled by RANKL, a key regulator of osteoclast function. Osteoclast-specific TFEB deletion results in impaired osteoclast function and increased bone mass, suggesting that TFEB regulates bone resorption in this tissue (Ferron et al., 2013).
In macrophages, depletion of both TFEB and TFE3 results in impaired expression and secretion of several pro-inflammatory cytokines in response to lipopolysaccharide (LPS) injection (Pastore et al., 2016), including interleukin (IL)-1β, IL-6, tumor necrosis factor (TNFα) and chemokine (C-C motif) ligand (CCL)-5, supporting a role for these transcription factors in modulating inflammatory and immune responses. In addition, combined depletion of TFEB and TFE3 in T cells leads to a defective T-cell-dependent antibody response (Huan et al., 2006), whereas TFEB controls antigen presentation by major histocompatibility complexes (MHCs) in dendritic cells (Samie and Cresswell, 2015), suggesting a broad role for TFEB in controlling the immune response.
Gain- and loss-of-function experiments in mouse skeletal muscle have shown that TFEB controls glucose homeostasis and energy balance in this tissue (G. Mansueto and A.B., unpublished data). These effects are associated with an unprecedented role of TFEB in modulating mitochondrial biogenesis in muscle (G. Mansueto and A.B., unpublished data).
In C. elegans, loss-of-function mutations of HLH-30, the worm TFEB orthologue, result in impaired expression of key enzymes required for proper fat catabolism and in impaired lipophagy, indicating that the role of TFEB in lipid catabolism is evolutionary conserved (O'Rourke and Ruvkun, 2013; Settembre et al., 2013a). Conversely, HLH-30 overexpression induces autophagy and lipid catabolism, and increases lifespan in several C. elegans longevity models, whereas nematodes lacking HLH-30 show reduced lifespan (Lapierre et al., 2013; Settembre et al., 2013a). Furthermore, HLH-30 has been shown to promote the expression of nearly 80% of genes that are responsible for immune response against Staphylococcus aureus infections (Visvikis et al., 2014), indicating that modulation of the immune response is another evolutionary conserved function of TFEB.
Finally, work in D. melanogaster has shown that the TFEB orthologue Mitf has an important role in the modulation of lysosomal genes, especially the subunits of the v-ATPase proton pump, as well as in the induction of intracellular clearance and reduction of protein aggregates, and in organ development (Bouche et al., 2016; Tognon et al., 2016; Zhang et al., 2015).
TFEB and the MiT family in tumorigenesis
Chromosomal translocations involving TFEB and TFE3 have been found in patients with clear cell renal cell carcinoma (RCC) (see poster) (Kauffman et al., 2014). In the case of TFEB, a 6p21/11q13 translocation results in the fusion of the TFEB coding region with the regulatory region of the non-coding MALAT1 gene (Davis et al., 2003; Kuiper et al., 2003). Owing to the strength of the MALAT1 promoter, this fusion event results in a significantly enhanced expression of a full-length TFEB protein (Inamura et al., 2012).
In addition to the MALAT1–TFEB translocation, several RCC-associated fusion events have also been found to occur between the TFE3 gene, located on chromosome X, and various other genes (Argani et al., 2003; Clark et al., 1997; Ladanyi et al., 2001; Sidhar et al., 1996, see poster). Interestingly, a TFE3–ASPL fusion has also been reported in alveolar soft part sarcoma (Ladanyi et al., 2001), indicating that upregulation of the MiT transcriptional network can drive tumorigenesis in a number of tissues. Accordingly, MITF amplification has been found in 10–20% of melanomas (Garraway et al., 2005) (see also Box 2).
Box 2. MiT proteins in tumorigenesis.
Dysregulation of MiT genes has been associated with several human solid tumors. MITF amplification accounts for about 20% of melanomas (Garraway et al., 2005; Stark and Hayward, 2007), and single-nucleotide mutations of MITF have been frequently found in human melanomas (Cronin et al., 2009). Importantly, MITF is both necessary and sufficient for melanoma cell growth (Garraway et al., 2005). Increased expression of MITF also drives tumor progression in soft tissue tumor clear cell sarcomas (Davis et al., 2006; Fujimura et al., 1996; Zucman et al., 1993). In this case, MITF is also necessary for clear cell sarcoma survival and proliferation (Davis et al., 2006). Chromosomal translocations involving TFEB and TFE3 occur in a significant amount of patients with RCCs (Argani et al., 2003; Clark et al., 1997; Kuiper et al., 2003; Sidhar et al., 1996), as well as in alveolar soft part sarcoma (Ladanyi et al., 2001) – see also main text. In addition, altered TFEB expression and/or activity has been associated with pancreatic cancer cell proliferation (Marchand et al., 2015) and non-small cell lung cancer motility (Giatromanolaki et al., 2015). Finally, constitutive activation of MiT family members promotes pancreatic tumorigenesis and is required for growth of pancreatic ductal adenocarcinomas (PDAs) (Perera et al., 2015). Although the mechanism by which MiT proteins promote tumorigenesis is still unclear, multiple factors are likely to contribute to MiT-induced cancer development. MiT members can promote cell proliferation and survival by direct upregulation of cell cycle mediators, such as cyclin D2, cyclin D3 and p21 (CDKN1A) (Medendorp et al., 2009; Muller-Hocker et al., 2008), as well as by induction of anti-apoptotic genes, including BCL2 and BIRC7 (Dynek et al., 2008; McGill et al., 2002). In addition, transcriptional upregulation of autophagy by MiT members fuels cancer metabolism and promotes PDA tumor progression (Perera et al., 2015). Finally, MiT-mediated upregulation of the endo-lysosomal system induces the activation of tumorigenic signaling pathways, such as Wnt signaling, with established roles in the initiation and progression of many types of cancer (Clevers and Nusse, 2012; Marchand et al., 2015; Ploper et al., 2015).
Taken together, overexpression of MiT members and upregulation of their transcriptional network appears to be sufficient to drive tumorigenesis in different tissues, although the specific genes involved and the associated mechanisms are still unclear.
TFEB as a therapeutic target for diseases
Owing to its involvement in intracellular clearance pathways, TFEB represents an appealing therapeutic target for many human diseases that are associated with autophagic or lysosomal dysfunction and the accumulation of toxic aggregates. Indeed, induction of TFEB activity has already been successfully used as a therapeutic strategy in several disease models.
A class of diseases that have been shown to benefit from TFEB overexpression are lysosomal storage disorders (LSDs), in which genetic defects in specific lysosomal proteins lead to accumulation of substrates in the lysosomal lumen (Parenti et al., 2015). Overexpression of TFEB in cellular and mouse models of several LSDs, including multiple sulfatase deficiency, mucopolysaccharidosis type IIIA, Batten disease, Pompe disease, Gaucher disease, Tay–Sachs disease and cystinosis, have been shown to be beneficial in reducing substrate accumulation; such overexpression also improved overall autophagy and lysosomal function, and ameliorated the severity of cellular and tissue phenotypes (Medina et al., 2011; Rega et al., 2016; Song et al., 2013; Spampanato et al., 2013). Although the mechanism by which TFEB reduces lysosomal storage and overcomes lysosomal malfunction in LSDs is not fully understood, it is likely that induction of lysosomal exocytosis and secretion of the undigested material plays a major beneficial role (Medina et al., 2011).
Another group of disorders, in which TFEB overexpression has been shown to ameliorate disease progression, are neurodegenerative diseases, including Parkinson's, Huntington's and Alzheimer's disease, as well as other tauopathies. Growing evidence has established the accumulation of protein aggregates and autophagic and/or lysosomal dysfunction as the major pathogenic mechanisms underlying such diseases (Menzies et al., 2015). Remarkably, heterozygous mutations in the gene encoding for the lysosomal enzyme glucocerebrosidase have been found in a substantial number of patients with Parkinson's disease (Sidransky et al., 2009), further supporting a role for lysosomal dysfunction in the pathogenesis of the disorder. Genetic or pharmacological activation of TFEB in cellular and mouse models of Parkinson's disease has been found to improve lysosomal function and ameliorate α-synuclein aggregation (Decressac et al., 2013; Dehay et al., 2010; Kilpatrick et al., 2015), which is a major hallmark of Parkinson's disease. Similarly, inducing TFEB activity in cellular and animal models of Hungtington's disease can reduce protein aggregation and improves neurological functions (Sardiello et al., 2009; Tsunemi et al., 2012). Furthermore, TFEB overexpression or its pharmacological activation in cellular and mouse models of Alzheimer's disease and other tauopathies can also reduce the amount of protein aggregates (Chauhan et al., 2015; Polito et al., 2014; Xiao et al., 2014, 2015), which results in a reduction of neurodegeneration and improvement of behavioral deficits. Therefore, promotion of intracellular clearance through the induction of TFEB activity might represent a common therapeutic strategy for neurodegenerative disorders.
TFEB overexpression has also shown therapeutic effects in mouse models of α1-antitrypsin deficiency, the most common genetic cause of liver disease (Pastore et al., 2013). In this case as well, enhanced intracellular clearance represents the major therapeutic mechanism, as induction of autophagy by TFEB reduces the levels of α1-antitrypsin accumulation and ameliorates liver injury. Finally, TFEB upregulation is also beneficial in reducing obesity and associated metabolic syndrome as it promotes lipophagy (Settembre et al., 2013a). Thus, upregulation of intracellular clearance by TFEB has proven to be beneficial in a vast number of disease models.
Concluding remarks
The identification of TFEB as a global modulator of intracellular clearance and energy metabolism, through the regulation of genes involved in the lysosomal–autophagic pathways, has provided new insights into the mechanism by which the cell responds to environmental cues such as nutrient availability. However, the mechanisms by which cells integrate multiple extra- and intra-cellular signals to modulate an appropriate response require further investigation. Furthermore, it remains unclear how different signals can activate particular MiT members and whether their specific homodimerization or heterodimerization promotes different responses.
Owing to the broad number of diseases that potentially benefit from promoting intracellular clearance, modulating the activity of TFEB and of other MiT members represents an appealing therapeutic strategy that, however, requires further investigation. Although acute induction of intracellular clearance ameliorates disease progression in several animal models, any long-term effects of such treatments have not been evaluated. Significantly and constitutively enhanced TFEB activity can lead to tumorigenesis, as in the case of clear cell carcinoma in the kidney (Davis et al., 2003; Kuiper et al., 2003). Furthermore, induction of autophagy by activation of MiT genes, including TFEB, has been found to have an important role in pancreatic cancer (Perera et al., 2015). Therefore, alternative strategies, such as inducing TFEB activity for only limited periods or enhancing only specific subsets of the TFEB-regulated gene network, should be considered when using TFEB as a therapeutic tool.
Acknowledgements
We apologize for not being able to discuss and cite the work of many researchers in the MiT field due to space limitations.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
We are grateful to the Fondazione Telethon; the Beyond Batten Disease Foundation; the European Research Council, the Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research); and the National Institutes of Health for their generous support. G.N. is supported by the DTI-IMPORT Marie Skłodowska-Curie COFUND program and by a Horizon 2020 Marie Skłodowska-Curie individual fellowship (IF) of the European commission. Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster and individual poster panels are available for downloading at http://jcs.biologists.org/lookup/doi/10.1242/jcs.146365.supplemental.
References
- Aksan I. and Goding C. R. (1998). Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol. Cell. Biol. 18, 6930-6938. 10.1128/MCB.18.12.6930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argani P., Lui M. Y., Couturier J., Bouvier R., Fournet J.-C. and Ladanyi M. (2003). A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23). Oncogene 22, 5374-5378. 10.1038/sj.onc.1206686 [DOI] [PubMed] [Google Scholar]
- Ballabio A. (2016). The awesome lysosome. EMBO Mol. Med. 8, 73-76. 10.15252/emmm.201505966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann H., Su L. K. and Kadesch T. (1990). TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 4, 167-179. 10.1101/gad.4.2.167 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Nichols J., Dietmann S., Corrin P. D., Paddison P. J. and Smith A. (2013). Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153, 335-347. 10.1016/j.cell.2013.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche V., Perez Espinosa A., Leone L., Sardiello M., Ballabio A. and Botas J. (2016). Drosophila Mitf regulates the V-ATPase and the lysosomal-autophagic pathway. Autophagy 12, 484-498. 10.1080/15548627.2015.1134081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S., Ahmed Z., Bradfute S. B., Arko-Mensah J., Mandell M. A., Won Choi S., Kimura T., Blanchet F., Waller A., Mudd M. H. et al. (2015). Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. Nat. Commun. 6, 8620 10.1038/ncomms9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J., Lu Y.-J., Sidhar S. K., Parker C., Gill S., Smedley D., Hamoudi R., Linehan W. M., Shipley J. and Cooper C. S. (1997). Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene 15, 2233-2239. 10.1038/sj.onc.1201394 [DOI] [PubMed] [Google Scholar]
- Clevers H. and Nusse R. (2012). Wnt/β-catenin signaling and disease. Cell 149, 1192-1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Cronin J. C., Wunderlich J., Loftus S. K., Prickett T. D., Wei X., Ridd K., Vemula S., Burrell A. S., Agrawal N. S., Lin J. C. et al. (2009). Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 22, 435-444. 10.1111/j.1755-148X.2009.00578.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciat C. M., Ohkawara B., Acebron S. P., Karaulanov E., Reinhard C., Ingelfinger D., Boutros M. and Niehrs C. (2010). Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science 327, 459-463. 10.1126/science.1179802 [DOI] [PubMed] [Google Scholar]
- Davis I. J., Hsi B.-L., Arroyo J. D., Vargas S. O., Yeh Y. A., Motyckova G., Valencia P., Perez-Atayde A. R., Argani P., Ladanyi M. et al. (2003). Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc. Natl. Acad. Sci. USA 100, 6051-6056. 10.1073/pnas.0931430100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I. J., Kim J. J., Ozsolak F., Widlund H. R., Rozenblatt-Rosen O., Granter S. R., Du J., Fletcher J. A., Denny C. T., Lessnick S. L. et al. (2006). Oncogenic MITF dysregulation in clear cell sarcoma: defining the MiT family of human cancers. Cancer Cell 9, 473-484. 10.1016/j.ccr.2006.04.021 [DOI] [PubMed] [Google Scholar]
- Decressac M., Mattsson B., Weikop P., Lundblad M., Jakobsson J. and Bjorklund A. (2013). TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc. Natl. Acad. Sci. USA 110, E1817-E1826. 10.1073/pnas.1305623110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B., Bove J., Rodriguez-Muela N., Perier C., Recasens A., Boya P. and Vila M. (2010). Pathogenic lysosomal depletion in Parkinson's disease. J. Neurosci. 30, 12535-12544. 10.1523/JNEUROSCI.1920-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynek J. N., Chan S. M., Liu J., Zha J., Fairbrother W. J. and Vucic D. (2008). Microphthalmia-associated transcription factor is a critical transcriptional regulator of melanoma inhibitor of apoptosis in melanomas. Cancer Res. 68, 3124-3132. 10.1158/0008-5472.CAN-07-6622 [DOI] [PubMed] [Google Scholar]
- Feng Y., Yao Z. and Klionsky D. J. (2015). How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 25, 354-363. 10.1016/j.tcb.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron M., Settembre C., Shimazu J., Lacombe J., Kato S., Rawlings D. J., Ballabio A. and Karsenty G. (2013). A RANKL-PKCβ-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 27, 955-969. 10.1101/gad.213827.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura Y., Ohno T., Siddique H., Lee L., Rao V. N. and Reddy E. S. (1996). The EWS-ATF-1 gene involved in malignant melanoma of soft parts with t(12;22) chromosome translocation, encodes a constitutive transcriptional activator. Oncogene 12, 159-167. [PubMed] [Google Scholar]
- Garraway L. A., Widlund H. R., Rubin M. A., Getz G., Berger A. J., Ramaswamy S., Beroukhim R., Milner D. A., Granter S. R., Du J. et al. (2005). Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436, 117-122. 10.1038/nature03664 [DOI] [PubMed] [Google Scholar]
- Giatromanolaki A., Kalamida D., Sivridis E., Karagounis I. V., Gatter K. C., Harris A. L. and Koukourakis M. I. (2015). Increased expression of transcription factor EB (TFEB) is associated with autophagy, migratory phenotype and poor prognosis in non-small cell lung cancer. Lung Cancer 90, 98-105. 10.1016/j.lungcan.2015.07.008 [DOI] [PubMed] [Google Scholar]
- Hallsson J. H., Haflidadóttir B. S., Stivers C., Odenwald W., Arnheiter H., Pignoni F. and Steingrímsson E. (2004). The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics 167, 233-241. 10.1534/genetics.167.1.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansdottir A. G., Pálsdóttir K., Favor J., Neuhäuser-Klaus A., Fuchs H., de Angelis M. H. and Steingrı´msson E. (2004). The novel mouse microphthalmia mutations Mitfmi-enu5 and Mitfmi-bcc2 produce dominant negative Mitf proteins. Genomics 83, 932-935. 10.1016/j.ygeno.2003.10.013 [DOI] [PubMed] [Google Scholar]
- Hemesath T. J., Steingrímsson E., McGill G., Hansen M. J., Vaught J., Hodgkinson C. A., Arnheiter H., Copeland N. G., Jenkins N. A. and Fisher D. E. (1994). microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8, 2770-2780. 10.1101/gad.8.22.2770 [DOI] [PubMed] [Google Scholar]
- Hodgkinson C. A., Moore K. J., Nakayama A., Steingrímsson E., Copeland N. G., Jenkins N. A. and Arnheiter H. (1993). Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74, 395-404. 10.1016/0092-8674(93)90429-T [DOI] [PubMed] [Google Scholar]
- Huan C., Kelly M. L., Steele R., Shapira I., Gottesman S. R. and Roman C. A. (2006). Transcription factors TFE3 and TFEB are critical for CD40 ligand expression and thymus-dependent humoral immunity. Nat. Immunol. 7, 1082-1091. 10.1038/ni1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamura K., Fujiwara M., Togashi Y., Nomura K., Mukai H., Fujii Y., Yamamoto S., Yonese J., Fukui I. and Ishikawa Y. (2012). Diverse fusion patterns and heterogeneous clinicopathologic features of renal cell carcinoma with t (6; 11) translocation. Am. J. Surg. Pathol. 36, 35-42. 10.1097/PAS.0b013e3182293ec3 [DOI] [PubMed] [Google Scholar]
- Kauffman E. C., Ricketts C. J., Rais-Bahrami S., Yang Y., Merino M. J., Bottaro D. P., Srinivasan R. and Linehan W. M. (2014). Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat. Rev. Urol. 11, 465-475. 10.1038/nrurol.2014.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick K., Zeng Y., Hancock T. and Segatori L. (2015). Genetic and chemical activation of TFEB mediates clearance of aggregated alpha-synuclein. PLoS ONE 10, e0120819 10.1371/journal.pone.0120819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper R. P., Schepens M., Thijssen J., van Asseldonk M., van den Berg E., Bridge J., Schuuring E., Schoenmakers E. F. P. M. and van Kessel A. G. (2003). Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum. Mol. Genet. 12, 1661-1669. 10.1093/hmg/ddg178 [DOI] [PubMed] [Google Scholar]
- Ladanyi M., Lui M. Y., Antonescu C. R., Krause-Boehm A., Meindl A., Argani P., Healey J. H., Ueda T., Yoshikawa H., Meloni-Ehrig A. et al. (2001). The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene 20, 48-57. 10.1038/sj.onc.1204074 [DOI] [PubMed] [Google Scholar]
- Lapierre L. R., De Magalhaes Filho C. D., McQuary P. R., Chu C. C., Visvikis O., Chang J. T., Gelino S., Ong B., Davis A. E., Irazoqui J. E. et al. (2013). The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4, 2267 10.1038/ncomms3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Handschin C. and Spiegelman B. M. (2005). Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1, 361-370. 10.1016/j.cmet.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Marchand B., Arsenault D. and Raymond-Fleury A. (2015). Glycogen synthase kinase-3 (GSK3) inhibition induces prosurvival autophagic signals in human pancreatic cancer cells. J. Biol. 290, 5592-5605. 10.1074/jbc.m114.616714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J. A. and Puertollano R. (2013). Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 200, 475-491. 10.1083/jcb.201209135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J. A., Chen Y., Gucek M. and Puertollano R. (2012). MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903-914. 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J. A., Diab H. I., Lishu L., Jeong-A L., Patange S., Raben N. and Puertollano R. (2014). The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 7, ra9 10.1126/scisignal.2004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J. A., Diab H. I., Brady O. A. and Puertollano R. (2016). TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 35, 479-495. 10.15252/embj.201593428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill G. G., Horstmann M., Widlund H. R., Du J., Motyckova G., Nishimura E. K., Lin Y.-L., Ramaswamy S., Avery W., Ding H.-F. et al. (2002). Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 109, 707-718. 10.1016/S0092-8674(02)00762-6 [DOI] [PubMed] [Google Scholar]
- Medendorp K., van Groningen J. J. M., Vreede L., Hetterschijt L., Brugmans L., van den Hurk W. H. and van Kessel A. G. (2009). The renal cell carcinoma-associated oncogenic fusion protein PRCCTFE3 provokes p21 WAF1/CIP1-mediated cell cycle delay. Exp. Cell Res. 315, 2399-2409. 10.1016/j.yexcr.2009.04.022 [DOI] [PubMed] [Google Scholar]
- Medina D. L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J. A., Sardiello M. et al. (2011). Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell 21, 421-430. 10.1016/j.devcel.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D. L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A. et al. (2015). Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17, 288-299. 10.1038/ncb3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies F. M., Fleming A. and Rubinsztein D. C. (2015). Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 16, 345-357. 10.1038/nrn3961 [DOI] [PubMed] [Google Scholar]
- Müller-Höcker J., Babaryka G., Schmid I. and Jung A. (2008). Overexpression of cyclin D1, D3, and p21 in an infantile renal carcinoma with Xp11.2 TFE3-gene fusion. Pathol. Res. Pract. 204, 589-597. 10.1016/j.prp.2008.01.010 [DOI] [PubMed] [Google Scholar]
- Nada S., Hondo A., Kasai A., Koike M., Saito K., Uchiyama Y. and Okada M. (2009). The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 28, 477-489. 10.1038/emboj.2008.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezich C. L., Wang C., Fogel A. I. and Youle R. J. (2015). MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol. 210, 435-450. 10.1083/jcb.201501002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke E. J. and Ruvkun G. (2013). MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 15, 668-676. 10.1038/ncb2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M. and Ballabio A. (2011). Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20, 3852-3866. 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- Parenti G., Andria G. and Ballabio A. (2015). Lysosomal storage diseases: from pathophysiology to therapy. Annu. Rev. Med. 66, 471-486. 10.1146/annurev-med-122313-085916 [DOI] [PubMed] [Google Scholar]
- Pastore N., Blomenkamp K., Annunziata F., Piccolo P., Mithbaokar P., Maria Sepe R., Vetrini F., Palmer D., Ng P., Polishchuk E. et al. (2013). Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in alpha-1-anti-trypsin deficiency. EMBO Mol. Med. 5, 397-412. 10.1002/emmm.201202046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore N., Brady O. A., Diab H. I., Martina J. A., Sun L., Huynh T., Lim J. A., Zare H., Raben N., Ballabio A., Puertollano R. (2016). TFEB and TFE3 Cooperate in the Regulation of the Innate Immune Response in Activated Macrophages. Autophagy [Epub] doi:10.1080/15548627.2016.1179405. 10.1080/15548627.2016.1179405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R. M., Stoykova S., Nicolay B. N., Ross K. N., Fitamant J., Boukhali M., Lengrand J., Deshpande V., Selig M. K., Ferrone C. R. et al. (2015). Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 524, 361-365. 10.1038/nature14587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploper D., Taelman V. F., Robert L., Perez B. S., Titz B., Chen H.-W., Graeber T. G., Von Euw E., Ribas A. and Robertis E. M. (2015). MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl. Acad. Sci. USA 112, E420-E429. 10.1073/pnas.1424576112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogenberg V., Ögmundsdóttir M. H., Bergsteinsdóttir K., Schepsky A., Phung B., Deineko V., Milewski M., Steingrímsson E. and Wilmanns M. (2012). Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev. 26, 2647-2658. 10.1101/gad.198192.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polito V. A., Li H., Martini-Stoica H., Wang B., Yang L., Xu Y., Swartzlander D. B., Palmieri M., di Ronza A., Lee V. M.-Y. et al. (2014). Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol. Med. 6, 1142-1160. 10.15252/emmm.201303671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rega L. R., Polishchuk E., Montefusco S., Napolitano G., Tozzi G., Zhang J., Bellomo F., Taranta A., Pastore A., Polishchuk R. et al. (2016). Activation of the transcription factor EB rescues lysosomal abnormalities in cystinotic kidney cells. Kidney Int. 89, 862-873. 10.1016/j.kint.2015.12.045 [DOI] [PubMed] [Google Scholar]
- Rehli M., Den Elzen N., Cassady A. I., Ostrowski M. C. and Hume D. A. (1999). Cloning and characterization of the murine genes for bHLH-ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics 56, 111-120. 10.1006/geno.1998.5588 [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Petit C. S., Froehlich F., Qian S., Ky J., Angarola B., Walther T. C. and Ferguson S. M. (2012). The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundy K., Kollhoff A., Eichwald E. J., Weis J. J. and Weis J. H. (1999). Microphthalmic mice display a B cell deficiency similar to that seen for mast and NK cells. J. Immunol. 163, 6671-6678. [PubMed] [Google Scholar]
- Samie M. and Cresswell P. (2015). The transcription factor TFEB acts as a molecular switch that regulates exogenous antigen-presentation pathways. Nat. Immunol. 16, 729-736. 10.1038/ni.3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L. and Sabatini D. M. (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496-1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S. and Sabatini D. M. (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290-303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S. et al. (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473-477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Sato S., Roberts K., Gambino G., Cook A., Kouzarides T. and Goding C. R. (1997). CBP/p300 as a co-factor for the Microphthalmia transcription factor. Oncogene 14, 3083-3092. 10.1038/sj.onc.1201298 [DOI] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V. A., Arencibia M. G., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P. et al. (2011). TFEB links autophagy to lysosomal biogenesis. Science 332, 1429-1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D. L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M. C. et al. (2012). A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095-1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., De Cegli R., Mansueto G., Saha P. K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T. J. et al. (2013a). TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15, 647-658. 10.1038/ncb2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Fraldi A., Medina D. L. and Ballabio A. (2013b). Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14, 283-296. 10.1038/nrm3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhar S. K., Clark J., Gill S., Hamoudi R., Crew A. J., Gwilliam R., Ross M., Linehan W. M., Birdsall S., Shipley J. et al. (1996). The t(X;1)(p11.2;q21.2) translocation in papillary renal cell carcinoma fuses a novel gene PRCC to the TFE3 transcription factor gene. Hum. Mol. Genet. 5, 1333-1338. 10.1093/hmg/5.9.1333 [DOI] [PubMed] [Google Scholar]
- Sidransky E., Nalls M. A., Aasly J. O., Aharon-Peretz J., Annesi G., Barbosa E. R., Bar-Shira A., Berg D., Bras J., Brice A. et al. (2009). Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N. Engl. J. Med. 361, 1651-1661. 10.1056/NEJMoa0901281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Wang F., Savini M., Ake A., di Ronza A., Sardiello M. and Segatori L. (2013). TFEB regulates lysosomal proteostasis. Hum. Mol. Genet. 22, 1994-2009. 10.1093/hmg/ddt052 [DOI] [PubMed] [Google Scholar]
- Spampanato C., Feeney E., Li L., Cardone M., Lim J.-A., Annunziata F., Zare H., Polishchuk R., Puertollano R., Parenti G. et al. (2013). Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 5, 691-706. 10.1002/emmm.201202176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M. and Hayward N. (2007). Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 67, 2632-2642. 10.1158/0008-5472.CAN-06-4152 [DOI] [PubMed] [Google Scholar]
- Stechschulte D. J., Sharma R., Dileepan K. N., Simpson K. M., Aggarwal N., Clancy J. Jr. and Jilka R. L. (1987). Effect of the mi allele on mast cells, basophils, natural killer cells, and osteoclasts in C57BI/6J mice. J. Cell. Physiol. 132, 565-570. 10.1002/jcp.1041320321 [DOI] [PubMed] [Google Scholar]
- Steingrimsson E., Tessarollo L., Reid S. W., Jenkins N. A. and Copeland N. G. (1998). The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development 125, 4607-4616. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E., Tessarollo L., Pathak B., Hou L., Arnheiter H., Copeland N. G. and Jenkins N. A. (2002). Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc. Natl. Acad. Sci. USA 99, 4477-4482. 10.1073/pnas.072071099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrímsson E., Copeland N. G. and Jenkins N. A. (2004). Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 38, 365-411. 10.1146/annurev.genet.38.072902.092717 [DOI] [PubMed] [Google Scholar]
- Tognon E., Kobia F., Busi I., Fumagalli A., De Masi F. and Vaccari T. (2016). Control of lysosomal biogenesis and Notch-dependent tissue patterning by components of the TFEB-V-ATPase axis in Drosophila melanogaster. Autophagy 12, 1-16. 10.1080/15548627.2015.1134080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T., Ashe T. D., Morrison B. E., Soriano K. R., Au J., Roque R. A., Lazarowski E. R., Damian V. A., Masliah E. and La Spada A. R. (2012). PGC-1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 4, 142ra97 10.1126/scitranslmed.3003799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T., Duchi S., Cortese K., Tacchetti C. and Bilder D. (2010). The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Development 137, 1825-1832. 10.1242/dev.045484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvikis O., Ihuegbu N., Labed S. A., Luhachack L. G., Alves A.-M. F., Wollenberg A. C., Stuart L. M., Stormo G. D. and Irazoqui J. E. (2014). Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40, 896-909. 10.1016/j.immuni.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q., Yan P., Ma X., Liu H., Perez R., Zhu A., Gonzales E., Burchett J. M., Schuler D. R., Cirrito J. R. et al. (2014). Enhancing astrocytic lysosome biogenesis facilitates Abeta clearance and attenuates amyloid plaque pathogenesis. J. Neurosci. 34, 9607-9620. 10.1523/JNEUROSCI.3788-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q., Yan P., Ma X., Liu H., Perez R., Zhu A., Gonzales E., Tripoli D. L., Czerniewski L., Ballabio A. et al. (2015). Neuronal-targeted TFEB accelerates lysosomal degradation of APP, reducing abeta generation and amyloid plaque pathogenesis. J. Neurosci. 35, 12137-12151. 10.1523/JNEUROSCI.0705-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagil Z., Hadad Erlich T., Ofir-Birin Y., Tshori S., Kay G., Yekhtin Z., Fisher D. E., Cheng C., Wong W. S., Hartmann K. et al. (2012). Transcription factor E3, a major regulator of mast cell-mediated allergic response. J. Allergy Clin. Immunol. 129, 1357-1366 e5. 10.1016/j.jaci.2011.11.051 [DOI] [PubMed] [Google Scholar]
- Young N. P., Kamireddy A., Van Nostrand J. L., Eichner L. J., Shokhirev M. N., Dayn Y. and Shaw R. J. (2016). AMPK governs lineage specification through Tfeb-dependent regulation of lysosomes. Genes Dev. 30, 535-552. 10.1101/gad.274142.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Zhou Q., Ogmundsdottir M. H., Moller K., Siddaway R., Larue L., Hsing M., Kong S. W., Goding C. R., Palsson A. et al. (2015). Mitf is a master regulator of the v-ATPase, forming a control module for cellular homeostasis with v-ATPase and TORC1. J. Cell Sci. 128, 2938-2950. 10.1242/jcs.173807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G. Q., Zhao Q., Zhou X., Mattei M. G. and de Crombrugghe B. (1993). TFEC, a basic helix-loop-helix protein, forms heterodimers with TFE3 and inhibits TFE3-dependent transcription activation. Mol. Cell. Biol. 13, 4505-4512. 10.1128/MCB.13.8.4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y. and Sabatini D. M. (2011). mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334, 678-683. 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucman J., Delattre O., Desmaze C., Epstein A. L., Stenman G., Speleman F., Fletchers C. D. M., Aurias A. and Thomas G. (1993). EWS and ATF-1 gene fusion induced by t(12;22) translocation in malignant melanoma of soft parts. Nat. Genet. 4, 341-345. 10.1038/ng0893-341 [DOI] [PubMed] [Google Scholar]