ABSTRACT
Neurofibromatosis type 1 (NF1) is a common genetic disorder characterized by the occurrence of nerve sheath tumors and considerable clinical heterogeneity. Some translational studies have been limited by the lack of animal models available for assessing patient-specific mutations. In order to test therapeutic approaches that might restore function to the mutated gene or gene product, we developed mice harboring NF1 patient-specific mutations including a nonsense mutation (c.2041C>T; p.Arg681*) and a missense mutation (c.2542G>C; p.Gly848Arg). The latter is associated with the development of multiple plexiform neurofibromas along spinal nerve roots. We demonstrate that the human nonsense NF1Arg681* and missense NF1Gly848Arg mutations have different effects on neurofibromin expression in the mouse and each recapitulates unique aspects of the NF1 phenotype, depending upon the genetic context when assessed in the homozygous state or when paired with a conditional knockout allele. Whereas the missense Nf1Gly848Arg mutation fails to produce an overt phenotype in the mouse, animals homozygous for the nonsense Nf1Arg681* mutation are not viable. Mice with one Nf1Arg681* allele in combination with a conditional floxed Nf1 allele and the DhhCre transgene (Nf14F/Arg681*; DhhCre) display disorganized nonmyelinating axons and neurofibromas along the spinal column, which leads to compression of the spinal cord and paralysis. This model will be valuable for preclinical testing of novel nonsense suppression therapies using drugs to target in-frame point mutations that create premature termination codons in individuals with NF1.
KEY WORDS: Neurofibromatosis type 1, Patient-derived mouse models, Nonsense mutation, Missense mutation
Summary: Two mouse models carrying NF1 patient-specific mutations have distinct effects on embryonic development, neurofibromin function and plexiform neurofibroma formation, with implications for therapeutic studies.
INTRODUCTION
Neurofibromatosis type 1 (NF1) is a common genetic disorder affecting ∼1 in 3000 individuals and over two million people worldwide (Friedman, 1999; Evans et al., 2010). NF1 is characterized principally by the occurrence of neurofibromas and café-au-lait macules (CALMs) (Boyd et al., 2009). Both children and adults with NF1 are at an increased risk for developing certain malignancies and display an assortment of benign and malignant lesions including neurofibromas (dermal and plexiform), pheochromocytomas, pilocytic astrocytomas, optic gliomas, bone abnormalities, leukemias and malignant peripheral nerve sheath tumors (Boyd et al., 2009). NF1 is caused by inherited or de novo pathogenic variants in the NF1 gene, whereas tumor development requires additional mutation of the other NF1 allele (loss of heterozygosity) (Garcia-Linares et al., 2011; Steinmann et al., 2009). To date, no strong genotype-phenotype correlation has been established for NF1 because of the varied disease expression observed in humans. Many individuals with NF1, even from the same family, show differences in the presentation or severity of the NF1 phenotype (Pasmant et al., 2012; Alkindy et al., 2012; Sabbagh et al., 2009). This is exemplified by the National Institutes of Health (NIH) diagnostic criteria for NF1, which require a minimum of two of the following: six or more CALMs, two or more cutaneous neurofibromas or one plexiform neurofibroma, axillary or inguinal freckling, optic pathway glioma, two or more Lisch nodules, bony dysplasia, or a first degree relative with NF1 (Ferner et al., 2007). Furthermore, there are few in vitro assays available to examine neurofibromin function, which is especially problematic when trying to assess the potential pathogenicity of missense variants.
Several mouse models have been developed that display some of the phenotypes seen in individuals with NF1, including learning difficulties, as well as the development of plexiform neurofibromas, optic gliomas and malignant peripheral nerve sheath tumors (Boyd et al., 2009). These existing animal models generally employ conditional knockout strategies that lead to loss of function of one or both Nf1 alleles, which have proven to be valuable tools to investigate the pathogenesis of NF1, and to evaluate several therapeutic strategies such as inhibitors of RAS or intercellular signaling (Yang et al., 2008; Robertson et al., 2012; Jessen et al., 2013). However, the underlying mutations used in current animal models are not sufficient to evaluate novel therapeutic approaches to target the mutated gene or gene product, including nonsense suppression and exon-skipping therapies. These therapies have been established in Duchenne muscular dystrophy and cystic fibrosis (Du et al., 2002, 2006; De Luca et al., 2008; Loufrani et al., 2004; Barton-Davis et al., 1999; Zhang et al., 2014; Xue et al., 2014; Mann et al., 2001; Lu et al., 2005; van Deutekom et al., 2001; Aartsma-Rus et al., 2003; Igreja et al., 2016). Thus, the use of NF1 mouse models harboring mutations identified in individuals with NF1 provides the ability to perform prospective investigations targeting a specific underlying mutation or mutation type.
To this end, we created and characterized two novel mouse models harboring mutations found in individuals with NF1, including a common nonsense mutation (nmNF1, exon 18 c.2041C>T; p.Arg681*) expected to produce complete loss of function, and a recurring missense mutation found in individuals with NF1 associated with multiple plexiform neurofibromas along spinal nerve roots (exon 21 c.2542G>C; p.Gly848Arg). We demonstrate that the nonsense c.2041C>T; p.Arg681* (hereafter called Nf1Arg681*) and missense c.2542G>C; p.Gly848Arg (hereafter called Nf1Gly848Arg) mutations in the mouse have different effects on neurofibromin expression, and each recapitulates unique aspects of NF1 depending upon the genetic context when assessed in the homozygous state, or when paired with a conditional knockout allele.
RESULTS
Establishing mouse models with NF1 patient mutations
Nonsense mutations make up ∼20% of characterized mutations (Messiaen and Wimmer, 2008). The Medical Genomics Laboratory at the University of Alabama at Birmingham has identified 19 of the most recurrent nonsense mutations found in individuals with NF1 (each responsible for at least 0.3% of NF1 cases). Among those is a nonsense mutation in exon 18, c.2041C>T; p.Arg681* observed in 49/8000 unrelated NF1-mutation-positive probands (0.61%). Of 39 individuals with phenotypic data available, 15 individuals were 0-2 years of age at the time of testing (mostly too young to fulfill clinical diagnostic criteria). Within these individuals, 2/15 fulfill NIH criteria with 2/15 individuals having <6 CALMs. The remaining 24 individuals are ≥3 years of age and all fulfill NIH criteria for NF1 diagnosis; although none possess symptomatic spinal neurofibromas (including half who were ≥19 years old), 8/24 individuals did have an externally visible plexiform neurofibroma.
In contrast, a missense mutation located in exon 21 (c.2542G>C; p.Gly848Arg), observed in 11/7800 unrelated NF1-mutation-positive probands (0.14%), is associated with symptomatic spinal nerve root tumors. Phenotypic data were obtained from 20 individuals from four different families with a p.Gly848Arg mutation. Three of these individuals are 0-2 years of age, with 1/3 fulfilling the NIH criteria and 2/3 having <6 CALMs, whereas 7/17 of the individuals aged ≥3 years fulfill the NIH criteria, with 4/17 having an externally visible plexiform neurofibroma. Symptomatic spinal nerve root tumors were found in 6/17 individuals, including 5/8 individuals who were ≥19 years old. For individuals 19 years or older, there is a significant difference in the frequency of spinal nerve root tumors between the c.2041C>T (0/12) vs c.2542G>A (5/8) mutation (2-tailed Fisher exact: P=0.0036).
To model each human mutation in the mouse, knock-in animals harboring the nonsense c.2041C>T; p.Arg681* mutation allele in exon 18 (Nf1Arg681*) and the missense c.2542G>C; p.Gly848Arg mutation allele in exon 21 (Nf1Gly848Arg) were created using conventional gene-targeting vectors and C57BL/6 embryonic stem cells, with chimeric mice and germ line transmission being confirmed (Table S1). Since the Nf1-floxed animals that have been used in previous studies were generated on a mixed genetic background (Zhu et al., 2002), we obtained mice from the International Knockout Mouse Consortium (IKMC) harboring a ‘knockout first’ allele for the Nf1 gene (Nf1tm1a(KOMP)Wtsi) in order to have all Nf1 alleles on the inbred C57BL/6 background. We generated a new conditional knockout ‘floxed’ allele of the Nf1 gene where exon 4 is flanked by loxP sites (hereafter referred to as Nf14F) after removal of the FRT-flanked LacZ/Neo cassettes by breeding to a FLP-recombinase line. Matings between heterozygous Nf1+/4F mice yielded homozygous floxed Nf14F/4F mice at the expected Mendelian ratio that were viable, fertile and did not show gross or histological abnormalities (data not shown). Heterozygous Nf1+/Δ4 mice harboring a null allele resulting from exon 4 deletion were created by breeding Nf1+/4F mice to an EIIaCre driver line [B6.FVB-Tg(EIIa-cre)C5379Lmgd/J strain #003724], thereby deleting the floxed sequence in all cells within the developing embryos.
Nonsense mutation NF1Arg681* recapitulated in the mouse is non-functional
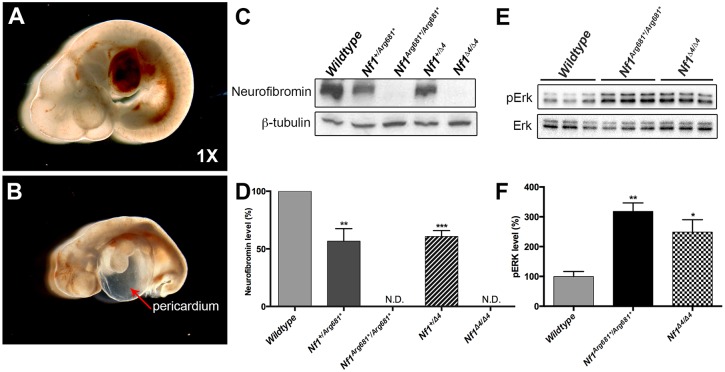
Previous studies have demonstrated that mouse embryos fail to develop beyond embryonic day (E)13.5 in the absence of Nf1 expression as a result of cardiovascular defects (Jacks et al., 1994; Brannan et al., 1994). To determine the functionality of each allele, we performed intercross breedings of Nf1+/Arg681* and Nf1+/Δ4 mice and examined embryos from E9.5 to E13.5. Embryos heterozygous for either the Nf1Arg681* or the Nf1Δ4 allele were viable and developed to term with no observable defects. At E9.5, all homozygous Nf1Arg681*/Arg681* and Nf1Δ4/Δ4 embryos were indistinguishable from their littermate controls; however, these homozygous embryos fail to develop past this point (Fig. S2). Although Nf1Arg681*/Arg681* mice display pericardial effusions at E10.5 (Fig. 1A,B), we were unable to ascertain the exact cause of death because of extensive tissue degradation.
Fig. 1.
Nf1Arg681*/Arg681* mice fail to develop after E9.5. (A,B) Morphology of representative wild-type (A) and Nf1Arg681*/Arg681* (B) embryos at E10.5. The red arrowhead in B indicates effusion into the pericardial space. The magnification is noted in the lower right corner. (C-F) Western blot of neurofibromin (C) and phosphorylated ERK (pERK) (E) in E9.5 MEFs of indicated genotypes. Results are representative of three independent experiments or samples. β-tubulin is shown as loading control. The western blot analysis of neurofibromin (D) and pERK (F) was quantified, and the results are expressed in histograms. *P<0.05, **P<0.01, ***P<0.001 vs wild type by one-way ANOVA. Data are presented as mean±s.e.m. MEFs, mouse embryonic fibroblasts. N.D., none detected.
To assess neurofibromin function in these models, we isolated and expanded mouse embryonic fibroblasts (MEFs) from E9.5 embryos carrying different allelic mutations, and performed western blot analyses. Neurofibromin was completely absent in Nf1Arg681*/Arg681* and Nf1Δ4/Δ4 MEFs, and was markedly reduced in Nf1+/Arg681* and Nf1+/Δ4 MEFs (Fig. 1C,D). Neurofibromin negatively regulates Ras signaling by accelerating conversion of activated Ras-GTP to inactive Ras-GDP, resulting in hyperactivation of phosphorylated ERK (pERK). We therefore compared pERK levels in wild-type, Nf1Arg681*/Arg681* and Nf1Δ4/Δ4 MEFs derived from E9.5 embryos, and observed significant elevations in pERK levels in Nf1Arg681*/Arg681* and Nf1Δ4/Δ4 MEFs as compared with wild-type controls (Fig. 1E,F). Both Nf1Arg681*/Arg681* and Nf1Δ4/Δ4 genotypes produced comparable alterations in neurofibromin and pERK expression. Taken together, these data provide evidence that Nf1Arg681* functions as a null allele.
Nonsense mutations create premature termination codons (PTCs) in genes that in turn trigger nonsense-mediated mRNA decay (NMD) to selectively and rapidly degrade PTC-bearing aberrant transcripts. To determine if Nf1Arg681* mutations induce NMD, we compared levels of wild-type versus mutant mRNA in Nf1+/Arg681* mice. It is expected that in the absence of NMD, the percentage ratio of mutant to wild-type mRNA would be 50:50; however, if NMD were occurring, the distribution of mRNA would be skewed. RNA was extracted from brain tissues of Nf1+/Arg681* mice and reverse transcription polymerase chain reaction (RT-PCR) was performed using primer sets flanking the nonsense mutation. The amplicons were then cloned into a TA vector for sequencing. Of twenty-two clones sequenced, only two (9.1%) were identified with the Nf1Arg681* mutation whereas the remaining twenty clones (90.9%) contained wild-type sequences. This skewed and unequal representation of wild-type versus mutant clones (2-tailed Fisher exact; P=0.0068) indicates the occurrence of NMD of the Nf1Arg681* allele transcripts. These findings indicate that both Nf1Arg681* and the novel Nf1Δ4 alleles significantly ablate Nf1 gene expression and function.
In the appropriate genetic context, the Nf1Arg681* mutation induces neurofibroma formation in mice
As described above, individuals with NF1 harboring nonsense NF1Arg681* mutations may develop plexiform neurofibromas; therefore, we hypothesized that we could recapitulate this phenotype in Nf1Arg681* mice in the appropriate genetic context. Using conditional knockout models, it has been established that neurofibroma development can be stimulated through Nf1 loss of heterozygosity within the Schwann cell lineage when driven by Cre recombinase expression (Wu et al., 2008). To investigate whether the Nf1Arg681* and the Nf14F allele can induce neurofibroma development in vivo, we generated two conditional knockout NF1 mouse models employing a DhhCre transgenic mouse line. In this model system, Cre recombinase is expressed from desert hedgehog regulatory sequences at E12.5, with previous demonstration that inactivation of the Nf1 gene in Nf131&32F/31&32F; DhhCre mice (with exons 31 and 32 flanked by loxP sites) elicits the formation of plexiform and subcutaneous neurofibromas (Wu et al., 2008).
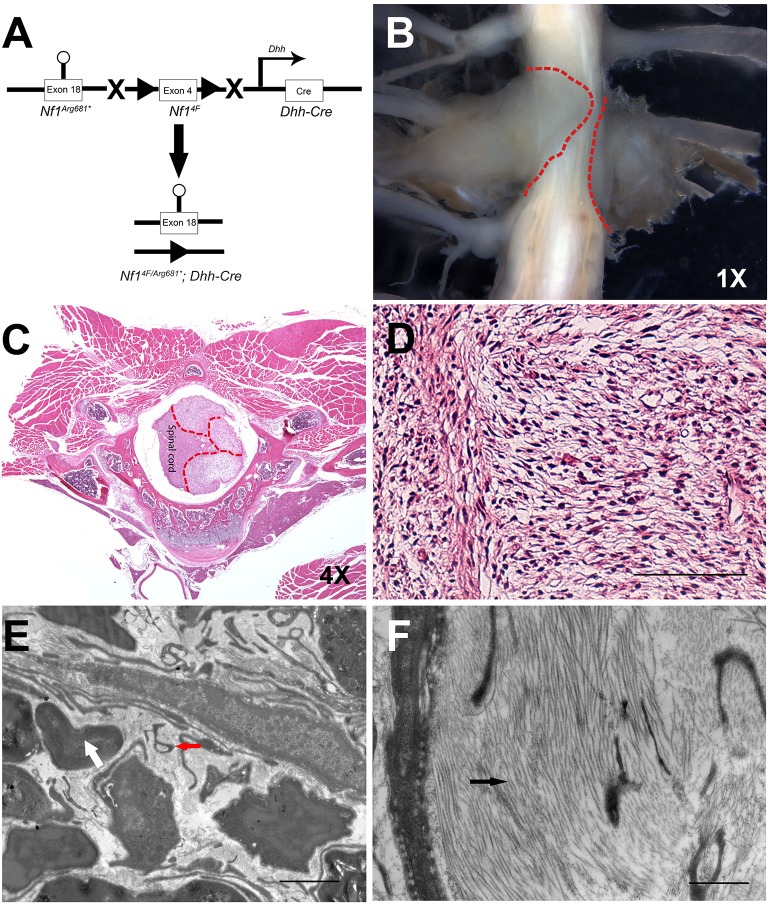
We bred the Nf1+/Arg681*, Nf14F/4F and DhhCre mice and generated Nf14F/Arg681*; DhhCre mice (Fig. 2A) as well as Nf14F/4F; DhhCre mice. Consistent with previous studies, both Nf14F/Arg681*; DhhCre and Nf14F/4F; DhhCre mice were viable and indistinguishable from their littermate controls at birth. However, we began to observe paralysis in one or both hind limbs in animals from both models at approximately four months of age. By five months of age, 55.6% (30/54) of our initial cohort of Nf14F/Arg681*; DhhCre mice exhibited paralysis, whereas 54.1% (13/24) of Nf14F/4F; DhhCre mice were paralyzed at the same age. In addition to paralysis, these mice also developed other symptoms including lethargy, weight loss, dermatitis and/or dehydration.
Fig. 2.
Nf14F/Arg681*; Dhh-cre mice develop neurofibromas along the spinal column and display disrupted ultrastructure. (A) Schematic illustration of the strategy used to breed the Nf14F/Arg681*; Dhh-Cre mice. The circle indicates the targeted mutation, triangles represent LoxP sites. (B-D) Gross dissection of spinal neurofibromas (B) and H&E staining (C) of neurofibromas along spinal cord of Nf1Arg681*/Arg681*; Dhh-Cre mice. Red dashed lines demarcate the neurofibroma boundaries. (D) A higher magnification demonstrating classic neurofibroma histology. (E,F) The aberrant ultrastructure of the neurofibromas as shown by electron microscopy of the Cre-negative control in (E) and the mutant in (F). The white arrow and red arrowhead indicate normal myelinated axons or disrupted nonmyelinating axons, respectively. The black arrowhead indicates collagen deposits. The magnifications are as indicated. Scale bars: 40 μm in D, 1 μm in E,F.
To determine the source of the paralysis, we performed gross dissection of each Nf14F/Arg681*; DhhCre or Nf14F/4F; DhhCre mouse. Tumors were observed in 81.5% (44/54) of Nf14F/Arg681*; DhhCre and 79.2% (19/24) of Nf14F/4F; DhhCre mice. All paralyzed mice were found to have visible tumors arising at the dorsal root ganglia resulting in compression of the spinal cord. Gross tumor numbers ranged from one to thirteen per mouse, with 90.9% detected in the cervical or thoracic spine and 9.1% detected in the lumbar spine (Fig. 2B, Fig. 3A). These tumors seemed to arise around the dorsal root ganglia, with invasion of the neural foramen, leading to compression of the spinal cord. In addition to these lesions, we also observed enlarged spinal nerve roots and peripheral nerves in the mice examined (data not shown). Thus, Nf14F/Arg681*; DhhCre mice develop neurofibromas with high efficiency.
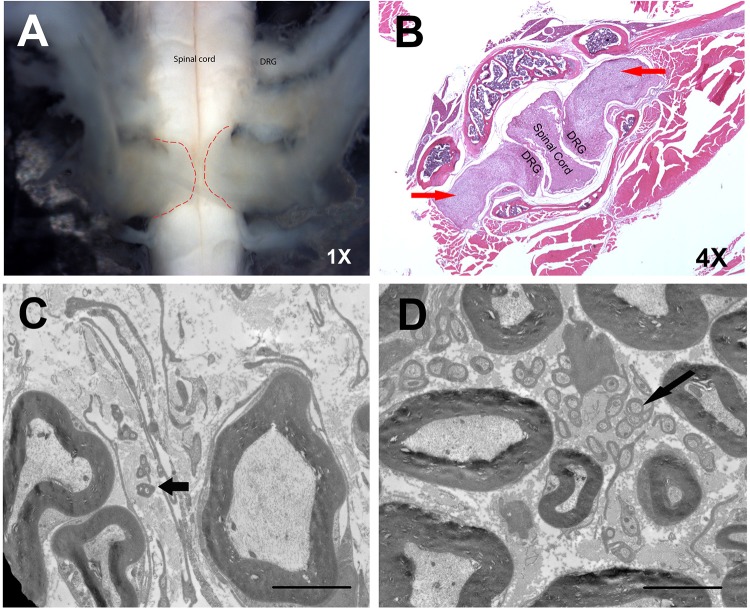
Fig. 3.
Nf14F/4F; DhhCre mice develop neurofibromas and sciatic nerve lesions. (A,B) Gross dissection (A) and H&E staining (B) of spinal tumors in Nf14F/4F; DhhCre mice. Red dashed lines demarcate the neurofibroma boundaries. Red arrows indicate neurofibromas. (C,D) The ultrastructure, as shown by electron microscopy, shows similar lesions in the Nf14F/Arg681*; DhhCre mice in both neurofibromas (C) and sciatic nerves (D). Black arrows indicate abnormal nonmyelinating axons. The magnifications are as indicated. Scale bars: 4 μm in C,D.
Upon histological analysis, neurofibromas are characterized by disorganized Remak bundles (the non-myelinated axon-Schwann cell unit) and enriched deposition of collagen. Similar lesions were seen from tumor preparations of both models at the ultrastructural level (Fig. 2C-F, Fig. 3B-D). These data indicate that Nf1Arg681* functions as a null allele, and recapitulates key characteristics of human disease in the proper genetic context.
Missense mutation NF1Gly848Arg recapitulated in the mouse retains function
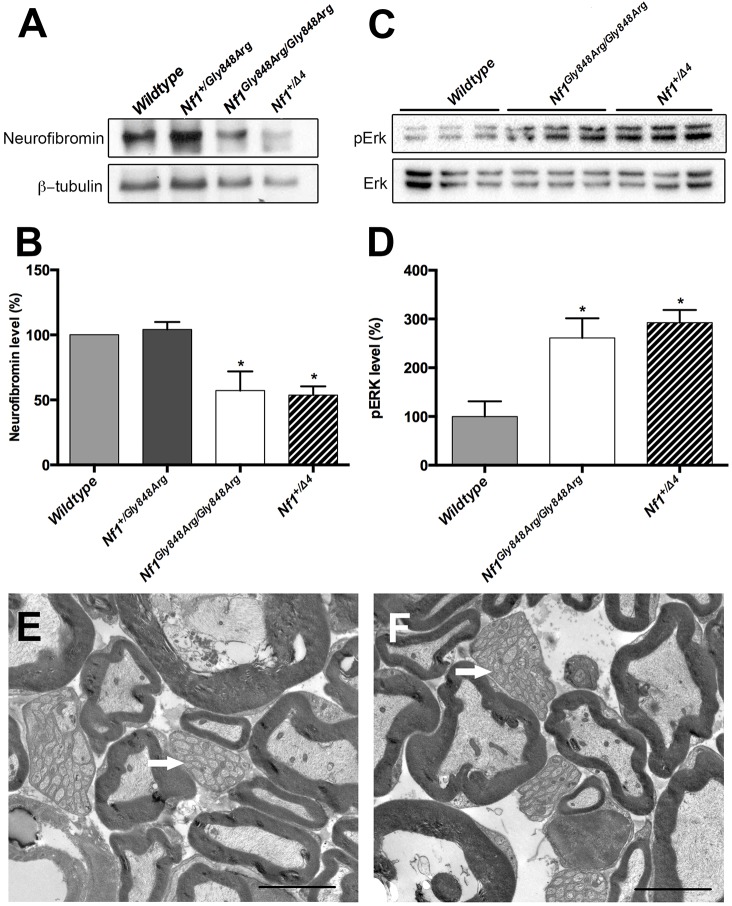
Mice homozygous for the Nf1Gly848Arg missense allele (Nf1Gly848Arg/Gly848Arg) are viable and are indistinguishable from their heterozygous (Nf1+/Gly848Arg) siblings. A cohort of 19 Nf1Gly848Arg/Gly848Arg mice were monitored throughout their lifetimes for tumor development. No overt phenotype was observed. All animals underwent gross dissection, and we were unable to detect tumors in the spinal columns or the peripheral nerves. The distinct manifestations between Nf1Gly848Arg and Nf1Arg681* might suggest different underlying Nf1 gene expression and function. We therefore derived MEFs from E13.5 embryos and assessed neurofibromin expression by western blot (Fig. 4A). The Nf1Gly848Arg allele impairs neurofibromin expression, as Nf1Gly848Arg/Gly848Arg MEFs show ∼50% less protein than cells from wild-type controls, which is similar to that observed in Nf1+/Δ4 samples (Fig. 4B). In response to neurofibromin reduction, pERK is hyperactivated in Nf1Gly848Arg/Gly848Arg MEFs to a level comparable with Nf1+/Δ4 samples, suggestive of increased Ras activity (Fig. 4C,D). Therefore, Nf1Gly848Arg only partially decreases Nf1 gene expression in mouse. Given that spinal neurofibromas form the primary manifestation of this mutation in humans, we examined the spinal nerve roots of Nf1Gly848Arg/Gly848Arg mice by electron microscopy for ultrastructural abnormalities. Neither myelinated nor non-myelinated axons were disrupted when compared with controls. (Fig. 4E,F).
Fig. 4.
The missense Nf1Gly848Arg allele impairs neurofibromin expression and function. (A-D) Western blots of neurofibromin (A) and pERK (C) in E13.5 MEFs from three independent experiments. β-tubulin is shown as loading control. The western blot analysis of neurofibromin (B) and pERK (D) was quantified and the results expressed in histograms. (E,F) Electron micrographs from spinal nerve roots of control (E) and Nf1Gly848Arg/Gly848Arg (F) mice. White arrows represent normal nonmyelinating axons. *P<0.05 vs wild type by one-way ANOVA. Data are presented as mean±s.e.m. Scale bars: 4 μm in E,F.
Individuals with NF1 carry second (somatic) mutations that are distinct from germline mutations. We hypothesized that Nf1Gly848Arg mice carrying a second null mutation in Nf1 might develop neurofibromas, which we tested by generating Nf1Δ4/Gly848Arg mice. Similar to Nf1Gly848Arg/Gly848Arg mice, these animals were viable and displayed no overt phenotype. We monitored ten Nf1Δ4/Gly848Arg mice for up to twenty-four months and still were unable to find tumors. Therefore, we conclude that the missense mutation Nf1Gly848Arg only partially impacts Nf1 gene function, and is unable to induce neurofibroma development in mice.
DISCUSSION
The factors that underlie the clinical heterogeneity seen in NF1 are still unclear. Assessment of the functional and physiological consequences of putative mutations is hampered by the lack of robust in vitro and in vivo assays, and limited knowledge of neurofibromin protein domains. In this study, two animal models were engineered to harbor NF1 mutations found in human patients, specifically nmNF1 (nonsense c.2041C>T; p.Arg681*) and a missense mutation of NF1 (missense c.2542G>C; p.Gly848Arg). This approach is possible given the greater than 98% amino acid sequence homology between the mouse and human neurofibromin protein (Bernards et al., 1993). Within the regions encoded by exons 18 and 21, the protein sequence homology is 98% and 96%, respectively (Fig. S3). The clinical features associated with these mutations in humans are distinct. Individuals with a p.Gly848Arg mutation suffer from multiple plexiform neurofibromas along the spinal cord but a paucity of dermal neurofibromas, whereas individuals with the c.2041C>T; p.Arg681* premature stop mutation also have dermal and plexiform neurofibromas but do not have multiple spinal nerve root tumors.
Examining the effects of these two different NF1 mutations in mouse models, we would like to make four points. Firstly, the two mutations have distinct effects on embryonic development and neurofibromin expression in the mouse. The Nf1Arg681* allele functions as a null allele with complete loss of Nf1 function, whereas Nf1Gly848Arg appears to retain some function. The Nf1Arg681* allele is embryonic lethal when homozygous, whereas Nf1Gly848Arg homozygous animals are viable. In accordance with embryonic lethality, complete ablation of neurofibromin expression was observed in Nf1Arg681*/Arg681* cells whereas neurofibromin expression in the Nf1Gly848Arg/Gly848Arg MEFs was reduced by ∼50%. Our data are consistent with a previous study on iPS cells and fibroblasts from individuals with NF1, which demonstrated that different germline NF1 mutations result in different degrees of neurofibromin expression (Anastasaki et al., 2015). However, in vitro analyses showed increases in pERK in MEFs from both homozygotes when compared with wild-type. This is comparable with what has been observed in neural progenitor cells from individuals with NF1, as individual NF1 mutations did not differentially affect Ras activation (Anastasaki et al., 2015). The observed changes in pERK suggest that the Nf1Gly848Arg allele could potentially function as a hypomorphic allele, though further studies would be necessary to confirm this hypothesis.
Secondly, the Nf1Gly848Arg allele failed to recapitulate the NF1 phenotype; whereas humans with NF1 suffer from multiple spinal plexiform neurofibromas, the mouse did not display an overt tumorigenic phenotype. A possible explanation for this discrepancy is the presence of modifier genes in the mouse impeding neurofibroma formation. In contrast, the Nf1Arg681* allele stimulates tumorigenesis when placed in context with a loss-of-function allele as demonstrated with the Nf14F/Arg681*; DhhCre mice. Similar observations were made in a parallel study where the same Nf1Arg681* allele in a different mouse model led to development of optic gliomas, but these tumors were not seen in Nf1Gly848Arg mice (Toonen et al., 2016).
Thirdly, the Nf14F/Arg681*; DhhCre model displays a robust tumor phenotype, with 81.5% of animals grossly developing neurofibromas throughout their lifetime. Previous studies using the Nf131&32F/31&32F; DhhCre model indicated that mice became paralyzed in one or both hind limbs in 28.6% of animals by six months of age (Wu et al., 2008); however, our data with the Nf14F/Arg681*; DhhCre model reveals an earlier onset of paralysis with tumorigenesis at 5 months of age in 55.6% of mice. These differences may either stem from the Nf1Arg681* allele, which provides a suitable environment for neurofibroma development, or from the conditional knockout of exon four versus the previously described exon 31 and 32, as the Nf14F/4F; DhhCre mice also appear to exhibit higher penetrance than the existing Nf131&32F/31&32F; DhhCre model. This hypothesis is further supported by the notable differences in embryonic lethality between the Nf1Δ4 allele, Nf1Arg681* and the previously published Nf131&32F allele. As previously reported, embryonic death of Nf1Δ31&32/Δ31&32 animals occurred from E11.5 to E13.5 as a result of defects in cardiac development (Jacks et al., 1994; Brannan et al., 1994). However, all embryos carrying the newly developed Nf1Δ4/Δ4 or Nf1Arg681*/Arg681* died around E10. Additionally, a parallel study observed that optic gliomas from mice harboring the Nf1Arg681* allele are more aggressive than those developing in mice with the traditional Nf1Δ31&32/Δ31&32 mutation, with more microglia and increased proliferative indices (Toonen et al., 2016). While it is difficult to make direct comparisons and draw conclusions across these studies because of the difference in null alleles utilized, collectively these data suggest the Nf1Δ4 and Nf1Arg681* alleles might yield a more severe phenotype.
Another possibility for the observed differences between Nf14F/4F; DhhCre and Nf131&32F/31&32F; DhhCre mice lies in the strains utilized in these studies; all mice in our experiments are on an inbred C57BL/6 background whereas previous studies used mice on a mixed 129/C57BL/6 background (Wu et al., 2008). Furthermore, Nf14F/Arg681*; DhhCre mice possess a different tumor microenvironment resulting from the nonsense mutation present in all somatic cells. Of note, the neurofibromas arising in Nf14F/Arg681*; DhhCre mice occur in a divergent location when compared with their human counterparts, as these animals develop tumors along the spinal column, whereas symptomatic spinal neurofibromas were absent in humans. This is likely due to the DhhCre driver line, which specifically promotes tumorigenesis along the spinal column per previous models (Wu et al., 2008).
Fourthly, there is a great need to develop non-surgical treatments for NF1 patients. To date, most therapeutic interventions that have been tested in preclinical models or clinical trials have been designed to regulate the downstream targets in the Ras signaling pathway or intercellular communication (Yang et al., 2008; Robertson et al., 2012; Jessen et al., 2013). Few studies have focused on restoring function of the defective NF1 alleles. The development of mutation-guided therapeutics provides new opportunities for treatment of NF1 caused by specific mutations. The robust phenotype of the nmNF1 mouse model and its parallels to human phenotype indicate that Nf14F/Arg681*; DhhCre mice will provide a valuable system for evaluating new therapies targeting the causative genetic mutation, such as nonsense suppression therapies. Nonsense mutations comprise ∼20% of mutations in individuals with NF1 (Messiaen and Wimmer, 2008). For other diseases, therapies (gentamicin, PTC124 and other compounds) have been developed that can suppress nonsense mutations to restore partial protein activity, with some now being used clinically (Du et al., 2002; Keeling et al., 2013; Sangkuhl et al., 2004; Yang et al., 2007; Guerin et al., 2008; Zilberberg et al., 2010). Assessment of nonsense suppression therapy in vivo is absolutely required prior to its assessment in individuals with nmNF1. Currently, premature stop mutation mouse models for other genes are being used to evaluate nonsense suppression therapy, including the Idua-W392X mouse model of human Hurler syndrome (mucopolysaccharidosis type I-H) and a cystic fibrosis transmembrane conductance regulator model (Zhang et al., 2014; Gunn et al., 2014; Wang et al., 2012). A fundamental question to address for any preclinical nonsense mutation model, including our newly established nmNF1 mouse, is whether sufficient protein activity can be rescued using nonsense suppression therapy approaches.
In summary, we have engineered and characterized the first NF1 patient-based germ line NF1 mutations modeled in mice, establishing that the Nf1Arg681* allele profoundly reduces neurofibromin expression and leads to plexiform neurofibroma formation. This study provides clear evidence that different mutations in NF1 can have unique consequences, and the germline mutation is an important factor underlying NF1 clinical heterogeneity. Further preclinical and translational studies of nonsense suppression therapy are underway utilizing the Nf1Arg681* allele. Future investigations of additional NF1 models harboring other human patient mutations will be used to better understand the NF1 phenotype, and will serve as translational models for therapeutic studies.
MATERIALS AND METHODS
Study approval
The patient data was collected after approval of a human subjects protocol (F080926009, Further Research on Samples Submitted for Molecular Genetic Testing) by the Institutional Review Board of the University of Alabama at Birmingham. All animal experiments were conducted after approval of an animal protocol (APN 130109835, Translational NF1 Mouse Models) by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham.
Generation of Nf1Arg681* and Nf1Gly848Arg mouse lines
We synthesized two targeting constructs that each contained a point mutation recapitulating human variant c.2041C>T; p.Arg681* (Nf1Arg681*) or c.2542G>C; p.Gly848Arg (Nf1Gly848Arg), a FRT-flanked neomycin selection cassette, and extended 5′ flanking regions homologous to intron 17 or 20, respectively (Genscript, Inc., Piscataway, NJ) (Fig. S1). The truncated versions of the two constructs were introduced into C57BL/6N-tac ES cells (PRX-B6N Primogenix, Laurie, MO) with correctly targeted clones identified by unique PCR fragments. Targeted clones were injected into blastocysts to generate chimeric founder animals that subsequently passed on the targeted alleles to offspring. The excision of the FRT-flanked neomycin cassette was carried out by breeding to a ubiquitously expressing FLP transgene mouse line [B6.129S4-Gt(Rosa)26Sortm1(FLP1), the Jackson Laboratory, #009086]. The new Nf1 alleles were designated as Nf1Arg681* and Nf1Gly848Arg. Mice harboring the ‘knockout first NF1 allele (Nf1tm1a(KOMP)Wtsi) were obtained from the Wellcome Trust Sanger Institute and bred to FLP mice [B6.129S4-Gt(Rosa)26Sortm1(FLP1), the Jackson Laboratory, #009086] to remove the FRT-flanked lacZ-Neo cassettes to produce animals with a conditional knockout allele of the NF1 gene with exon 4 flanked by loxP sites (i.e. Nf1tm1c, hereafter referred to as Nf14F).
Mouse breeding
All animals were maintained on an inbred C57BL/6 background using a temperature- and humidity-controlled vivarium on a 12 h dark-light cycle with free access to food and water. All homozygous Nf1Gly848Arg/Gly848Arg and Nf14F/4F mice were viable, fertile and did not display any detectable embryonic and postnatal phenotypes. Equal numbers of male and female animals were analyzed throughout the study. Nf1Arg681*/Arg681* and Nf1Δ4/Δ4 embryos failed to develop beyond E9.5. A DhhCre transgenic mouse line [FVB(Cg)-Tg(Dhh-cre)1Mejr/J] was kindly provided by Dr Dies Meijer (Erasmus University Medical Center, Netherlands) to breed with the Nf14F/4F and Nf1+/Arg681* strains. Cre-negative Nf14F/Arg681* or Nf14F/4F littermates were used for controls. For analysis of embryos, we established timed matings, E0.5 was designated as noon of the day a vaginal plug was detected.
MEF generation & culture
To generate MEFs, embryos were isolated in cold PBS and the heads and visceral organs were removed. Each embryo was minced into small pieces in trypsin solution and placed at 37°C for 5-10 min. The trypsin solution was then neutralized with two volumes of MEF medium (10% heat-inactivated fetal bovine serum in DMEM). After centrifugation, the supernatant was discarded and the pellet resuspended in fresh MEF medium. The cell suspensions were seeded onto gelatin-coated culture vessels, and incubated at 37°C and 5% CO2 in a culture incubator until the cells were confluent and ready for passage. These cells were designated as passage zero (P0), only those P4 or less were used in the study.
Western blot analysis
Frozen MEFs in culturing vessels or liquid nitrogen-frozen samples of adult brains were lysed in M-PER or T-PER protein extraction reagents containing protease and phosphatase inhibitors (all from Thermo Scientific, Waltham, MA). Protein samples (30 μg) were separated by reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to PVDF membrane. The membranes were then immunoblotted with indicated primary antibodies, followed by incubation with a horseradish peroxidase-coupled anti-rabbit IgG antibody (1:20,000; Jackson ImmunoResearch, West Grove, PA; 711-035-152). Protein bands were visualized with Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore, Billerica, MA). Primary antibodies used for immunoblotting were phospho ERK1/2 and ERK1/2 (1:5000; Cell Signaling Technology, Danvers, MA; 9101 and 9102), β-tubulin (1:5000; Santa Cruz Biotechnology, Santa Cruz, CA; H-235) and neurofibromin (1:500; Santa Cruz Biotechnology; sc-67). Western signal band intensity was quantified using ImageJ Software (National Institutes of Health, Bethesda, MD).
Nonsense-mediated mRNA decay assay
Total RNA was extracted from brain tissues of adult Nf1+/Arg681* mice using TRIzol reagent (Thermo Scientific). Polyadenylated RNA was reversed transcribed into cDNA in a 20 µl reaction containing 1 μg of total RNA using SuperScript® III First-Strand Synthesis kit (Thermo Scientific). Nf1 transcripts were amplified around the region of interest using 5 PRIME MasterMix (5 PRIME, Gaithersburg, MD) with 2 µl of the transcribed cDNA as the PCR template. A fragment of 417 bp was amplified with primers forward: 5′-GTTCCTGCGTGCCTGTACTC and reverse: 5′-TGTTCCCACTTTGCATGTGT. Three standard PCR steps were carried out for 30 cycles with 94°C denaturing, 55°C annealing and 72°C extension. The resulting PCR amplicons were cloned into pCR4-TOP vector from the TOP TA cloning kit (Thermo Scientific). Twenty-two individual clones were sequenced using the T7 primer.
Electron microscopy
Mice of indicated genotypes were intracardially perfused with Karnovsky's fixation solution (3% paraformaldehyde and 3% glutaraldehyde in 0.1 mol/l phosphate buffer, pH 7.4 to 7.6). Sciatic nerves and spinal nerve roots were dissected and post-fixed overnight. Tissues were then transferred to 0.175 mol/l cacodylate buffer, osmicated, dehydrated and embedded. Semi-thin sections were stained in uranyl acetate and lead citrate, and viewed on a FEI Tecnai T12 microscope.
Histological and immunological analyses
Mice were intracardially perfused with 4% paraformaldehyde (PFA, w/v) in PBS. Neurofibromas, sciatic nerves, spinal nerve roots and other tissues were dissected and post-fixed overnight, followed by cryoprotection and frozen sectioning. Serial sections of 12 μm thickness were prepared using a cryostat (CM3050 S; Leica). Hematoxylin and Eosin (H&E) staining was performed according to standard protocols.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA). Statistical comparisons were analyzed with one-way ANOVA followed by a Bonferroni post-test. A value of P<0.05 was considered statistically significant. Data are presented as mean±s.e.m.
Acknowledgements
The authors are grateful for the support of many organizations, programs, and sponsors to accomplish these studies. For assistance creating the knock-in mouse models, we would like to thank Stacey Kelpke, Jennifer Zhang, Judy Kesterson, Jinju Zhang and Larry Johnson and the UAB Transgenic & Genetically Engineered Models Systems (TGEMS) Core Facility. Services obtained from the UAB TGEMS Core Facility (RAK) are supported by the National Institutes of Health [awards P30 CA13148, P30 AR048311, P30 DK074038, P30 DK05336 and P60 DK079626]. Services obtained from the UAB CCC Small Animal Imaging Shared Facility (KRZ) are supported by the National Institutes of Health [award P30 CA13148]. We would like to acknowledge the UAB Animal Resources Program and Comparative Pathology Laboratory for their technical assistance and support of these studies. The authors thank Amber K. O'Connor, PhD. of akoWriting LLC for editorial services.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
K.L. and A.N.T. performed experiments with M.C., T.R.S. and C.A.; K.L., A.N.T. and S.N.B. wrote the main manuscript text, analyzed data and prepared all figures; M.C. and R.A.K. designed and created the animal models; K.L. and A.N.T. conducted genotyping and animal husbandry, prepared reagents and performed western blot experiments; R.A.K., D.M.B., K.R.Z., D.H.G. and B.R.K. designed experiments and interpreted data; R.A.K., L.M.M. and B.R.K. conceived the project, designed experiments and interpreted data. All authors discussed the results and implications and commented on the data at all stages.
Funding
This work was supported by the University of Alabama at Birmingham Neurofibromatosis Program, through generous philanthropic gifts and by the University of Alabama at Birmingham Department of Genetics. K.L. was supported by a Children's Tumor Foundation Young Investigator Award [grant number 203-01-029].
Supplementary information
Supplementary information available online athttp://dmm.biologists.org/lookup/doi/10.1242/dmm.025783.supplemental
References
- Aartsma-Rus A., Janson A. A. M., Kaman W. E., Bremmer-Bout M., den Dunnen J. T., Baas F., van Ommen G.-J. B. and van Deutekom J. C. T. (2003). Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum. Mol. Genet. 12, 907-914. 10.1093/hmg/ddg100 [DOI] [PubMed] [Google Scholar]
- Alkindy A., Chuzhanova N., Kini U., Cooper D. N. and Upadhyaya M. (2012). Genotype-phenotype associations in neurofibromatosis type 1 (NF1): an increased risk of tumor complications in patients with NF1 splice-site mutations? Hum. Genomics 6, 12 10.1186/1479-7364-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasaki C., Woo A. S., Messiaen L. M. and Gutmann D. H. (2015). Elucidating the impact of neurofibromatosis-1 germline mutations on neurofibromin function and dopamine-based learning. Hum. Mol. Genet. 24, 3518-3528. 10.1093/hmg/ddv103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton-Davis E. R., Cordier L., Shoturma D. I., Leland S. E. and Sweeney H. L. (1999). Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J. Clin. Invest. 104, 375-381. 10.1172/JCI7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A., Snijders A. J., Hannigan G. E., Murthy A. E. and Gusella J. F. (1993). Mouse neurofibromatosis type 1 cDNA sequence reveals high degree of conservation of both coding and non-coding mRNA segments. Hum. Mol. Genet. 2, 645-650. 10.1093/hmg/2.6.645 [DOI] [PubMed] [Google Scholar]
- Boyd K. P., Korf B. R. and Theos A. (2009). Neurofibromatosis type 1. J. Am. Acad. Dermatol. 61, 1-14; quiz 5-6 10.1016/j.jaad.2008.12.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan C. I., Perkins A. S., Vogel K. S., Ratner N., Nordlund M. L., Reid S. W., Buchberg A. M., Jenkins N. A., Parada L. F. and Copeland N. G. (1994). Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 8, 1019-1029. 10.1101/gad.8.9.1019 [DOI] [PubMed] [Google Scholar]
- De Luca A., Nico B., Rolland J.-F., Cozzoli A., Burdi R., Mangieri D., Giannuzzi V., Liantonio A., Cippone V., De Bellis M. et al. (2008). Gentamicin treatment in exercised mdx mice: identification of dystrophin-sensitive pathways and evaluation of efficacy in work-loaded dystrophic muscle. Neurobiol. Dis. 32, 243-253. 10.1016/j.nbd.2008.07.009 [DOI] [PubMed] [Google Scholar]
- Du M., Jones J. R., Lanier J., Keeling K. M., Lindsey R. J., Tousson A., Bebök Z., Whitsett J. A., Dey C. R., Colledge W. H. et al. (2002). Aminoglycoside suppression of a premature stop mutation in a Cftr-/- mouse carrying a human CFTR-G542X transgene. J. Mol. Med. 80, 595-604. 10.1007/s00109-002-0363-1 [DOI] [PubMed] [Google Scholar]
- Du M., Keeling K. M., Fan L., Liu X., Kovacs T., Sorscher E. and Bedwell D. M. (2006). Clinical doses of amikacin provide more effective suppression of the human CFTR-G542X stop mutation than gentamicin in a transgenic CF mouse model. J. Mol. Med. 84, 573-582. 10.1007/s00109-006-0045-5 [DOI] [PubMed] [Google Scholar]
- Evans D. G., Howard E., Giblin C., Clancy T., Spencer H., Huson S. M. and Lalloo F. (2010). Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am. J. Med. Genet. A 152A, 327-332. 10.1002/ajmg.a.33139 [DOI] [PubMed] [Google Scholar]
- Ferner R. E., Huson S. M., Thomas N., Moss C., Willshaw H., Evans D. G., Upadhyaya M., Towers R., Gleeson M., Steiger C. et al. (2007). Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J. Med. Genet. 44, 81-88. 10.1136/jmg.2006.045906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. M. (1999). Epidemiology of neurofibromatosis type 1. Am. J. Med. Genet. 89, 1-6. [DOI] [PubMed] [Google Scholar]
- Garcia-Linares C., Fernandez-Rodriguez J., Terribas E., Mercade J., Pros E., Benito L., Benavente Y., Capella G., Ravella A., Blanco I. et al. (2011). Dissecting loss of heterozygosity (LOH) in neurofibromatosis type 1-associated neurofibromas: importance of copy neutral LOH. Hum. Mutat. 32, 78-90. 10.1002/humu.21387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin K., Gregory-Evans C. Y., Hodges M. D., Moosajee M., Mackay D. S., Gregory-Evans K. and Flannery J. G. (2008). Systemic aminoglycoside treatment in rodent models of retinitis pigmentosa. Exp. Eye Res. 87, 197-207. 10.1016/j.exer.2008.05.016 [DOI] [PubMed] [Google Scholar]
- Gunn G., Dai Y., Du M., Belakhov V., Kandasamy J., Schoeb T. R., Baasov T., Bedwell D. M. and Keeling K. M. (2014). Long-term nonsense suppression therapy moderates MPS I-H disease progression. Mol. Genet. Metab. 111, 374-381. 10.1016/j.ymgme.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igreja S., Clarke L. A., Botelho H. M., Marques L. and Amaral M. D. (2016). Correction of a cystic fibrosis splicing mutation by antisense oligonucleotides. Hum. Mutat. 37, 209-215. 10.1002/humu.22931 [DOI] [PubMed] [Google Scholar]
- Jacks T., Shih T. S., Schmitt E. M., Bronson R. T., Bernards A. and Weinberg R. A. (1994). Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat. Genet. 7, 353-361. 10.1038/ng0794-353 [DOI] [PubMed] [Google Scholar]
- Jessen W. J., Miller S. J., Jousma E., Wu J., Rizvi T. A., Brundage M. E., Eaves D., Widemann B., Kim M.-O., Dombi E. et al. (2013). MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J. Clin. Invest. 123, 340-347. 10.1172/JCI60578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling K. M., Wang D., Dai Y., Murugesan S., Chenna B., Clark J., Belakhov V., Kandasamy J., Velu S. E., Baasov T. et al. (2013). Attenuation of nonsense-mediated mRNA decay enhances in vivo nonsense suppression. PLoS ONE 8, e60478 10.1371/journal.pone.0060478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loufrani L., Dubroca C., You D., Li Z., Levy B., Paulin D. and Henrion D. (2004). Absence of dystrophin in mice reduces NO-dependent vascular function and vascular density: total recovery after a treatment with the aminoglycoside gentamicin. Arterioscler. Thromb. Vasc. Biol. 24, 671-676. 10.1161/01.ATV.0000118683.99628.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q. L., Rabinowitz A., Chen Y. C., Yokota T., Yin H., Alter J., Jadoon A., Bou-Gharios G. and Partridge T. (2005). Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc. Natl. Acad. Sci. USA 102, 198-203. 10.1073/pnas.0406700102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann C. J., Honeyman K., Cheng A. J., Ly T., Lloyd F., Fletcher S., Morgan J. E., Partridge T. A. and Wilton S. D. (2001). Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc. Natl. Acad. Sci. USA 98, 42-47. 10.1073/pnas.011408598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messiaen L. and Wimmer K. (2008). NF1 mutational spectrum. In Monographs in Human Genetics: Neurofibromatoses (ed. Kaufmann D.), pp. 63-77. Basel: Karger. [Google Scholar]
- Pasmant E., Vidaud M., Vidaud D. and Wolkenstein P. (2012). Neurofibromatosis type 1: from genotype to phenotype. J. Med. Genet. 49, 483-489. 10.1136/jmedgenet-2012-100978 [DOI] [PubMed] [Google Scholar]
- Robertson K. A., Nalepa G., Yang F.-C., Bowers D. C., Ho C. Y., Hutchins G. D., Croop J. M., Vik T. A., Denne S. C., Parada L. F. et al. (2012). Imatinib mesylate for plexiform neurofibromas in patients with neurofibromatosis type 1: a phase 2 trial. Lancet Oncol. 13, 1218-1224. 10.1016/S1470-2045(12)70414-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagh A., Pasmant E., Laurendeau I., Parfait B., Barbarot S., Guillot B., Combemale P., Ferkal S., Vidaud M., Aubourg P. et al. ; members of the NFFN. (2009). Unravelling the genetic basis of variable clinical expression in neurofibromatosis 1. Hum. Mol. Genet. 18, 2768-2778. 10.1093/hmg/ddp212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangkuhl K., Schulz A., Rompler H., Yun J., Wess J. and Schoneberg T. (2004). Aminoglycoside-mediated rescue of a disease-causing nonsense mutation in the V2 vasopressin receptor gene in vitro and in vivo. Hum. Mol. Genet. 13, 893-903. 10.1093/hmg/ddh105 [DOI] [PubMed] [Google Scholar]
- Steinmann K., Kluwe L., Friedrich R. E., Mautner V.-F., Cooper D. N. and Kehrer-Sawatzki H. (2009). Mechanisms of loss of heterozygosity in neurofibromatosis type 1-associated plexiform neurofibromas. J. Invest. Dermatol. 129, 615-621. 10.1038/jid.2008.274 [DOI] [PubMed] [Google Scholar]
- Toonen J. A., Anastasaki C., Smithson L. J., Gianino S. M., Li K., Kesterson R. A. and Gutmann D. H. (2016). NF1 germline mutation differentially dictates optic glioma formation and growth in neurofibromatosis-1. Hum. Mol. Genet. [Epub ahead of print] doi:10.1093/hmg/ddw039. 10.1093/hmg/ddw039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom J. C. T., Bremmer-Bout M., Janson A. A. M., Ginjaar I. B., Baas F., den Dunnen J. T. and van Ommen G.-J. B. (2001). Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum. Mol. Genet. 10, 1547-1554. 10.1093/hmg/10.15.1547 [DOI] [PubMed] [Google Scholar]
- Wang D., Belakhov V., Kandasamy J., Baasov T., Li S.-C., Li Y.-T., Bedwell D. M. and Keeling K. M. (2012). The designer aminoglycoside NB84 significantly reduces glycosaminoglycan accumulation associated with MPS I-H in the Idua-W392X mouse. Mol. Genet. Metab. 105, 116-125. 10.1016/j.ymgme.2011.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Williams J. P., Rizvi T. A., Kordich J. J., Witte D., Meijer D., Stemmer-Rachamimov A. O., Cancelas J. A. and Ratner N. (2008). Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell 13, 105-116. 10.1016/j.ccr.2007.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Mutyam V., Tang L., Biswas S., Du M., Jackson L. A., Dai Y., Belakhov V., Shalev M., Chen F. et al. (2014). Synthetic aminoglycosides efficiently suppress cystic fibrosis transmembrane conductance regulator nonsense mutations and are enhanced by ivacaftor. Am. J. Respir. Cell Mol. Biol. 50, 805-816. 10.1165/rcmb.2013-0282OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Feng J., Song W., Wang J., Tsai B., Zhang Y., Scaringe W. A., Hill K. A., Margaritis P., High K. A. et al. (2007). A mouse model for nonsense mutation bypass therapy shows a dramatic multiday response to geneticin. Proc. Natl. Acad. Sci. USA 104, 15394-15399. 10.1073/pnas.0610878104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F.-C., Ingram D. A., Chen S., Zhu Y., Yuan J., Li X., Yang X., Knowles S., Horn W., Li Y. et al. (2008). Nf1-dependent tumors require a microenvironment containing Nf1+/−- and c-kit-dependent bone marrow. Cell 135, 437-448. 10.1016/j.cell.2008.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Ranganath N. K., Skinner D., Bedwell D. M., Buckley-Lanier J. A., Sorscher E. J. and Woodworth B. A. (2014). Marked repression of CFTR mRNA in the transgenic Cftr(tm1kth) mouse model. J. Cyst. Fibrosis 13, 351-352. 10.1016/j.jcf.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Ghosh P., Charnay P., Burns D. K. and Parada L. F. (2002). Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science 296, 920-922. 10.1126/science.1068452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberberg A., Lahav L. and Rosin-Arbesfeld R. (2010). Restoration of APC gene function in colorectal cancer cells by aminoglycoside- and macrolide-induced read-through of premature termination codons. Gut 59, 496-507. 10.1136/gut.2008.169805 [DOI] [PubMed] [Google Scholar]