Abstract
The protocadherins Fat4 and Dchs1 act as a receptor-ligand pair to regulate many developmental processes in mice and humans, including development of the vertebrae. Based on conservation of function between Drosophila and mammals, Fat4-Dchs1 signalling has been proposed to regulate planar cell polarity (PCP) and activity of the Hippo effectors Yap and Taz, which regulate cell proliferation, survival and differentiation. There is strong evidence for Fat regulation of PCP in mammals but the link with the Hippo pathway is unclear. In Fat4−/− and Dchs1−/− mice, many vertebrae are split along the midline and fused across the anterior-posterior axis, suggesting that these defects might arise due to altered cell polarity and/or changes in cell proliferation/differentiation. We show that the somite and sclerotome are specified appropriately, the transcriptional network that drives early chondrogenesis is intact, and that cell polarity within the sclerotome is unperturbed. We find that the key defect in Fat4 and Dchs1 mutant mice is decreased proliferation in the early sclerotome. This results in fewer chondrogenic cells within the developing vertebral body, which fail to condense appropriately along the midline. Analysis of Fat4;Yap and Fat4;Taz double mutants, and expression of their transcriptional target Ctgf, indicates that Fat4-Dchs1 regulates vertebral development independently of Yap and Taz. Thus, we have identified a new pathway crucial for the development of the vertebrae and our data indicate that novel mechanisms of Fat4-Dchs1 signalling have evolved to control cell proliferation within the developing vertebrae.
KEY WORDS: Fat4, Dchs1, Vertebrae, Somite, Chondrocyte, Yap1, Taz (Wwtr1), Mouse
Summary: The protocadherins Fat4 and Dchs1 promote proliferation in the developing mouse somites, independently of both the planar cell polarity pathway and the Yap/Taz Hippo effectors.
INTRODUCTION
Fat4 and Dchs1 are protocadherins that act as a receptor-ligand pair to play key roles during embryonic development. Gene inactivation of either Fat4 or Dchs1 in mice results in a wide spectrum of phenotypes ranging from branching and cystic defects in the kidney, altered neuronal proliferation and migration, to a change in sternum shape (Saburi et al., 2008, 2012; Mao et al., 2011, 2015, 2016; Zakaria et al., 2014; Bagherie-Lachidan et al., 2015; Badouel et al., 2015). FAT4 and DCHS1 are also essential in humans, and compound mutations result in Van Maldergem syndrome, which is characterised, in part, by intellectual disability and altered craniofacial development (Cappello et al., 2013); in some individuals vertebral defects have also been reported (Mansour et al., 2012). Mutation in FAT4 has also been shown to be responsible in a subset of Hennekam syndrome patients (Alders et al., 2014). The prime feature of Hennekam syndrome is congenital lymphedema and lymphangiectasia, but some individuals also have features overlapping with Van Maldergem syndrome.
Fat4 and Dchs1 are the vertebrate orthologs of Drosophila Fat and Dachsous (Ds), respectively, which, together with the novel Golgi kinase Four-jointed (Fj), regulate planar cell polarity (PCP) and growth via the Hippo pathway (Staley and Irvine, 2012; Matis and Axelrod, 2013). PCP is classically described as the coordinated cell polarity within an epithelium, but in vertebrates has also been shown to control cellular rearrangements and behaviours within mesenchymal tissues (Goodrich and Strutt, 2011). The Hippo pathway includes the Hippo (Mst1/2, or Stk4/3) and Warts (Lats1/2) kinases, which act through the transcriptional co-factor Yorkie (Yap/Taz) (Yu and Guan, 2013). In Drosophila, when Fat signalling is active, the transcriptional co-factor Yorkie is phosphorylated and retained in the cytoplasm where it is inactive. In the absence of Fat, Yorkie is dephosphorylated and moves to the nucleus, where it binds to the transcription factor Scalloped to promote cell proliferation and survival. Ds can also directly regulate Hippo signalling, but in this case Fat-to-Ds signalling inhibits the Hippo kinase, resulting in the activation of Yorkie (Degoutin et al., 2013).
There is evidence that Fat-Ds regulation of both the Hippo and PCP pathways is conserved in vertebrates. Fat regulation of PCP has been clearly demonstrated in the kidney, hindbrain and sternum. Fat4−/− mutants show defects in orientated cell divisions in the extending kidney tubules (Saburi et al., 2008), and Fat4 and Dchs1 are essential for the collective neuronal polarity and migration of facial branchiomotor neurons within the hindbrain (Zakaria et al., 2014). Fat-PCP also controls cell intercalation movements during the early development of the sternum (Mao et al., 2016).
In mammals, Fat4 regulation of the Hippo pathway has been implicated in the cerebral cortex and kidney (Cappello et al., 2013; Das et al., 2013). In the cerebral cortex, loss of Fat4 and Dchs1 is associated with increased neuronal proliferation and increased nuclear Yap/Taz activity (Cappello et al., 2013). Fat4 regulation of Yap intracellular localisation within the pre-nephrogenic precursors has also been observed (Das et al., 2013). However, other studies have reported that Fat4 does not regulate Yap localisation and activity in the kidney (Mao et al., 2015; Bagherie-Lachidan et al., 2015).
We have previously reported that in Fat4 and Dchs1 mouse mutants the lumbar vertebrae are malformed, which may be consistent with the vertebral defects reported in humans (Mao et al., 2011; Mansour et al., 2012). The vertebrae in P0 Fat4 and Dchs1 mutants are also wider, raising the possibility that Fat-PCP alters the relative dimensions of the vertebrae during early development. Vertebrae, together with axial muscles and tendons, arise from the epithelial somites, which are produced sequentially and regularly from the pre-somitic mesoderm (Hubaud and Pourquie, 2014). In response to Shh and Bmp antagonists from the adjacent tissues, the ventral somite undergoes an epithelial-to-mesenchymal transformation to form the sclerotome (Fan and Tessier-Lavigne, 1994; Johnson et al., 1994; Hirsinger et al., 1997; McMahon et al., 1998; Stafford et al., 2011). The sclerotomal cells express Sox9, a marker of chondrogenic commitment, and subsequently migrate to surround the dorsal neural tube and notochord, forming the neural arches and the vertebral body of the vertebrae, respectively.
Vertebral development requires the precise integration of several signalling cascades, including Notch, FGFs, Shh, Wnt and Bmps (Murtaugh et al., 1999; Maroto et al., 2012; Hubaud and Pourquie, 2014; Wahi et al., 2016). Here, we determine the role of Fat4 and Dchs1 signalling during vertebral development and whether these effects are mediated through alterations in the Hippo pathway or PCP. We show a crucial role for Fat4-Dchs1 signalling during the regulation of sclerotome proliferation that is independent of Yap and Taz.
RESULTS
Fat4 and Dchs1 control vertebral development
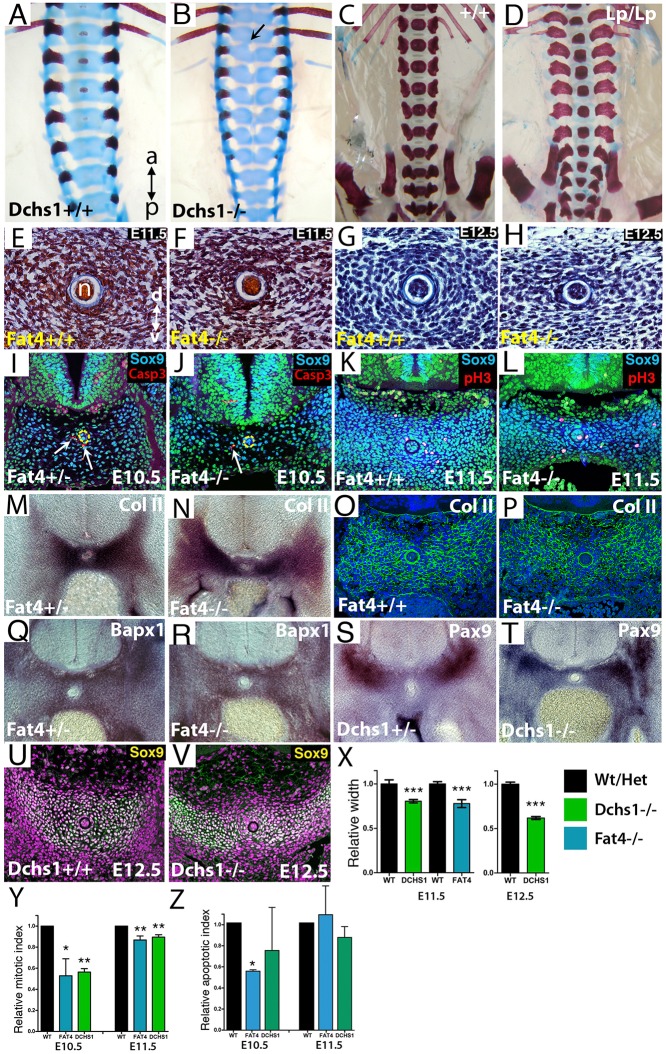
Our previous analysis revealed that development of the thoracic and lumbar vertebrae is affected in Dchs1 and Fat4 mouse mutants (Mao et al., 2011). There is a clear split in the midline of lumbar and posterior thoracic vertebrae and some vertebrae are also fused along the anterior-posterior axis (Fig. 1A-C) (Mao et al., 2011). To fully understand the severity of the defects and how they occur, we first examined each of the vertebrae in P0 wild type and Dchs1−/− and Fat4−/− mutants. In thoracic vertebrae T6-13 and lumbar vertebrae L1-L5 the vertebral body is wider and flatter, typically with dual ossification centres. Patterning and identity of the vertebrae are not affected and the lamina and transverse processes are present, although the spinous process of T8 is bifid (Fig. 1). The more rostral vertebrae are normal. No vertebral defects were observed in heterozygous mice.
Fig. 1.
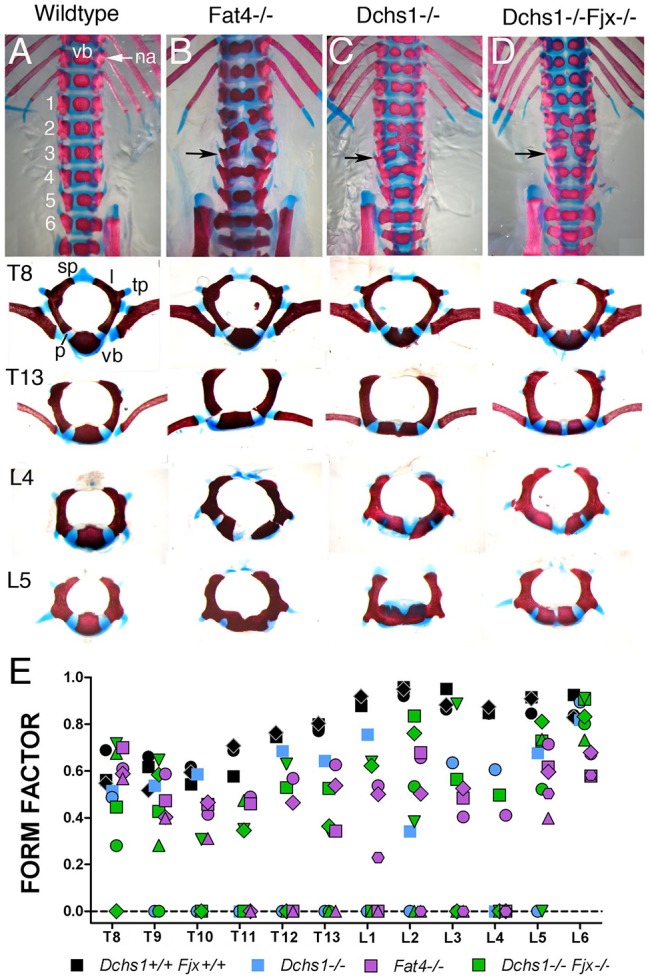
Characterisation of vertebral defects in Fat4 and Dchs1 mouse mutants. (A-D) Ventral view of the lumbar and posterior thoracic vertebrae of P0 skeletons of the indicated genotypes stained with Alcian Blue and Alizarin Red. Lumbar vertebrae are numbered. The panels beneath show individual vertebrae: T, thoracic; L, lumbar. (E) Form factors of individual vertebrae in the wild type and mutants. Each symbol indicates an individual mouse. The lower the form factor the more the shape of the ossification centre within the vertebra deviates from a perfect circle. A split vertebra, i.e. a vertebra with dual ossification centres, has a score of 0. na, neural arch; l, lamina; p, pedicle; tp, transverse process; sp, spinous process; vb, vertebral body.
To analyse the severity of the defects in each vertebra, and if particular vertebrae are more affected than others, we determined whether they were split and/or fused across the anterior-posterior axis. Vertebrae were also imaged from the dorsal side, and the shape (form factor) of the ossification centres in wild type, Fat4−/− and Dchs1−/− mutants was compared. Form factor is calculated as 4×π×area/perimeter2 and equals 1 for a circle. The split vertebrae were given a score of 0. We found that the pattern and severity of the defects in particular vertebrae varied between individual mutants, but L2-L4 were the most frequently affected, i.e. split across the midline and/or fused across the anterior-posterior axis (Fig. 1E, Table S1). The range and severity of defects within the lumbar vertebrae are very similar in Fat4−/− and Dchs1−/− mutants indicating that, here, they operate as a receptor-ligand pair (Fig. 1A-C,E, Table S1).
In Drosophila, Fat, Ds and Fj operate as a signalling cassette, and in fat mutants Fj expression is increased (Yang et al., 2002). Likewise, expression of Fjx1, the mammalian Fj homologue, is increased in the kidneys of Fat4 mutants (Saburi et al., 2008). Fjx1 is expressed in the developing vertebrae in regions of Fat4 and Dchs1 expression (Fig. 2A-F) (Rock et al., 2005), raising the possibility that increased levels of Fjx1 might contribute to the vertebral phenotype in Dchs1−/− and Fat4−/− mutants. To examine this, we analysed vertebral development in Dchs1−/−;Fjx1−/− double mutants. We found that simultaneous loss of Fjx1 neither rescues the vertebral abnormalities nor enhances their severity in Dchs1−/− mutants (Fig. 1D,E; n=3), consistent with an independent study that compared Fat4−/− and Fat4−/−;Fjx1−/− mutants (Saburi et al., 2012).
Fig. 2.
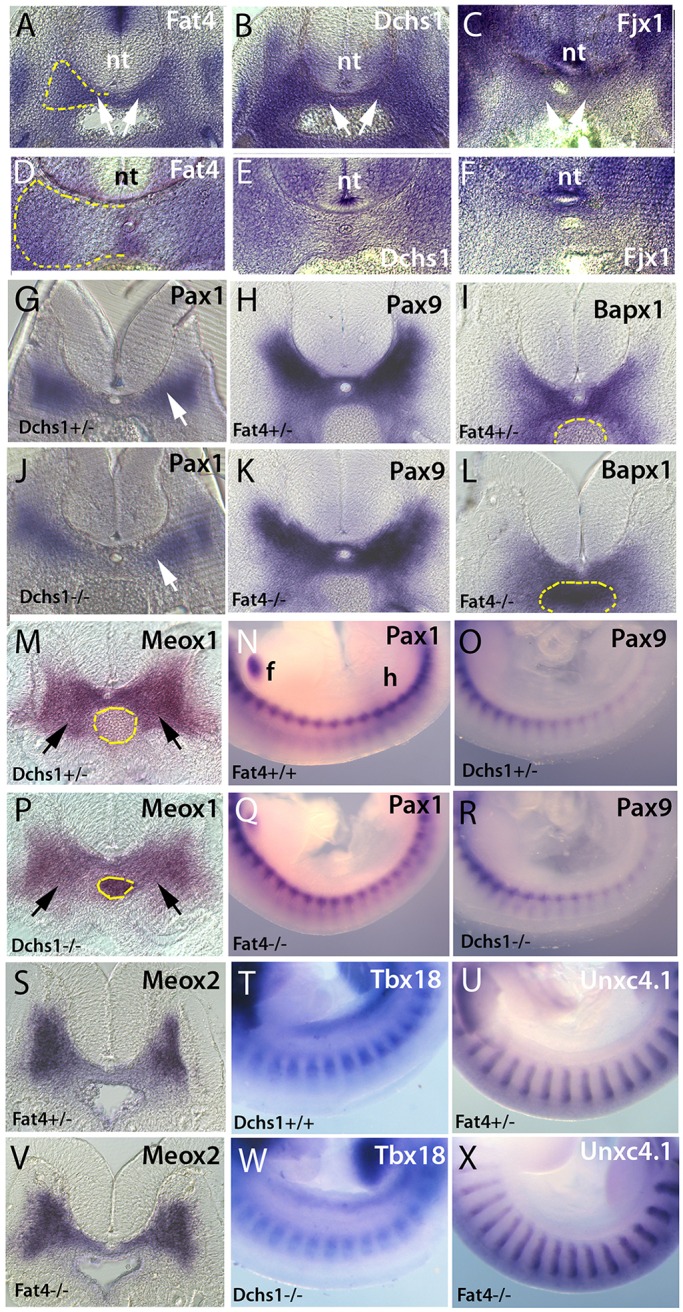
Sclerotome specification is unaffected in Fat4 and Dchs1 mutants. In situ hybridisation showing expression of Fat4 (A,D), Dchs1 (B,E) and Fjx1 (C,F) in the E10.5 (A-C) and E11.5 (D-F) wild-type sclerotome (arrows, A-C). (G-X) Transverse sections (G-L,M,P,S,V) and lateral views (N,O,Q,R,T,U,W,X) of in situ hybridisations of E10.5 wild-type/heterozygous (G-I,M-O,S-U), Dchs1−/− (J,P,R,W) and Fat4−/− (K,L,Q,V,X) embryos showing expression of the indicated genes. Arrows (G,J,M,P) indicate the developing sclerotome. Dashed yellow lines outline one half of the developing vertebral body to show the region of the sclerotome analysed for cell proliferation and apoptosis (A,D) or the aorta (I,L,M,P) to show the sclerotomal expression more clearly. In N, the forelimb (f) and hindlimb (h) have been labelled to indicate the orientation of the embryos in lateral views. nt, neural tube.
The sclerotome is specified appropriately in Fat4 and Dchs1 mutants
The sclerotome is specified at E9.5 and gives rise to the anlagen of the vertebral body by E12.5. To understand potential mechanisms, and when Fat4 and Dchs1 may control development of the vertebrae, we analysed the expression of Fat4 and Dchs1 between E10.5 and E13.5 by in situ hybridisation to whole-mounts and tissue sections. We have previously shown that Fat4 and Dchs1 are expressed in the early sclerotome at E9.5 (Mao et al., 2011). We found that Fat4 and Dchs1 are still present throughout the sclerotome at E10.5 (Fig. 2A,B), but by E11.5 they are expressed at higher levels along the dorsoventral midline and the lateral edges of the developing vertebral body (Fig. 2D,E). By E13.5, Fat4 and Dchs1 are expressed in the cells around the differentiating vertebral chondrocytes (data not shown) (Rock et al., 2005). These expression data indicate that the vertebral defect most likely arises during the early steps of sclerotome development and morphogenesis between E9.5 and E12.5.
To determine if the vertebral defects are due to an alteration in factors that are crucial for sclerotome development, we carried out whole-mount in situ hybridisation of E10.5 and/or E11.5 embryos. Specifically, we analysed the expression of the transcription factors Pax1, Pax9, Bapx1 (Nkx3-2), Meox1 and Meox2. We also determined the expression of Uncx4.1 (Uncx) and Tbx18, which specify the posterior and anterior regions of the developing sclerotome, respectively (Mansouri et al., 2000; Bussen et al., 2004). All these genes were found to be expressed in their normal domains (Fig. 2G-X, Fig. 3Q-T, Fig. S1). The data indicate that the somites and sclerotome are specified appropriately, and that the sclerotome is not obviously smaller at E10.5. The data also rule out the possibility that potential alterations in the segmentation clock contribute to these defects.
Fig. 3.
Fat4 and Dchs1 are required for establishment of the early vertebrae. (A-D) Dorsal views of Alcian Blue and Alizarin Red stained E15.5 (A,B) and E16.5 (C,D) wild-type (A,C), Dchs1−/− (B) and Vangl2Lp/Lp (D) embryos. A fused vertebra along the anterior-posterior axis is arrowed in B. (E-H) H&E (E,F) or Mallory's triple (G,H) stained transverse tissue sections through the developing vertebrae of E11.5 (E,F) and E12.5 (G,H) wild type (E,G) and Fat4−/− mutants (F,H). (I-L,U,V) Immunostaining of Sox9 (blue in I-L, yellow/white in U,V) and active caspase 3 (red in I,J) or phospho-histone H3 (pH3) (red/pink in K,L) of E10.5 (I,J), E11.5 (K,L) and E12.5 (U,V) wild-type (I,K,U), Fat4−/− (J,L) and Dchs1−/− (V) developing lumbar vertebral bodies. Arrows (I,J) indicate apoptotic cells; the notochord is outlined by a yellow dashed line. (M,N,Q-T) In situ hybridisations showing the expression of collagen type II (M,N), Bapx1 (Q,R) and Pax9 (S,T) in the developing vertebral body of heterozygous (M,Q,S), Fat4−/− (N,R) and Dchs1−/− (T) E11.5 embryos. (E-T) Dorsal is uppermost. (O,P) Immunostaining of collagen type II expression in wild-type (O) and Fat4−/− (P) E11.5 embryos. (X) The relative width across the dorsoventral axis at E11.5 and E12.5 of the developing lumbar vertebral body of Fat4−/− and Dchs1−/− embryos as compared with wild-type/heterozygous littermates. (Y) The relative percentage of proliferating cells as determined by pH3 immunostaining at E10.5 and E11.5; the percentage proliferation in wild-type/heterozygous embryos is standardised to 100%. (Z) The relative percentage of apoptotic cells as determined by immunostaining for active caspase 3 at E10.5 and E11.5; the percentage apoptosis in wild-type/heterozygous embryos is standardised to 100%. Mean plus s.d. *P<0.05, **P<0.005, ***P<0.001. a, anterior; d, dorsal; n, notochord; p, posterior; v, ventral.
To determine if there are more subtle changes in the expression of these somitic markers, and to quantify their expression, we carried out qPCR of E10.5 lumbar and posterior thoracic somites. Here, as a control, we also included Pax3, which is essential for the development of the myogenic lineage (Tajbakhsh et al., 1997). We found no significant change in the expression levels of these markers (Fig. S2). These data again indicate that dysregulation of these transcription factors is unlikely to contribute to the vertebral defects and also indicate that the transcriptional network between Bapx1, Pax1 and Pax9, which drives chondrocyte commitment, is intact (Fig. S2).
Early morphogenesis of the vertebrae is affected in Fat4 and Dchs1 mutants
The above analyses showed that early sclerotome development occurs normally in Fat4 and Dchs1 mouse mutants. To determine when the midline defect arises, we made skeletal preparations and performed histological and immunolocalisation analyses at E10.5-E14.5. Histological analysis showed that in Dchs1−/− and Fat4−/− embryos the sclerotomal cells had started to condense around the notochord at E11.5, but by E12.5 the sclerotomal cells appeared to be more disorganised (Fig. 3E-H). The vertebrae were clearly split by E13.5 (data not shown; Fat4−/−, n=3) and by E14.5 some vertebrae were fused along the anterior-posterior axis (Fig. 3A,B for E15.5; data not shown; Dchs1−/−, n=2; Fat4−/−, n=7).
To determine if the sclerotomal cells differentiate appropriately, we analysed the expression of Sox9, which marks chondrogenic precursors and committed chondrocytes (Lefebvre and Bhattaram, 2010), by immunolocalisation between E10.5 and E12.5. This revealed the presence of Sox9-expressing cells throughout the sclerotome at E10.5 (Fig. 3I,J) and E11.5 (Fig. 3K,L), but by E12.5 a subset of cells at the midline did not express Sox9 in Fat4 and Dchs1 mutants (Fig. 3U,V; data not shown). The developing sclerotome was also found to be significantly narrower along the dorsoventral axis of the developing vertebral body at E11.5 and E12.5 (Fig. 3U,V,X; P<0.001). We also analysed the expression of collagen type II, a direct target of Sox9 (Lefebvre and Bhattaram, 2010), by in situ hybridisation (E10.5 and E11.5) and immunolocalisation (E11.5) to determine potential alterations in early chondrocyte differentiation. We found that collagen type II is expressed normally (Fig. 3M-P; E10.5, Fat4−/−, n=2; E11.5, Fat4−/−, n=6; E11.5, Dchs1−/−, n=3). As the vertebrae are fused across the anterior-posterior axis, we also analysed the development of the intervertebral disc. Histological analysis of E13 embryos showed that the intervertebral discs are initially formed appropriately (Fig. S3; n=6).
The neural tube is wider in P0 Fat4 and Dchs1 mutants (Saburi et al., 2008; Mao et al., 2011). To rule out the possibility that altered neural tube development contributes to the development of the hemivertebrae, we analysed skeletal preparations of E16.5 Vangl2Lp/Lp mice. The Vangl2Lp/Lp mutant is characterised by a wide neural tube with a broad floor plate and also has vertebral abnormalities (Greene et al., 1998; Stein and Mackensen, 1957). Consistent with previous analysis, we found that the lumbar vertebral bodies are not split, although a few split anterior thoracic vertebrae that are also fused along the anterior-posterior axis were observed (Fig. 3D; data not shown) (see also Greene et al., 1998; Stein and Mackensen, 1957).
To confirm that Fat4-Dchs1 signalling acts intrinsically within the mesoderm and that the vertebral defects are not secondary to altered neural tube development, we generated Dermo1CreFat4f/f mutants to inactivate expression within the mesoderm. We found that the Dermo1CreFat4f/f mice are also characterised by hemivertebrae, demonstrating an intrinsic requirement for Fat4 and Dchs1 within the mesoderm (Fig. S4).
Cell polarity is unaffected in Fat4 and Dchs1 mutants
The previous analyses indicated that the sclerotome is specified appropriately but that it is narrower along the dorsoventral axis of the developing vertebral body at E11.5. This might be due to alterations in cell migration/rearrangements, survival or proliferation. As Fat-Ds controls polarised cell arrangements in Drosophila and mammals, and vertebral defects are also found in Vangl2Lp/Lp mutants, we next addressed the possibility that Fat4 and Dchs1 might control polarised cell behaviours during vertebra formation.
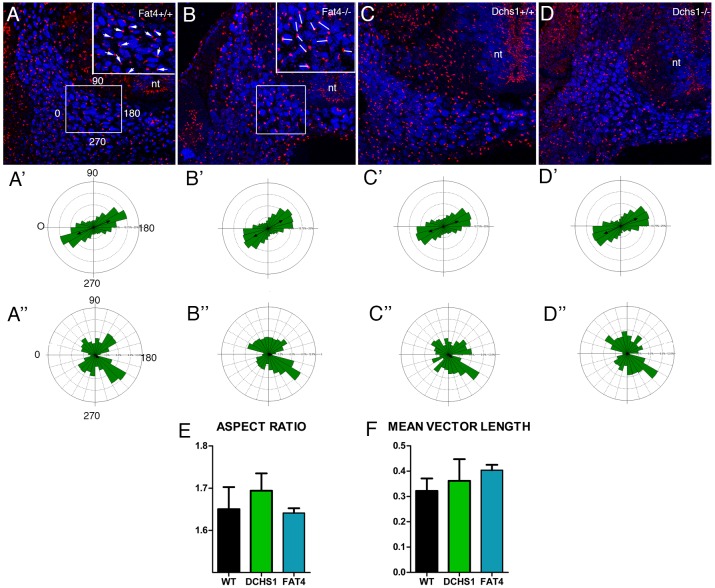
Cell orientation and polarity were determined at E10.5, a time point when sclerotomal cells are starting to surround the notochord and just prior to the formation of the narrower sclerotome at E11.5. First we determined the orientation of the long axis of the nucleus, the shape of the nuclei and the collective organisation of cells within the sclerotome. Second, the angle of the Golgi complex relative to the mediolateral axis of the embryo was measured.
Analysis of cell orientation indicated that in wild-type embryos (n=7) the sclerotomal cells are collectively polarised (Fig. 4A,A′,C,C′; P<0.001). The sclerotomal cells are also polarised in Fat4−/− (n=4; P=0.031) and Dchs1−/− (n=4; P=0.00003) embryos and there is no significant difference in the degree of polarisation compared with wild-type embryos (Mardia-Whatson-Wheeler test; P=0.245 for Fat4−/− versus wild type and P=0.284 for Dchs1−/− versus wild type). Furthermore, the length of the mean vector, which indicates how closely the observations are clustered around the mean value of the angle, was analysed. We again found no differences in the overall organisation and alignment of nuclei between wild-type, Fat4−/− (P=0.0954) and Dchs1−/− (P=0.4507) embryos (Fig. 4F). We also examined the relative shape and elongation of nuclei, which is presented as a ratio of the longer to shorter axis of the nucleus (the aspect ratio). The aspect ratio averaged 1.65 for wild-type, 1.64 for Fat4−/− and 1.69 for Dchs1−/− embryos and, therefore, no differences in cell shape (as indicated by nuclear shape) were identified (Fig. 4E).
Fig. 4.
Cell polarity analysis within the developing sclerotome. (A-D) Transverse sections through E10.5 wild-type (A,C), Fat4−/− (B) and Dchs1−/− (D) developing lumbar vertebrae showing immunostaining of Sox9 (blue) and the Golgi (red). Boxes (A,B) indicate the region of the sclerotome analysed, as shown at higher magnification in the insets. Arrows (A) indicate the angle of the Golgi complex. Lines (B) show the direction of the long axis of the nuclei. (A′-D′) Rose plots of nuclear alignment, where 0-180° is the alignment across the mediolateral axis of the embryo. (A″-D″) Orientation of the Golgi complex in the sclerotome relative to the mediolateral axis of the embryo. (E) The relative elongation of nuclei within the sclerotome, presented as a ratio of the longer to shorter axis of the nucleus (the aspect ratio). (F) The length of the mean vector, which indicates how closely the observations are clustered around the mean value of the angle. WT, wild type; DCHS1, Dchs1−/−; FAT4, Fat4−/−. Mean+s.e.m.
Analysis of the position of the Golgi apparatus revealed far more variation and less coordinated polarity compared with the orientation of the nuclei. However, there was significant bias and the Rayleigh uniformity test reported P<0.05 for both wild-type and mutant embryos. This indicates that although the distribution of angles is not uniform and there is a large variation, a clear bias can be observed. Again no difference in the bias/overall polarity of the Golgi apparatus was detected between wild-type, Fat4−/− (P=0.245) and Dchs1−/− (P=0.284) embryos (Fig. 4A″-D″).
Fat4 and Dchs1 regulate proliferation within the developing sclerotome
The above studies showed that the developing vertebral body is thinner across the dorsoventral axis at E11.5 and splits by E13.5, but there are no alterations in cell polarity. To examine if changes in cell mitosis and apoptosis contribute to the vertebral defects, we carried out immunolocalisation of phospho-histone H3 (pH3) and activated caspase 3. In E10.5 heterozygous/wild-type controls, pH3 staining ranged between 1.2% and 3.1% (n=10), whereas in the Fat4−/− and Dchs1−/− mutants this proliferation marker ranged between 0.76% and 2.67% (Dchs1−/−, n=5; Fat4−/−, n=3). Proliferation was consistently decreased in the mutant compared with heterozygous/wild-type littermates. This decrease was significant at E10.5 and E11.5 in both Fat4−/− and Dchs1−/− mutants when compared with the control embryos (Fig. 3Y; E10.5, Dchs1, n=5, P<0.005; E10.5, Fat4, n=3, P<0.01; E11.5, Dchs1, n=4, P<0.005; E11.5, Fat4, n=6, P<0.005).
A significant reduction in the number of apoptotic cells was also seen in Fat4−/−, but not Dchs1−/−, mutants when compared with wild-type/heterozygous littermates (Fig. 3Z; P<0.01). The percentage of apoptotic cells ranged between 1.2% and 2.7% in control embryos and between 0.69% and 2.15% in mutant embryos at E10.5 (Fig. 3Z). At E11.5, the levels of apoptosis were very similar between wild-type and mutant embryos, averaging 1.22% for control embryos versus 1.06% for Dchs1 mutants and 0.68% for controls versus 0.73% for Fat4 mutants (n=3 for each). Therefore, decreased cell mitosis, and not increased cell death, contributes to the narrowing of the sclerotome in Fat4−/− and Dchs1−/− embryos.
Fat4 and Dchs1 do not regulate Yap/Taz activity in the developing vertebrae
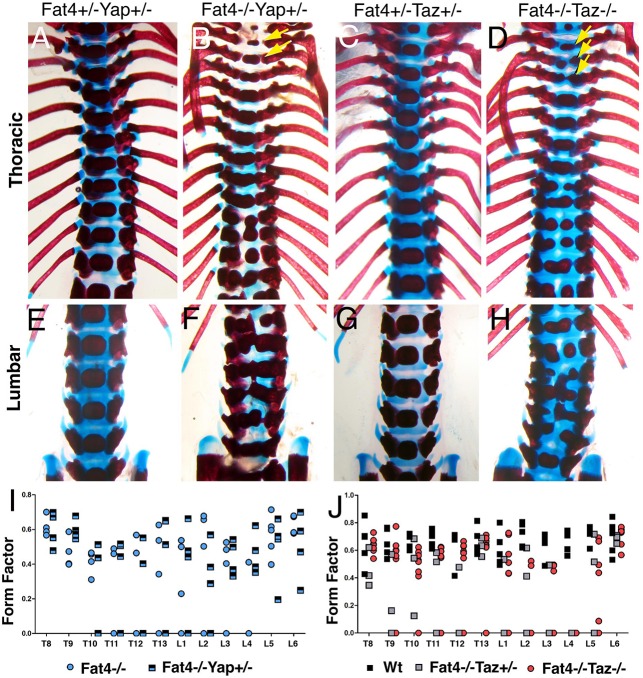
Fat signalling regulates Yorkie in Drosophila and Fat4-Dchs1 has also been reported to inhibit Yap/Taz activity in mammals (Cappello et al., 2013; Das et al., 2013). In Drosophila fat mutants, the growth phenotype can be rescued by simultaneous knockdown of yorkie (Reddy and Irvine, 2008). Likewise, in mice knockdown of Yap rescues the neuronal proliferation defects caused by reduction in Fat4 and Dchs1 expression (Cappello et al., 2013). To determine any potential genetic interaction between Fat4/Dchs1 and Yap (Yap1)/Taz (Wwtr1), we analysed the form factor of individual vertebrae in P0 Fat4−/−;Taz−/−, Fat4−/−;Taz+/− and Fat4−/−;Yap+/− mutants. This revealed that neither homozygous/heterozygous loss of Taz nor heterozygous loss of Yap can rescue the severity and range of posterior thoracic and lumbar vertebral defects in Fat4−/− mutants (Fig. 5A-J). Instead, the defects were extended to include the anterior thoracic vertebrae, which were smaller and malformed, particularly in Fat4−/−;Taz−/− double mutants (Fig. 5B,D). L6 was also affected in two of four Fat4−/−;Yap+/− mice.
Fig. 5.
Loss of Yap or Taz does not rescue the Fat4−/− vertebral defects. (A-H) Alcian Blue and Alizarin Red staining of P0 thoracic (A-D) and lumbar (E-H) vertebrae of mice of the indicated genotypes. In Fat4−/−;Yap+/− and Fat4−/−;Taz−/− mice, some of the anterior thoracic vertebrae, which are normal in Fat4−/− mice, are now also affected (arrowed). (I,J) Form factors of vertebrae T8-L6 of mice of the indicated genotypes. Each symbol indicates an individual mouse.
Another possibility raised by the identification of Fat-to-Ds activation of Yorkie in Drosophila (Degoutin et al., 2013), is that Dchs1 acts as the receptor and that loss of Fat4-to-Dchs1 signalling decreases Yap/Taz activity. This decreased activity could be expected to be responsible for decreased sclerotomal proliferation. To test this potential link genetically, we analysed vertebral development in Fat4+/−;Taz+/− and Fat+/−;Yap+/− P0 mice. However, we found that vertebral development was normal (Fig. 5A,C). The expression of the Yap/Taz target Ctgf in E10.5 mutant somites was also analysed by qPCR and found to be similar to wild-type levels (Fig. S2). Together, the data indicate that decreased Yap or Taz activity alone is unlikely to contribute to the vertebral defects in Fat4−/− and Dchs1−/− mutants.
DISCUSSION
The Fat4-Dchs1 pathway is crucial for the development of many organs, but in general the cellular and intracellular signalling mechanisms that are regulated by Fat4-Dchs1 are unknown. Based on potential conservation between Drosophila and mammals it has been proposed that Fat4-Dchs1 regulates PCP and the Hippo pathway. Conservation of PCP has clearly been shown in the hindbrain and kidney but conservation of Fat4-Hippo signalling is controversial (Cappello et al., 2013; Das et al., 2013; Zakaria et al., 2014; Mao et al., 2015; Bagherie-Lachidan et al., 2015). We have previously reported that Fat4 and Dchs1 mutants are characterised by wider vertebrae that are also fused along the anterior-posterior axis, raising the possibility that alterations in polarised cell behaviours contribute to this phenotype (Mao et al., 2011). We find, however, that cell polarity is unperturbed and that the early sclerotome is specified appropriately. Instead, Fat4-Dchs1 signalling acts cell-autonomously within the somitic mesoderm to regulate cell proliferation at E10.5 and E11.5, time points when cell proliferation and sclerotomal expansion are at their highest (Gasser, 1979). In the absence of Fat4 or Dchs1, the developing vertebral body anlagen is smaller and at the midline does not reach the critical size to condense appropriately. Despite the altered cell proliferation, we find no evidence of Fat4-Dchs1 regulation of Yap/Taz activity, suggesting novel mechanisms of Fat4-Dchs1 signalling.
Vertebral development involves complex signalling interactions that must be regulated both temporally and spatially to first specify the somites from the presomitic mesoderm in a regular manner and then to specify the somite into its various somitic cell lineages, including chondrocytes, muscle and dermis (Maroto et al., 2012). Here, we have added another signalling pathway to this network, namely the Fat4-Dchs1 signalling pathway, which is essential for vertebral development in mice and humans (our data; Mansour et al., 2012). In Fat4 and Dchs1 mutants, the vertebrae are narrower along the dorsoventral and anterior-posterior axes. They are also split at the midline and are fused along the anterior-posterior axis. The neural arches are unaffected. These defects resemble the vertebral anomalies that occur in the absence of Pax1, Pax9, Meox1/Meox2 and Bapx1, a transcriptional network that promotes sclerotome cell proliferation and drives chondrogenesis (Wallin et al., 1994; Lettice et al., 1999; Murtaugh et al., 1999; Peters et al., 1999; Tribioli and Lufkin, 1999; Mankoo et al., 2003; Rodrigo et al., 2003). This transcriptional complex and the regulatory network that drives Sox9 expression and differentiation appear to be intact in Fat4 and Dchs1 mutants. Although we cannot rule out later roles of Fat4/Dchs1 in the maintenance of chondrocyte identity, the early role of Fat4-Dchs1 signalling during vertebral development is in contrast to the role of the related proteins Fat3 and Dchs2 in the developing pharyngeal cartilages of zebrafish. In the pharyngeal cartilages, Fat3-Dchs2 have been shown to control chondrocyte development via the regulation of Sox9a (Le Pabic et al., 2014). Downregulation of either Fat3 or Dchs2 results in decreased Sox9a expression and the failure of the developing chondrocytes to intercalate appropriately, ultimately resulting in dysmorphic pharyngeal cartilages (Le Pabic et al., 2014).
The key defect in mouse Fat4 and Dchs1 mutants is the reduction in the number of sclerotomal cells due to decreased cell proliferation, although it is possible that there are also minor changes in sclerotomal specification that might contribute to the formation of a smaller vertebral body. Fat4 and Dchs1 are expressed in the sclerotome between E9.5 and E11.5 (Mao et al., 2011; this study). The decreased proliferation at E10.5 results in a smaller sclerotome at E11.5, and cells at the centre of the developing vertebrae do not reach the critical cell mass for chondrogenesis to proceed around the notochord (Hall and Miyake, 2000). As a consequence, two cartilage condensations develop adjacent to the notochord and the vertebral bodies are split.
In both Drosophila and mammals, loss of Fat signalling has been linked to increased Yorkie and Yap/Taz signalling and knockdown of Yorkie, Yap or Taz in Drosophila and the mammalian cerebral cortex can rescue the defects that result from the loss of Fat/Fat4 signalling (Reddy and Irvine, 2008; Cappello et al., 2013). Analysis of Fat4−/−;Taz−/−, Fat4−/−;Taz+/− and Fat4−/−;Yap+/− double mutants suggests that the vertebral defects are not mediated by increased Yap/Taz signalling. In fact, in Fat4−/−;Taz−/− and, to a lesser extent, in Fat4−/−;Yap+/− double mutants the vertebral defects are extended to encompass the anterior thoracic vertebrae, suggesting that they operate in parallel pathways. This lack of interaction between Fat4 and Yap/Taz has also been observed in the kidney (Mao et al., 2015; Bagherie-Lachidan et al., 2015), indicating distinct mechanisms of Fat4-Dchs1 signalling during vertebrate development. Indeed, there is currently very little, if any, evidence of a direct link between Fat4 and Hippo signalling in vertebrates. How Fat4 might regulate Yap/Taz activity is also unclear, as the intracellular region of Fat4 either lacks or shows limited sequence conservation in the domains that are crucial for Fat-Hippo signalling in Drosophila (Bossuyt et al., 2014). Consistent with this limited homology, the intracellular domain of human FAT4 can rescue the PCP, but not the growth, defects in Drosophila fat mutants (Pan et al., 2013).
As Ds has been shown to act cell-autonomously as a receptor to increase Yorkie signalling in Drosophila (Degoutin et al., 2013), we also considered the reverse possibility that there is decreased Yap/Taz activity in Fat4−/− or Dchs1−/− mouse mutants. This decreased Yap/Taz activity would be anticipated to result in decreased cell proliferation, consistent with our data. However, neither Taz−/−, Yap+/−, Fat4+/−;Taz+/− or Fat4+/−;Yap+/− mice have vertebral defects. Also, expression of Ctgf, a direct transcriptional target of Yap/Taz, is unaffected in Fat4 and Dchs1 mutants. Therefore, we also conclude that loss of Yap/Taz signalling is unlikely to be responsible for the vertebral malformations in Fat4 and Dchs1 mutants. It is not yet clear if Fat4/Dchs1 regulate cell proliferation directly via a transcriptional mechanism and/or modulate the activity of another signalling pathway required for cell proliferation, such as Shh (Fan et al., 1995; Marcelle et al., 1999).
PCP classically controls cellular organisation across an epithelial tissue. In vertebrates, the Fz-PCP pathway has also been shown to control coordinated cellular behaviours within mesenchymal tissues. Examples include cell intercalation during gastrulation, cell orientation in the developing limb bud, and cell-to-cell interactions within migratory neural crest streams (Goodrich and Strutt, 2011; Gao and Yang, 2013; Mayor and Theveneau, 2014; Sokol, 2015). As there are vertebral defects in both Fz-PCP and the Fat4 and Dchs1 mutants, we considered the possibility that Fat4-Dchs1 might regulate polarised cell behaviours and rearrangements during sclerotomal morphogenesis. An alteration in how the cells are organised could contribute to both the formation of the hemivertebrae and the fusion of the vertebrae along the anterior-posterior axis. The vertebrae of P0 Fat4 and Dchs1 mutants are also wider, which again raised the possibility that cells are not arranged appropriately during early morphogenesis. We found that in wild-type embryos the sclerotomal cells are polarised and the orientation of the Golgi complex, another indicator of polarity, also reveals a polarity bias. However, we found no evidence that this polarised cell behaviour is disrupted in Fat4 and Dchs1 mutants. The width of the developing vertebral body at E10.5-E11.5 along the mediolateral axis also appears to be initially the same in Fat4 and Dchs1 mutants when compared with wild-type embryos. The increased width of the vertebrae and neural arches in P0 Fat4 and Dchs1 mutants therefore occurs later and is secondary to changes in the development of the split vertebral bodies and the increased width of the spinal cord (Saburi et al., 2008; Mao et al., 2011).
Fat4-Dchs1 signalling controls the development of many organs in both mice and humans, but in general the cellular mechanisms of Fat4 signalling are unknown. Fat/Fat4 regulation of cell behaviours through PCP has been shown to be conserved (Saburi et al., 2008; Mao et al., 2011, 2016; Zakaria et al., 2014). Evidence linking Fat4/Dchs1 to the Hippo pathway is less clear (Cappello et al., 2013; Das et al., 2013; Mao et al., 2015; Bagherie-Lachidan et al., 2015). Here we show that Fat4-Dchs1 controls cell proliferation within the developing sclerotome. This Fat4-Dchs1 promotion of cell proliferation has also been observed in the kidney mesenchyme and ureteric distal tips (Mao et al., 2011) and is in stark contrast to Fat4-Dchs1 inhibition of cell proliferation in the cerebral cortex (Cappello et al., 2013). We conclude that novel mechanisms of Fat4-Dchs1 signalling have evolved to regulate cell proliferation, probably through a Hippo-independent pathway. As with other signalling pathways, there are pleiotropic roles for Fat4-Dchs1 signalling that range from the regulation of polarised cell behaviours, proliferation and differentiation to cell survival (Saburi et al., 2008; Mao et al., 2011, 2015; Cappello et al., 2013; Das et al., 2013; Zakaria et al., 2014). Determining the exact roles of Fat4-Dchs1 signalling during organogenesis and how these distinct cellular outcomes are established will be essential to understand the basis of developmental abnormalities in Van Maldergem and Hennekam syndromes.
MATERIALS AND METHODS
Mice and genotyping
The Fat4−/−, Dchs1−/−, Dchs1f/f, Fat4f/f, Taz−/− (Wwtr1−/−), Yapf/f and Dermo1Cre (Dermo1 is also known as Twist1) mouse lines have been described previously (Humphreys et al., 2010; Kobayashi et al., 2008; Mao et al., 2011, 2015; Saburi et al., 2008; Yu et al., 2003). Noon of the day of the vaginal plug was assigned E0.5. DNA was isolated using Direct PCR-Ear Lysis Reagent (PeqLab) or the Promega Hot Start 2 Kit and genotyping was performed as previously described (Mao et al., 2011). All mouse procedures were approved by the IACUC either at Rutgers University or King's College London in agreement with established guidelines for animal care.
Histology and immunohistological analyses
Embryos were fixed overnight in 4% paraformaldehyde and processed into wax. Sections (10 µm) through the developing lumbar vertebrae were stained with either Haematoxylin and Eosin (H&E), Mallory's triple stain or immunostained with primary antibodies anti-Sox9 (rabbit, Millipore, AB5535, 1:400; or goat, Santa Cruz, SC-17341, 1:50), rabbit anti-human cleaved caspase 3 (Cell Signaling Technology, 9661S, 1:200), mouse anti-phospho-Ser10 histone H3 (Millipore, 9706S, 1:100), rabbit anti-Golgi (Abcam, 24586, 1:100) or collagen type II (Abcam, ab34712, 1:200) following antigen retrieval in citrate buffer. The region of the developing vertebral body analysed is indicated by the dashed yellow lines in Fig. 2A,D and is shown by the Sox9 immunostaining in Fig. 3I-L. At least six sections from each embryo, 60 µm apart, or all sections spanning L4-T12 were analysed.
For the apoptosis analysis at least 870 cells were analysed (range 870-6000/embryo at E10.5 and at least 2000-9000 cells/embryo at E11.5). The total number of cells analysed were as follows: E10.5, Dchs1+/+ 14,510, Dchs1−/− 16,326, Fat4+/+ 8074, Fat4−/− 5699; E11.5, Dchs1+/+ 12,162, Dchs1−/− 10,758, Fat4+/+ 40,491, Fat4−/− 24,942. For the proliferation analysis, at least 500 cells were counted (range 523-9844 at E10.5 and at least 2000-9844 at E11.5). The total number of cells analysed were as follows: E10.5, Dchs1+/+ 15,277, Dchs1−/− 13,196, Fat4+/+ 11,339, Fat4−/− 7182; E11.5, Dchs1+/+ 12,162, Dchs1−/− 10,758, Fat4+/+ 57,919, Fat4−/− 45,718. The following numbers of mutant embryos were analysed and compared with wild-type/heterozygous littermates: pH3 at E10.5, Fat4 n=3, Dchs1 n=7; pH3 at E11.5, Fat4 n=6, Dchs1 n=4; active caspase 3 at E10.5, Fat4 n=3, Dchs1 n=4; active caspase 3 at E11.5, Fat4 n=3, Dchs1 n=4; collagen type II at E11.5, Fat4 n=2, Dchs1 n=2. Skeletal staining was performed as described by Huang et al. (2002).
In situ hybridisation
In situ hybridisation was performed on 10 µm sections of paraformaldehyde-fixed tissue processed into wax or whole embryos according to standard protocols, using DIG-labelled probes. Embryos were processed in 20% gelatin in PBS for vibratome sectioning (40 µm). The following numbers of embryos were analysed: E10.5, for Pax1, Fat4 n=3, Dchs1 n=3; for Pax9, Fat4 n=3, Dchs1 n=4; for Bapx1, Fat4 n=3, Dchs1 n=3; for Meox1, Fat4 n=2, Dchs1 n=2; for Meox2, Fat4 n=3, Dchs1 n=2; for Tbx18, Fat4 n=2, Dchs1 n=2; for Uncx4.1, Fat4 n=3, Dchs1 n=2; for collagen type II, Fat4 n=2; E11.5, for Pax9, Fat4 n=3, Dchs1 n=1; for Bapx1, Fat4 n=3, Dchs1 n=2; for collagen type II, Fat4 n=4, Dchs1 n=1.
qPCR analysis
RNA was isolated from lumbar and posterior thoracic somites of E10.5 wild-type, Dchs1−/− and Fat4−/− embryos using TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. cDNA was prepared from 200 ng total RNA as a template using the Precision nanoScript Reverse Transcription Kit (Primer Design). qPCR for mRNA expression was performed in triplicate using Precision Real-Time PCR MasterMix with SYBR Green (Primer Design) according to the manufacturer's instructions. Primers were designed using Primer-BLAST (NCBI) (see Table S1). qPCR was performed and results analysed using a Qiagen Rotor-Gene Q machine and software package. Data were quantified by the ΔΔCT method using Gapdh as a control. Significance of the data was analysed by Student's t-test.
Image analysis and statistics
Immunolocalisation images were collected using a laser-scanning confocal microscope (Leica) and analysis performed using AxioVision (Zeiss) for Golgi orientation and CellProfiler 2.0 (Broad Institute) for nuclei parameters. Circular analysis of Golgi complex orientation and nuclear alignment was performed using Oriana4.0 (Kovach) (Rayleigh uniformity test and Mardia-Whatson-Wheeler nonparametric test). Graphs were prepared and linear statistical analysis performed using GraphPad (Prism). P0 skeletons stained with Alcian Blue and Alizarin Red were imaged from the ventral side and individual vertebral bodies segmented using CellProfiler 2.0. For each vertebral body, a form factor was automatically calculated as 4×π×area/perimeter2. Form factor equals 1 for a perfectly circular object, while 0 was assigned for split vertebral bodies.
Acknowledgements
We thank Helen McNeill for Fat4−/− and Fat4f/f mice and David Ornitz for the Dermo1Cre mouse line.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
P.H.F.-W., K.D.I. and B.M. designed the study; P.H.F.-W. wrote the manuscript with contributions from A.K. and B.J.; A.K., B.B., Y.M., D.W., C.F.d.S., T.M., I.C.-E., P.E., P.H.F.-W. and S.Z. carried out the research and analysed the data.
Funding
The research is funded by the Biotechnology and Biological Sciences Research Council (BBSRC) [BB/G021074/1, BB/K001671/1 and BB/K008668/1 to P.H.F.-W.]; a King's College London Scholarship (S.Z.); and the Howard Hughes Medical Institute (K.D.I.). Deposited in PMC for release after 6 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.131037.supplemental
References
- Alders M., Al-Gazali L., Cordeiro I., Dallapiccola B., Garavelli L., Tuysuz B., Salehi F., Haagmans M. A., Mook O. R., Majoie C. B. et al. (2014). Hennekam syndrome can be caused by FAT4 mutations and be allelic to Van Maldergem syndrome. Hum. Genet. 133, 1161-1167. 10.1007/s00439-014-1456-y [DOI] [PubMed] [Google Scholar]
- Badouel C., Zander M. A., Liscio N., Bagherie-Lachidan M., Sopko R., Coyaud E., Raught B., Miller F. D. and McNeill H. (2015). Fat1 interacts with Fat4 to regulate neural tube closure, neural progenitor proliferation and apical constriction during mouse brain development. Development 142, 2781-2791. 10.1242/dev.123539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagherie-Lachidan M., Reginensi A., Pan Q., Zaveri H. P., Scott D. A., Blencowe B. J., Helmbacher F. and McNeill H. (2015). Stromal Fat4 acts non-autonomously with Dachsous1/2 to restrict the nephron progenitor pool. Development 142, 2564-2573. 10.1242/dev.122648 [DOI] [PubMed] [Google Scholar]
- Bossuyt W., Chen C.-L., Chen Q., Sudol M., McNeill H., Pan D., Kopp A. and Halder G. (2014). An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene 33, 1218-1228. 10.1038/onc.2013.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussen M., Petry M., Schuster-Gossler K., Leitges M., Gossler A. and Kispert A. (2004). The T-box transcription factor Tbx18 maintains the separation of anterior and posterior somite compartments. Genes Dev. 18, 1209-1221. 10.1101/gad.300104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello S., Gray M. J., Badouel C., Lange S., Einsiedler M., Srour M., Chitayat D., Hamdan F. F., Jenkins Z. A., Morgan T. et al. (2013). Mutations in genes encoding the cadherin receptor-ligand pair DCHS1 and FAT4 disrupt cerebral cortical development. Nat. Genet. 45, 1300-1308. 10.1038/ng.2765 [DOI] [PubMed] [Google Scholar]
- Das A., Tanigawa S., Karner C. M., Xin M., Lum L., Chen C., Olson E. N., Perantoni A. O. and Carroll T. J. (2013). Stromal-epithelial crosstalk regulates kidney progenitor cell differentiation. Nat. Cell Biol. 15, 1035-1044. 10.1038/ncb2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degoutin J. L., Milton C. C., Yu E., Tipping M., Bosveld F., Yang L., Bellaiche Y., Veraksa A. and Harvey K. F. (2013). Riquiqui and minibrain are regulators of the hippo pathway downstream of Dachsous. Nat. Cell Biol. 15, 1176-1185. 10.1038/ncb2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C.-M. and Tessier-Lavigne M. (1994). Patterning of mammalian somites by surface ectoderm and notochord: evidence for sclerotome induction by a hedgehog homolog. Cell 79, 1175-1186. 10.1016/0092-8674(94)90009-4 [DOI] [PubMed] [Google Scholar]
- Fan C.-M., Porter J. A., Chiang C., Chang D. T., Beachy P. A. and Tessier-Lavigne M. (1995). Long-range sclerotome induction by sonic hedgehog: direct role of the amino-terminal cleavage product and modulation by the cyclic AMP signaling pathway. Cell 81, 457-465. 10.1016/0092-8674(95)90398-4 [DOI] [PubMed] [Google Scholar]
- Fleming A., Kishida M. G., Kimmel C. B. and Keynes R. J. (2015). Building the backbone: the development and evolution of vertebral patterning. Development 142, 1733-1744. 10.1242/dev.118950 [DOI] [PubMed] [Google Scholar]
- Gao B. and Yang Y. (2013). Planar cell polarity in vertebrate limb morphogenesis. Curr. Opin. Genet. Dev. 23, 438-444. 10.1016/j.gde.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser R. F. (1979). Evidence that sclerotomal cells do not migrate medially during normal embryonic development of the rat. Am. J. Anat. 154, 509-524. 10.1002/aja.1001540406 [DOI] [PubMed] [Google Scholar]
- Goodrich L. V. and Strutt D. (2011). Principles of planar polarity in animal development. Development 138, 1877-1892. 10.1242/dev.054080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene N. D. E., Gerrelli D., Van Straaten H. W. M. and Copp A. J. (1998). Abnormalities of floor plate, notochord and somite differentiation in the loop-tail (Lp) mouse: a model of severe neural tube defects. Mech. Dev. 73, 59-72. 10.1016/S0925-4773(98)00029-X [DOI] [PubMed] [Google Scholar]
- Hall B. K. and Miyake T. (2000). All for one and one for all: condensations and the initiation of skeletal development. BioEssays 22, 138-147. [DOI] [PubMed] [Google Scholar]
- Hirsinger E., Duprez D., Jouve C., Malapert P., Cooke J. and Pourquie O. (1997). Noggin acts downstream of Wnt and Sonic Hedgehog to antagonize BMP4 in avian somite patterning. Development 124, 4605-4614. [DOI] [PubMed] [Google Scholar]
- Huang Y., Roelink H. and McKnight G. S. (2002). Protein kinase A deficiency causes axially localized neural tube defects in mice. J. Biol. Chem. 277, 19889-19896. 10.1074/jbc.M111412200 [DOI] [PubMed] [Google Scholar]
- Hubaud A. and Pourquie O. (2014). Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol. 15, 709-721. 10.1038/nrm3891 [DOI] [PubMed] [Google Scholar]
- Johnson R. L., Laufer E., Riddle R. D. and Tabin C. (1994). Ectopic expression of Sonic hedgehog alters dorsal-ventral patterning of somites. Cell 79, 1165-1173. 10.1016/0092-8674(94)90008-6 [DOI] [PubMed] [Google Scholar]
- Le Pabic P., Ng C. and Schilling T. F. (2014). Fat-Dachsous signaling coordinates cartilage differentiation and polarity during craniofacial development. PLoS Genet. 10, e1004726 10.1371/journal.pgen.1004726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V. and Bhattaram P. (2010). Vertebrate skeletogenesis. Curr. Top. Dev. Biol. 90, 291-317. 10.1016/S0070-2153(10)90008-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice L. A., Purdie L. A., Carlson G. J., Kilanowski F., Dorin J. and Hill R. E. (1999). The mouse bagpipe gene controls development of axial skeleton, skull, and spleen. Proc. Natl. Acad. Sci. USA 96, 9695-9700. 10.1073/pnas.96.17.9695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankoo B. S., Skuntz S., Harrigan I., Grigorieva E., Candia A., Wright C. V. E., Arnheiter H. and Pachnis V. (2003). The concerted action of Meox homeobox genes is required upstream of genetic pathways essential for the formation, patterning and differentiation of somites. Development 130, 4655-4664. 10.1242/dev.00687 [DOI] [PubMed] [Google Scholar]
- Mansour S., Swinkels M., Terhal P. A., Wilson L. C., Rich P., Van Maldergem L., Zwijnenburg P. J. G., Hall C. M., Robertson S. P. and Newbury-Ecob R. (2012). Van Maldergem syndrome: further characterisation and evidence for neuronal migration abnormalities and autosomal recessive inheritance. Eur. J. Hum. Genet. 20, 1024-1031. 10.1038/ejhg.2012.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A., Voss A. K., Thomas T., Yokota Y. and Gruss P. (2000). Uncx4.1 is required for the formation of the pedicles and proximal ribs and acts upstream of Pax9. Development 127, 2251-2258. [DOI] [PubMed] [Google Scholar]
- Mao Y., Mulvaney J., Zakaria S., Yu T., Morgan K. M., Allen S., Basson M. A., Francis-West P. and Irvine K. D. (2011). Characterization of a Dchs1 mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development. Development 138, 947-957. 10.1242/dev.057166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Francis-West P. and Irvine K. D. (2015). A Fat4-Dchs1 signaling between stromal and cap mesenchyme cells influences nephrogenesis and ureteric bud branching. Development 142, 2574-2585. 10.1242/dev.122630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Kuta A., Crespo-Enriquez I., Whiting D., Martin T., Mulvaney J., Irvine K. D. and Francis-West P. (2016). Dchs1-Fat4 regulation of polarized cell behaviours during skeletal morphogenesis. Nat. Commun. 7, 11469 10.1038/ncomms11469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelle C., Ahlgren S. and Bronner-Fraser M. (1999). In vivo regulation of somite differentiation and proliferation by Sonic Hedgehog. Dev. Biol. 214, 277-287. 10.1006/dbio.1999.9389 [DOI] [PubMed] [Google Scholar]
- Maroto M., Bone R. A. and Dale J. K. (2012). Somitogenesis. Development 139, 2453-2456. 10.1242/dev.069310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matis M. and Axelrod J. D. (2013). Regulation of PCP by the Fat signaling pathway. Genes Dev. 27, 2207-2220. 10.1101/gad.228098.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R. and Theveneau E. (2014). The role of the non-canonical Wnt-planar cell polarity pathway in neural crest migration. Biochem. J. 457, 19-26. 10.1042/BJ20131182 [DOI] [PubMed] [Google Scholar]
- McMahon J. A., Takada S., Zimmerman L. B., Fan C.-M., Harland R. M. and McMahon A. P. (1998). Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 12, 1438-1452. 10.1101/gad.12.10.1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh L. C., Chyung J. H. and Lassar A. B. (1999). Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 13, 225-237. 10.1101/gad.13.2.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G., Feng Y., Ambegaonkar A. A., Sun G., Huff M., Rauskolb C. and Irvine K. D. (2013). Signal transduction by the Fat cytoplasmic domain. Development 140, 831-842. 10.1242/dev.088534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H., Wilm B., Sakai N., Imai K., Maas R. and Balling R. (1999). Pax1 and Pax9 synergistically regulate vertebral column development. Development 126, 5399-5408. [DOI] [PubMed] [Google Scholar]
- Reddy B. V. V. G. and Irvine K. D. (2008). The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development 135, 2827-2838. 10.1242/dev.020974 [DOI] [PubMed] [Google Scholar]
- Rock R., Schrauth S. and Gessler M. (2005). Expression of mouse dchs1, fjx1, and fat-j suggests conservation of the planar cell polarity pathway identified in Drosophila. Dev. Dyn. 234, 747-755. 10.1002/dvdy.20515 [DOI] [PubMed] [Google Scholar]
- Rodrigo I., Hill R. E., Balling R., Munsterberg A. and Imai K. (2003). Pax1 and Pax9 activate Bapx1 to induce chondrogenic differentiation in the sclerotome. Development 130, 473-482. 10.1242/dev.00240 [DOI] [PubMed] [Google Scholar]
- Saburi S., Hester I., Fischer E., Pontoglio M., Eremina V., Gessler M., Quaggin S. E., Harrison R., Mount R. and McNeill H. (2008). Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat. Genet. 40, 1010-1015. 10.1038/ng.179 [DOI] [PubMed] [Google Scholar]
- Saburi S., Hester I., Goodrich L. and McNeill H. (2012). Functional interactions between Fat family cadherins in tissue morphogenesis and planar polarity. Development 139, 1806-1820. 10.1242/dev.077461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol S. Y. (2015). Spatial and temporal aspects of Wnt signaling and planar cell polarity during vertebrate embryonic development. Semin. Cell Dev. Biol. 42, 78-85. 10.1016/j.semcdb.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford D. A., Brunet L. J., Khokha M. K., Economides A. N. and Harland R. M. (2011). Cooperative activity of noggin and gremlin 1 in axial skeleton development. Development 138, 1005-1014. 10.1242/dev.051938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley B. K. and Irvine K. D. (2012). Hippo signaling in Drosophila: recent advances and insights. Dev. Dyn. 241, 3-15. 10.1002/dvdy.22723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein K. F. and Mackensen J. A. (1957). Abnormal development of the thoracic skeleton in mice homozygous for the gene for looped-tail. Am. J. Anat. 100, 205-223. 10.1002/aja.1001000203 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Rocancourt D., Cossu G. and Buckingham M. (1997). Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89, 127-138. 10.1016/S0092-8674(00)80189-0 [DOI] [PubMed] [Google Scholar]
- Tribioli C. and Lufkin T. (1999). The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development 126, 5699-5711. [DOI] [PubMed] [Google Scholar]
- Wahi K., Bochter M. S. and Cole S. E. (2016). The many roles of Notch signaling during vertebrate somitogenesis. Semin. Cell Dev. Biol. 49, 68-75. 10.1016/j.semcdb.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Wallin J., Wilting J., Koseki H., Fritsch R., Christ B. and Balling R. (1994). The role of Pax-1 in axial skeleton development. Development 120, 1109-1121. [DOI] [PubMed] [Google Scholar]
- Yang C.-H., Axelrod J. D. and Simon M. A. (2002). Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell 108, 675-688. 10.1016/S0092-8674(02)00658-X [DOI] [PubMed] [Google Scholar]
- Yu F.-X. and Guan K.-L. (2013). The Hippo pathway: regulators and regulations. Genes Dev. 27, 355-371. 10.1101/gad.210773.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E. N., Towler D. A. and Ornitz D. M. (2003). Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130, 3063-3074. 10.1242/dev.00491 [DOI] [PubMed] [Google Scholar]
- Zakaria S., Mao Y., Kuta A., Ferreira de Sousa C., Gaufo G. O., Mcneill H., Hindges R., Guthrie S., Irvine K. D. and Francis-West P. H. (2014). Regulation of neuronal migration by dchs1-fat4 planar cell polarity. Curr. Biol. 24, 1620-1627. 10.1016/j.cub.2014.05.067 [DOI] [PMC free article] [PubMed] [Google Scholar]