Abstract
Hmx1 encodes a homeodomain transcription factor expressed in the developing lateral craniofacial mesenchyme, retina and sensory ganglia. Mutation or mis-regulation of Hmx1 underlies malformations of the eye and external ear in multiple species. Deletion or insertional duplication of an evolutionarily conserved region (ECR) downstream of Hmx1 has recently been described in rat and cow, respectively. Here, we demonstrate that the impact of Hmx1 loss is greater than previously appreciated, with a variety of lateral cranioskeletal defects, auriculofacial nerve deficits, and duplication of the caudal region of the external ear. Using a transgenic approach, we demonstrate that a 594 bp sequence encompassing the ECR recapitulates specific aspects of the endogenous Hmx1 lateral facial expression pattern. Moreover, we show that Hoxa2, Meis and Pbx proteins act cooperatively on the ECR, via a core 32 bp sequence, to regulate Hmx1 expression. These studies highlight the conserved role for Hmx1 in BA2-derived tissues and provide an entry point for improved understanding of the causes of the frequent lateral facial birth defects in humans.
KEY WORDS: Hmx1, Evolutionarily conserved region (ECR), Enhancer, Craniofacial mesenchyme, Pinna, Mouse
Summary: The transcription factors Hoxa2, Meis and Pbx act cooperatively on an evolutionarily conserved region downstream of Hmx1 to regulate Hmx1 expression and craniofacial development.
INTRODUCTION
Mutation of the homeodomain transcription factor Hmx1 results in malformation of the external ear and eye. This condition has been termed oculo-auricular syndrome (OAS) in humans (Schorderet et al., 2008; Gillespie et al., 2015), ‘dumbo’ and/or ‘misplaced ears’ in mice, ‘dumbo’ in rats (Munroe et al., 2009; Kuramoto et al., 2010; Quina et al., 2012a), and ‘crop ear’ in cattle (Koch et al., 2013). Previous work has also shown that Hmx1 is essential for the development of sensory neurons in the geniculate/facial ganglion (Quina et al., 2012b).
OAS in humans and the dumbo phenotype in mice are both recessive phenotypes that result from loss-of-function mutations in the Hmx1 coding region (Schorderet et al., 2008; Munroe et al., 2009; Vaclavik et al., 2011; Gillespie et al., 2015). However, in rats, the recessive dumbo allele consists of a 5777 bp deletion ∼80 kb downstream of Hmx1. This deletion is associated with a loss of Hmx1 expression only in the first and second branchial arch (BA1 and BA2) mesenchyme (Quina et al., 2012a), suggesting that the deletion removes an important regulatory element driving Hmx1 expression in the lateral face. This deleted sequence contains a ∼600 bp evolutionarily conserved region (ECR). Remarkably, the recessive crop ear allele in cattle also involves this ECR, specifically a 76 bp duplication of the central core (Koch et al., 2013).
The dumbo and OAS phenotypes suggest that the role of Hmx1 is distinct from that of classic homeobox transcription factors, such as Hoxa2, which act early in the patterning of the branchial arch region. Patients with mutations in HOXA2 display severe microtia, middle ear deformities and hearing loss (Alasti et al., 2008). By contrast, OAS patients and dumbo mice have dysmorphic external ears but the middle ear and hearing appear to be unaffected (Schorderet et al., 2008; Munroe et al., 2009; Gillespie et al., 2015).
In the present study, we show that the 594 bp distal ECR functions as a strong and highly dynamic lateral facial enhancer that recapitulates specific aspects of the endogenous Hmx1 expression pattern. Moreover, we provide evidence that the upstream patterning factors Hoxa2, Meis and Pbx act cooperatively on sequences within the ECR to regulate Hmx1. We also describe an array of previously unappreciated lateral cranioskeletal anomalies in dumbo mice and in outbred dumbo rats. Taken together, our findings support the hypothesis that Hmx1 plays an important evolutionarily conserved role in lateral facial mesenchyme differentiation, downstream of embryonic patterning genes, providing an entry point into understanding the regulatory network underpinning common lateral facial birth defects.
RESULTS
Hmx1 mutant mice exhibit craniofacial and cranioskeletal anomalies
To undertake a thorough assessment of the dumbo phenotype, we first transferred the mutation on to a pure C57Bl/6N background by more than ten backcrosses. Mendelian transmission of the dumbo allele was observed between embryonic day (E) 10.5 and E14.5 (42 Hmx1+/+, 105 Hmx1+/dm and 37 Hmx1dm/dm embryos; χ2 P=0.1391). However, similar to the previous study on a mixed genetic background (Munroe et al., 2009), we observed non-Mendelian transmission of the dumbo allele in post-weaning animals; specifically, 49 Hmx1+/+, 86 Hmx1+/dm and 21 Hmx1dm/dm mice (31.4%, 55.1% and 13.5%, respectively; χ2 P=0.0029), suggesting ∼57% perinatal lethality of homozygous mutants, which appeared to be equally distributed between the sexes. In contrast to the earlier report (Munroe et al., 2009), only post-weaning Hmx1dm/dm male mice showed a significant reduction in weight, on average ∼23% lighter than controls (control: 17.28±0.494 g, Hmx1dm/dm: 13.29±1.102 g). Female Hmx1dm/dm mice were comparable in weight to controls (control: 13.59±0.762 g, Hmx1dm/dm: 13.11±0.440 g).
Imaging of embryos using optical projection tomography (OPT) demonstrated that auricular anomalies were evident in Hmx1dm/dm animals as early as E11.5 (data not shown). By E13.5, all mutants could be recognized by a distinct bifurcation of the developing lobe and a failure or delay in rotation of the developing auricle, giving it a ventrally displaced appearance (Fig. 1A,B), which became more pronounced during later embryonic stages (Fig. 1C,D). Postnatally, this bifurcation in dumbo mice developed into an ectopic cartilaginous flap (caudal duplication) of the pinna (Fig. 1E-H).
Fig. 1.
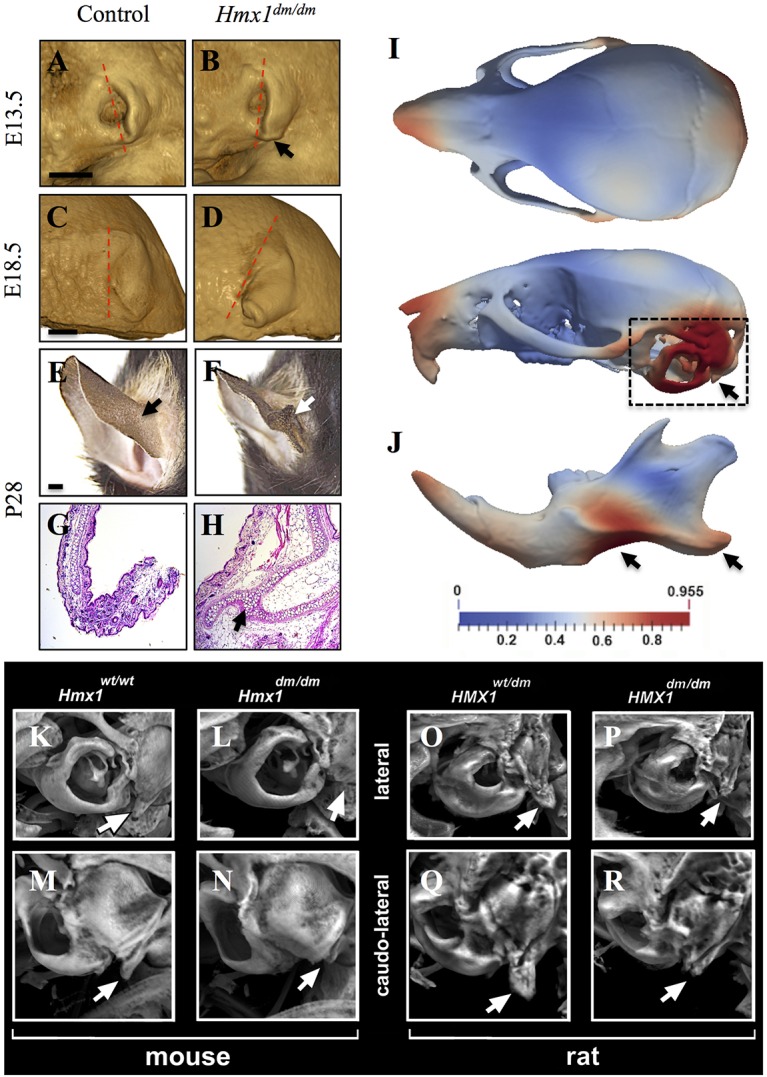
Characterization of craniofacial defects and shape changes in Hmx1dm/dm mutant animals. (A-D) OPT imaging of E13.5 (A,B; control n=4, Hmx1dm/dm mutant n=5) and E18.5 (C,D; control n=2, Hmx1dm/dm mutant n=2) control (A,C) and Hmx1dm/dm mutant (B,D) embryos show morphological differences in the external ear (arrow). The most commonly observed pinna morphologies are displayed. Dashed line indicates pinna orientation. (E-H) Representative posterior view (E,F) and Hematoxylin & Eosin-stained sections (G,H) through a P28 control (E,G) and Hmx1dm/dm mutant (F,H) ear highlight the malformation present in Hmx1dm/dm mutant mice (E,F,H, arrows; control n=28, Hmx1dm/dm mutant n=17). (I,J) Deformational analysis of P28 Hmx1dm/dm skulls and mandibles. Heat maps are projected onto a standard control skull and highlight the relative mean shape differences (control n=24, Hmx1dm/dm n=17). Dashed box and arrows highlight the highest magnitude of shape differences present in the skull (I) and mandible (J) of Hmx1dm/dm animals. (K-R) MicroCT analysis of control (K,M) and Hmx1dm/dm mutant (L,N) mouse skulls and heterozygote (O,Q) and homozygote (P,R) dumbo rat skulls. Marked hypoplasia of the paraoccipital process is evident in Hmx1dm/dm mutant mice and rats (compare K with L, and O with P, arrows for lateral view; compare M with N, and Q with R, arrows for caudo-lateral view). Control Hmx1+/+ mice (n=28) and Hmx1dm/dm mutant mice (n=17). Hmx1dm/+ rats (n=2) and Hmx1dm/dm mutant (n=2). Scale bars: 1 mm (A-F).
Two of nine E14.5 Hmx1dm/dm imaged embryos, both males, presented with an anterior head malformation overlying the anlage of the pre-tectum, caudal to the epiphysis and pineal recess, at the junction between the diencephalon and the mesencephalon (Fig. S1A-D′). This may represent a mild neural tube defect related to the exencephaly previously documented on the mixed background (Munroe et al., 2009) and might contribute to the occasional perinatal lethality of homozygous mutant embryos.
Postnatal day (P) 28 Hmx1dm/dm and control mice were then examined using high-resolution microCT imaging. Although Hmx1dm/dm mice had smaller skulls (Fig. S1E,F′), these were deemed to be proportional to body size after correcting for differences in body mass or long bone length (data not shown). Despite this, Hmx1dm/dm mice did exhibit a number of specific cranial malformations. In two male Hmx1dm/dm mutants, but no female mutants or controls of either sex, a frontal bone/posterior frontal suture anomaly was noted (Fig. S1E-F), which could be related to the anterior head malformation shown in Fig. S1A-D′. However, in all mutants (both sexes) but no controls, premature fusion of the squamosal and parietomastoid sutures was observed (Fig. S1E′-F′). In addition, all Hmx1dm/dm mice exhibited hypoplasia of the temporal processes of the occipital bone (Fig. S1E′-F′) as well as marked hypoplasia of each paraoccipital process (Fig. 1K-N). Notably, microCT imaging of a small number of outbred adult dumbo rats and their littermate controls revealed that the dumbo rats also exhibited marked hypoplasia of the paraoccipital processes, mirroring that seen in the Hmx1dm/dm mice (Fig. 1O-R). Using a deformable registration-based tool to assess skull shape objectively (Rolfe et al., 2013), we also found that the difference in skull shape between Hmx1dm/dm mice and background-matched controls extended beyond these structures. For example, this quantitative analysis showed significant differences over the snout, more broadly over the otic region and back of the skull (Fig. 1I), as well as over the mandibular body and angular process (Fig. 1J). The mandibular and otic region phenotypes showed relatively little variability (as determined by an assessment of the standard deviation of each shape dataset), consistent with these lateral facial features being robust and penetrant.
Hmx1dm/dm mutants exhibit abnormal nerve branching and organization surrounding the pinna
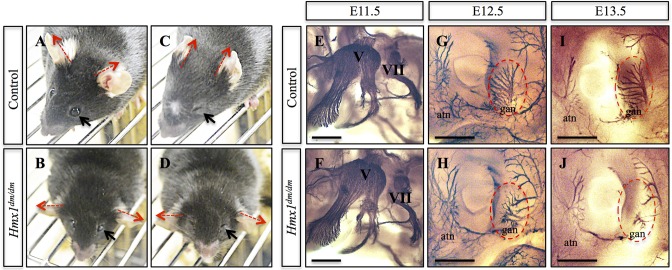
Given the nature of the lateral cranioskeletal dysmorphology, and the prior finding of a geniculate ganglion (sensory nerve) defect in dumbo mice (Quina et al., 2012b), we considered the possibility that these arise as a result of nerve and/or muscle defects. To begin to investigate this, we performed a facial stimulus test involving exposure to a 10 s puff of air (Arenkiel et al., 2004; Tvrdik and Capecchi, 2006). Although mutant and control mice both responded to the puff by closing their eyes, only the control mice tucked their ears back against their heads: the ears of Hmx1dm/dm mice remained perpendicular to the head (compare Fig. 2A,C with 2B,D).
Fig. 2.
Hmx1dm/dm animals show abnormal response to an aversive facial stimulus and defects in sensory nerve branches. (A-D) Representative images of control (A,C) and Hmx1dm/dm (B,D) mice before (A,B) and during the air puff test (C,D) demonstrate that Hmx1dm/dm mice, in contrast to controls, do not move their ears in response to having air blown on their face (compare red dashed arrows in A with C, and B with D; control n=8, Hmx1dm/dm n=7), although Hmx1dm/dm mice can effectively close their eyes (compare black arrow in A with C, and B with D). (E,F) Neurofilament staining of E11.5 embryos using a 2H3 antibody demonstrate that the trigeminal (V) ganglion is intact in Hmx1dm/dm embryos (control n=6, Hmx1dm/dm n=5). (G-J) Noticeable defects can be seen by 2H3 staining in the great auricular nerve (gan) and the auriculotemporal nerve (atn) that surround and innervate the pinna of Hmx1dm/dm embryos at both E12.5 (compare G with H, red dashed oval; control n=6, Hmx1dm/dm n=4) and E13.5 (compare I with J, red dashed oval; control n=8, Hmx1dm/dm n=2). Scale bars: 500 µm (E-J).
In order to determine whether Hmx1 directly contributes to either muscle or nerve development and organization, we performed immunohistochemistry for Isl1, a known marker for muscle precursors in BA2 (Nathan et al., 2008; Gopalakrishnan et al., 2015), and for neurofilament. Using dmECR-driven lacZ activity as a proxy for endogenous Hmx1 (see below), we found that Hmx1 was not present in muscle precursor cells (Fig. S2B). With the neurofilament staining, we consistently observed abnormalities in the branching and organization of the nerves in and surrounding the pinna in Hmx1dm/dm embryos at both E12.5 and E13.5 (Fig. 2G-J). Most prominently, the leaf-like branching of the great auricular nerve that penetrates the caudal aspects of the pinna was markedly reduced in Hmx1dm/dm embryos compared with controls (compare Fig. 2G,I with Fig. 2H,J). Differences in the auriculotemporal nerve, a somatosensory branch of the mandibular nerve, were also apparent (Fig. 2G-J). By contrast, the trigeminal (V) nerve was of normal appearance in mutants (Fig. 2E,F).
A 594 bp sequence containing an ECR and present within the region deleted in dumbo rats is sufficient to drive expression in lateral craniofacial tissue
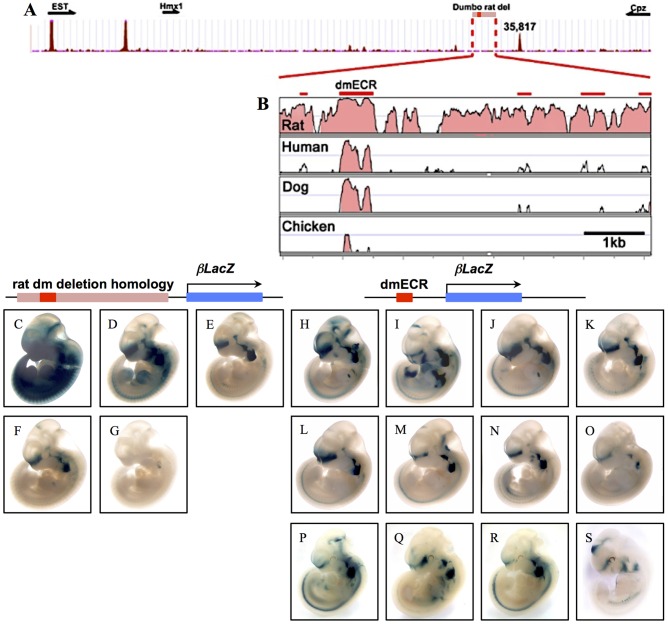
Previously, we showed that the deleted interval in the dumbo rat was associated with specific loss of Hmx1 expression in BA1 and BA2 mesenchyme (Quina et al., 2012a,b). This data supported the notion that the region harbors a putative regulatory element required for Hmx1 expression in lateral facial mesenchyme, accounting for the convergence of the mouse and rat dumbo ear phenotypes. To test whether this region functions as a cis-regulatory element (enhancer) sufficient to drive Hmx1 expression in the developing ear, we independently tested both a 6094 bp sequence from C57Bl/6 mouse genomic DNA that was homologous to the region deleted in the dumbo rat, as well as a smaller 594 bp fragment that just contained the ECR (dmECR) for enhancer activity using a lacZ reporter in transient transgenesis (Fig. 3C-O). All five transgenic embryos produced from the larger construct showed β-galactosidase staining in BA2 at E11.5 (Fig. 3C-G). Three of these embryos also exhibited staining in a strip of mesenchyme across the frontonasal region, in the dorsal BA1 mesenchyme and dorsal side of the eye (Fig. 3D-F). All eight of the transient transgenic embryos generated from the small dmECR fragment showed remarkably consistent staining (Fig. 3H-O) in the same regions observed in the embryos created using the larger construct: specifically, strong staining in BA2, the dorsal aspect of BA1, the mesenchyme dorsal to the eye, and in a strip of tissue across the frontonasal region. Thus, the 594 bp sequence encompassing the ECR is sufficient to direct expression to these key regions relevant to the observed craniofacial deformities in Hmx1dm/dm mice and rats.
Fig. 3.
Transient transgenic analysis identifies a functional ECR conferring facial-specific expression. (A) CTCF binding at 35,817 kb identifies an ‘insulator’ sequence separating Hmx1 regulatory sequences from regulatory elements in Cpz. (B) VISTA plot of the rat dumbo deletion region (5777 bp) showing the dmECR (thick red bar) and additional regions less strongly conserved across mammalian species (red bars). (C-G) E11.5 transient transgenic embryos generated from a 6094 bp construct, encompassing the 5777 bp rat deletion region, show staining in BA2 at E11.5. (H-S) E11.5 transient transgenic (H-O) and stable transgenic (P-S) lines generated from a 594 bp sequence, encompassing the ECR, show strong staining in the frontonasal region, in addition to the craniofacial mesenchyme of dorsal BA1, and completely throughout BA2.
We then generated stable transgenic lines for the dmECR-lacZ reporter. Staining of E11.5 embryos from four independent founders revealed a common pattern consistent with the results of the transient transgenesis experiments (Fig. 3P-S). In addition, each transgenic line exhibited a few additional regions of staining, possibly due to insertion site effects. These included the limbs, brain, heart, dorsal root ganglia and derivatives of the lateral plate mesoderm (Fig. 3P-S).
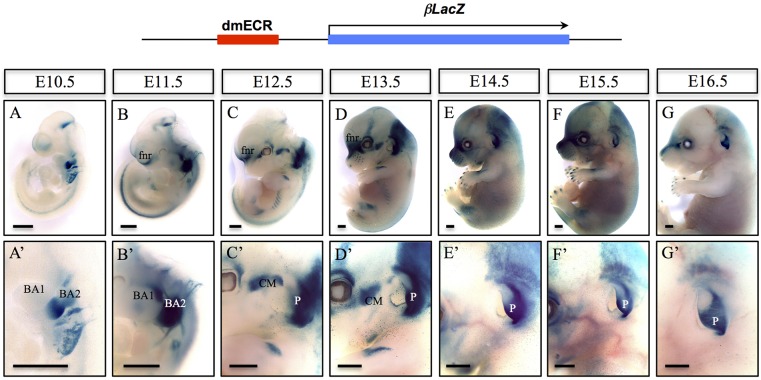
The activity of the dmECR-lacZ transgene was then analyzed at developmental stages ranging from E9.5 to E16.5 using the line displayed in Fig. 3P, although similar results were observed for the line shown in Fig. 3R (data not shown). This assessment showed minimal activity at E9.5 (data not shown). By E10.5, activation of the dmECR was readily observed in the frontonasal region and throughout BA2 (Fig. 4A,A′). At E11.5, expression expanded to include the dorsal region of BA1 (Fig. 4B,B′). From E12.5 to E13.5, the dmECR drove transgene expression in the frontonasal region and lateral facial mesenchyme, including tissue surrounding the eye and in a band of tissue at the back of the head that included the posterior region of the developing pinna (Fig. 4C-D′). The dmECR-lacZ transgene continued to be expressed in two broader domains within the head and face that became more regionally restricted and separate from one another between E14.5 and E16.5 (Fig. 4E,F,G); specifically, the rostral face, including the frontonasal region and tissue surrounding the eye, and a separate expression domain in the caudal region of the head, extending from the posteriorly restricted staining in the pinna to the back of the head (Fig. 4E-G′). This particular transgenic line also showed expression in the midbrain, hindbrain, zeugopod, both the fore- and hindlimb digits, ribs and the developing neural tube, which were not observed in all of the lines characterized, and may or may not be related to dmECR function.
Fig. 4.
dmECR-driven reporter gene activity shows dynamic expression from E10.5 to E16.5. (A-G′) Developmental series of E10.5 (A,A′), E11.5 (B,B′), E12.5 (C,C′), E13.5 (D,D′), E14.5 (E,E′), E15.5 (F,F′) and E16.5 (G,G′) embryo staining from the 594 bp dmECR transgenic line shown in Fig. 3P. CM, craniofacial mesenchyme; fnr, frontal nasal region; P, pinna. Scale bars: 1 mm.
To validate that the dmECR activity reflected endogenous Hmx1 expression, we sectioned E10.5, E11.5 and E14.5 dmECR-lacZ transgenic embryos and performed Hmx1 immunohistochemistry following X-gal staining. Analysis of the two-color staining patterns revealed that the dmECR and Hmx1 expression domains strongly overlapped in both the anterior (Fig. 5G,G′) and posterior (Fig. 5H) regions of BA2 at E10.5. At E11.5, β-galactosidase staining overlapped endogenous Hmx1 expression in the rostral zone of the lateral craniofacial mesenchyme (Fig. 5J), but was absent from the caudal zone of endogenous Hmx1 expression. At the level of the trigeminal ganglion, dmECR activity overlapped Hmx1 staining in the dorsal region of BA1 (Fig. 5K). As previously reported, endogenous Hmx1 is readily detectable in the trigeminal, geniculate and statoacoustic ganglia (Quina et al., 2012b), but the dmECR is not active in these regions (Fig. 5C,E,K), consistent with the retention of Hmx1 expression in the cranial sensory ganglia in the dumbo rat (Quina et al., 2012a). At the level of the embryonic eye, dmECR activity is present in the anterior mesoderm, whereas endogenous Hmx1 is expressed in the eye itself (Fig. 5B,D,L-M), consistent with the dumbo rat findings (Quina et al., 2012a). However, in the anterior craniofacial mesenchyme of BA2 at E11.5, dmECR activity and endogenous Hmx1 expression overlapped only in a band of cells at the interface between the expression domains (Fig. 5N). Moving posteriorly within the transverse plane, we found an increase in the cell populations that displayed both dmECR activity and Hmx1, although this was regionally restricted to the dorsal domain of BA2 (Fig. 5O). The restricted overlap between dmECR activity and Hmx1 in BA2 at E11.5 could represent an example of perdurance; that is, persistence of β-galactosidase activity from an earlier time point (i.e. precursor cell population) when endogenous Hmx1 was expressed but has since turned off.
Fig. 5.
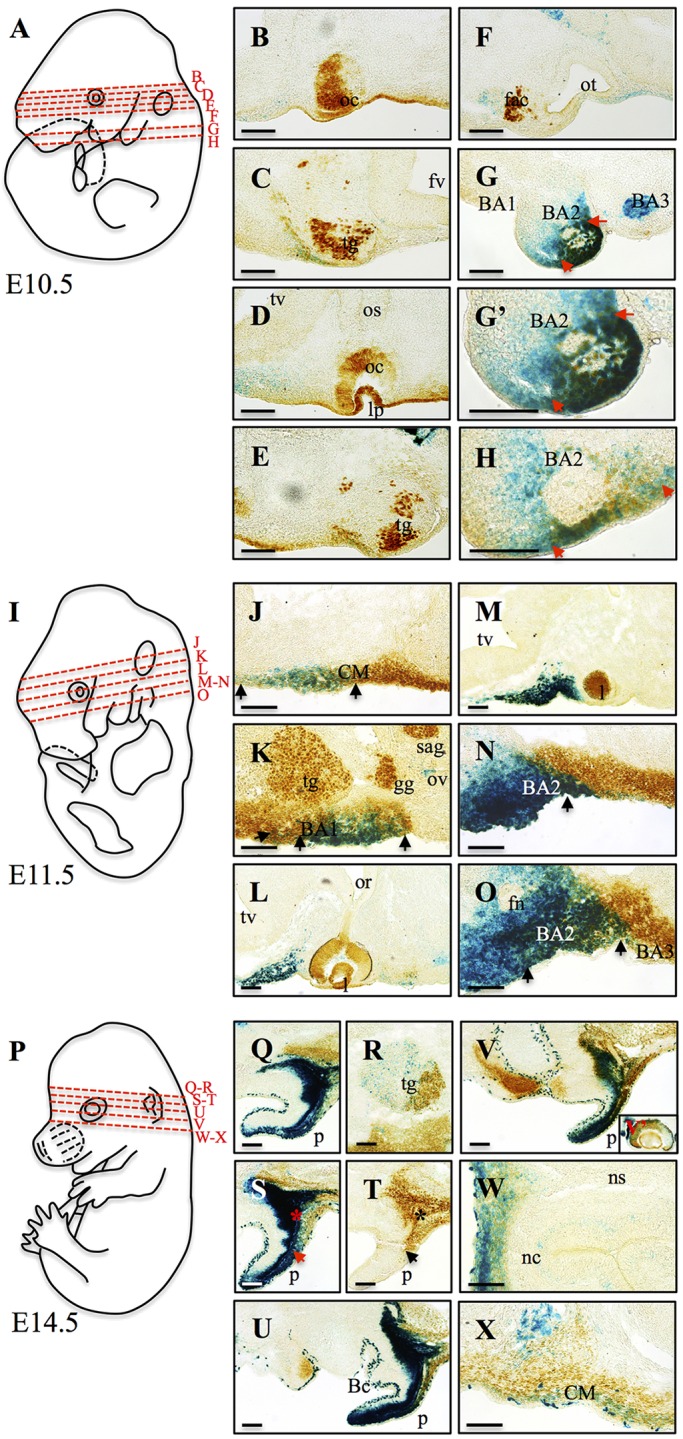
dmECR enhancer activity overlaps endogenous Hmx1 protein localization. (A,I,P) Diagrams of E10.5 (A), E11.5 (I) and E14.5 (P) mouse embryos highlighting the different transverse sections displayed in B-H (E10.5), J-O (E11.5) and Q-X (E14.5). (B-H,J-O,Q-X) Comparison of endogenous mouse Hmx1 expression (brown) with dmECR enhancer activity, visualized by staining with X-gal (blue), in transverse cryosections at E10.5, E11.5 and E14.5. (G-H) dmECR and Hmx1 expression domains overlap in the anterior (G,G′) and posterior (H) regions of the exterior portion of BA2 at E10.5 (red arrows). (J) At E11.5, sectioning showed overlap between dmECR staining in the craniofacial mesenchyme (CM) of the lateral face with the rostral CM expression of endogenous Hmx1 (black arrows). (K) E11.5 dmECR staining overlaps endogenous Hmx1 in dorsal BA1 (black arrows). (N) Few cells share dmECR and Hmx1 expression domains in the CM of the anterior region of BA2 (black arrow) at E11.5. (O) Posteriorly, dmECR and Hmx1 expression domains were regionally restricted to the dorsal domain of BA2 at E11.5 (black arrows). (Q,S-V) Adjacent to the ear, overlapping dmECR and Hmx1 staining is evident (asterisks). In the ear, the ventral dmECR activity was almost distinct from the small dorsal region of endogenous Hmx1 expression (arrows). (R) Overlap in dmECR activity and endogenous Hmx1 was evident in the caudal region of the trigeminal ganglion (tg). (V′-X) Overlap in dmECR and Hmx1 expression was also apparent in the eye (V′), the frontonasal region (W) and CM of the lateral face (X). BA1-3, branchial arches 1-3; Bc, branchial cleft; fac, facial acoustic complex; fn, facial nerve; fv, fourth ventricle; gg, geniculate ganglion; l, lens; lp, lens placode; nc, nasal capsule; ns, nasal septum; oc, optic cup; or, optic recess; os, optic stalk; ot, otic vesicle; ov, otic vesicle; p, pinna; sag, statoacoustic ganglion; tv, telencephalic vesicle. Scale bars: 100 µm.
Similar to that observed at E10.5 and E11.5, the dmECR pattern of activity at E14.5 overlapped with that of endogenous Hmx1, particularly in the pinna (Fig. 5Q-V) and frontonasal region (Fig. 5W). We also detected discordant expression of dmECR and Hmx1 in the trigeminal ganglion (Fig. 5R), eye (Fig. 5V′) and craniofacial mesenchyme of the lateral face (Fig. 5X), which could be a result of ectopic dmECR activity. Despite these regions of discordance, the distribution of the dmECR-driven reporter overlapped with endogenous Hmx1 in the craniofacial mesenchyme that contributes to structures that are abnormal in the dumbo rat (Fig. 1O-R) (Quina et al., 2012a).
Expansion of dmECR activity and endogenous Hmx1 expression in dumbo mutants
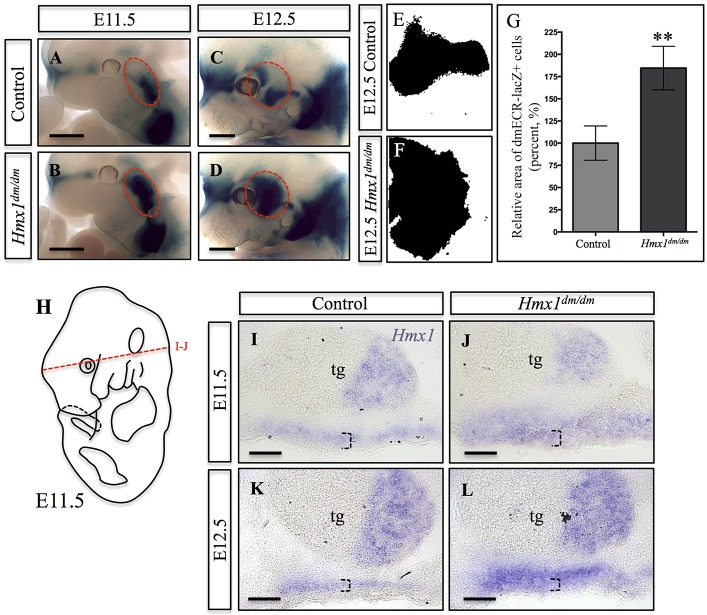
When we introduced the dmECR-lacZ reporter onto the dumbo (Hmx1dm/dm) background, we noticed a consistent expansion of the dmECR-driven β-galactosidase staining in the mesenchyme between the eye and ear of E11.5 to E14.5 Hmx1dm/dm embryos that was not seen in controls (Fig. 6A-D; data not shown). Quantification of this dmECR-stained region in E12.5 embryos (Fig. 6E,F) revealed an expansion of dmECR-lacZ+ tissue in mutant embryos to almost double that seen in controls (Fig. 6G). In situ hybridization in E11.5 and E12.5 Hmx1dm/dm embryos and controls (Fig. 6I-L) revealed that endogenous Hmx1 mRNA expression was also expanded (compare Fig. 6I,K with 6J,L), confirming that this was not just a phenomenon associated with the dmECR and suggested that Hmx1 negatively regulates its own expression via the dmECR. Proliferation (Ki67) and cell death (cleaved caspase 3) remained comparable between control and Hmx1dm/dm embryos in this region (Fig. S3A-I).
Fig. 6.
dmECR enhancer activity and Hmx1 expression are expanded in Hmx1dm/dm mutants. (A-D) dmECR staining in control (A,C) and Hmx1dm/dm mutant (B,D) embryos at E11.5 (A,B; control n=5, Hmx1dm/dm mutant n=7) and E12.5 (C,D; control n=4, Hmx1dm/dm mutant n=4) shows an expansion of the dmECR staining pattern in Hmx1dm/dm mutants (red dashed circle). (E,F) Representative binary particle area diagrams from ImageJ displaying dmECR-lacZ+ cells present in the E12.5 face of controls (E) and Hmx1dm/dm mutant (F) embryos. (G) Quantification of the area displayed in E and F for E12.5 controls (n=4) and Hmx1dm/dm mutant (n=4) embryos (plotted values are the mean±s.e.m., **P=0.0016). (H) Diagram of an E11.5 mouse embryo highlighting the transverse plane displayed in I and J. (I-L) Hmx1 in situ on control (I,K) and Hmx1dm/dm mutant (J,L) E11.5 (I,J; n=2) and E12.5 (K,L; n=2) transverse sections show an expansion of Hmx1 transcripts in Hmx1dm/dm mutants (compare I with J, and K with L, black dashed brackets). tg, trigeminal ganglion. Scale bars: 1 mm (A-D); 100 µm (I-L).
Hox, Pbx and Meis function cooperatively to activate the dmECR
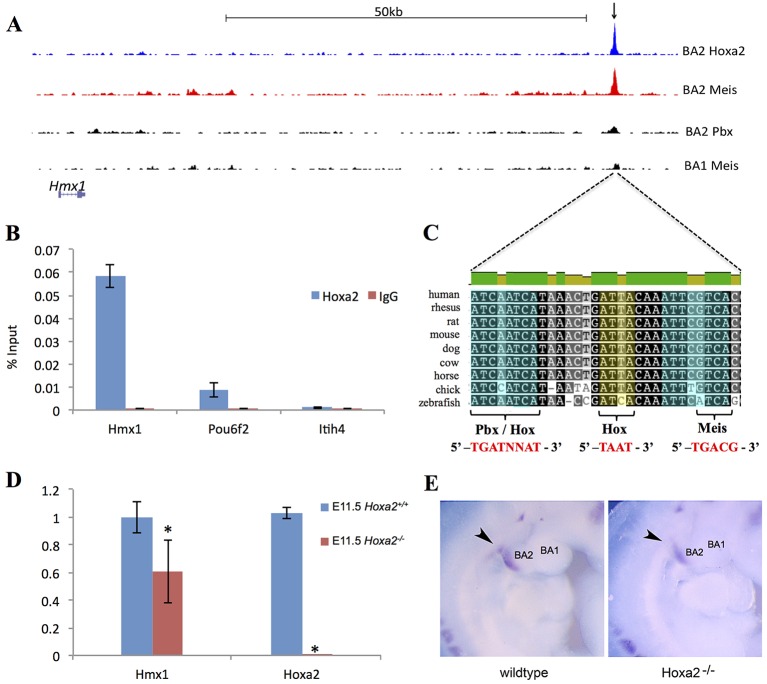
Examination of the dmECR sequence revealed consensus binding site sequences for the combined Hox-Pbx complex (Amin et al., 2015) within the most highly conserved part of the dmECR (Fig. 7). Hoxa2 and Pbx1, as well as their partner Meis proteins (Gendron-Maguire et al., 1993; Rijli et al., 1993; Selleri et al., 2001) are known to be expressed in the early lateral facial mesenchyme and contribute to BA2 morphogenesis. To investigate whether the dmECR is occupied by the Hox-Pbx-Meis complex in vivo, we examined Hoxa2 and Meis binding surrounding the Hmx1 gene, in chromatin isolated from E11.5 BA1 and BA2 tissue, as described previously (Amin et al., 2015). Hoxa2 and Meis binding profiles in BA2 showed overlapping prominent peaks ∼80 kb downstream of the Hmx1 gene (Fig. 7A), corresponding to the location of the dmECR. Enhanced Meis binding was observed in BA2 tissue relative to the Hox-negative BA1, a feature associated with Hoxa2 functional targets (Fig. 7A) (Amin et al., 2015). Chromatin immunoprecipitation (ChIP)-qPCR on BA2 chromatin revealed nearly tenfold greater enrichment of the Hmx1 dmECR enhancer region (Fig. 7B) than the established positive control (Amin et al., 2015), confirming Hoxa2 binding to the core dmECR sequence. Moreover, analysis of microarray data obtained from E11.5 Hoxa2 mutant and control BA2 tissue (Donaldson et al., 2012) showed a 1.6-fold downregulation of Hmx1 expression (P<0.005), similar to that of Meis2, a known target of Hoxa2 (Amin et al., 2015). This suppression of Hmx1 expression in E11.5 Hoxa2 mutant embryos was validated by qRT-PCR (Fig. 7D). In situ hybridization for Hmx1 on E10.5 embryos also supported reduced caudal BA2 expression in Hoxa2 mutants compared with controls (Fig. 7E).
Fig. 7.
Hoxa2 binds to the dmECR in vivo and controls Hmx1 expression. (A) ChIP-seq binding profile of Hoxa2, Meis2 and Pbx at Hmx1 gene in chromatin isolated from E11.5 first (BA1) and second (BA2) branchial arches. Black arrow highlights the binding region tested by qPCR in B. (B) Analysis of Hoxa2 binding to Hmx1 enhancer by ChIP-qPCR. Enrichment is calculated as percent input. IgG is a negative control antibody. Pou6f2 is a positive control and Itih4 is a negative control. (C) Consensus binding sites within the dmECR are indicated on the 32 bp multispecies alignment, which includes multiple mammalian species as well as chick and zebrafish, and highlights potential Hox-Pbx (left, blue), Hox (yellow) and Meis (right, blue) binding sites (Amin et al., 2015). (D) qRT-PCR validates diminished Hmx1 expression in E11.5 Hoxa2 mutant embryos (five technical replicates; Hmx1 P=0.0155, Hoxa2 P=0.0001). (E) Whole-mount in situ hybridization for Hmx1 on E10.5 embryos shows reduced caudal BA2 expression in Hoxa2 mutants (right panel, arrowhead) compared with controls (left panel, arrowhead). Data are mean±s.d.
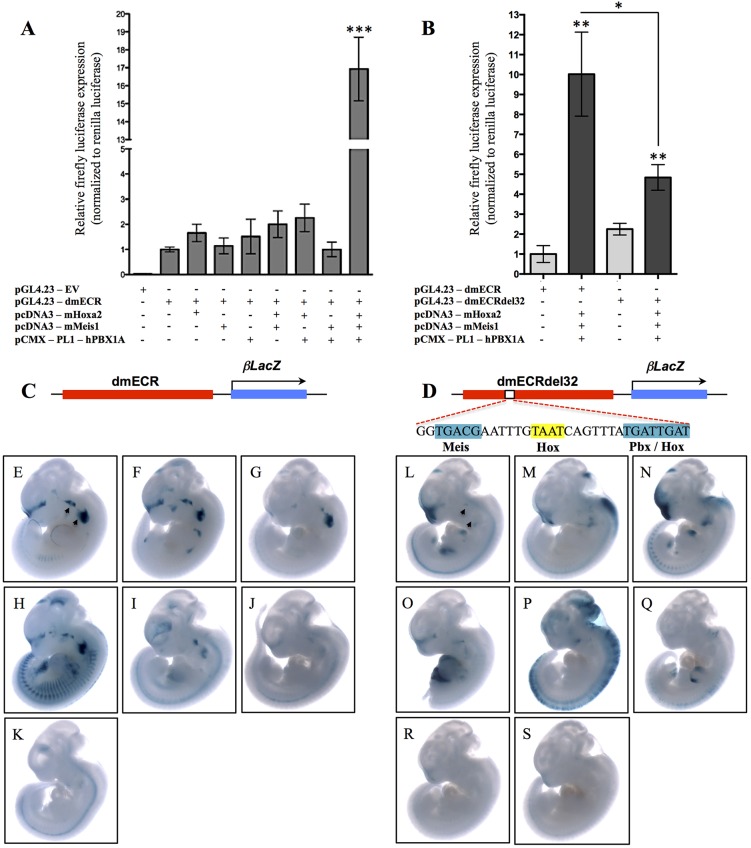
To assess whether the Hox-Pbx-Meis complex could transactivate the dmECR in a heterologous cell type, we transiently transfected COS-1 cells with a construct, pGL4.23-dmECR, containing a luciferase reporter cassette in which the dmECR was inserted upstream of the minimal promoter. The dmECR reporter was co-transfected with expression vectors encoding mHoxa2, mMeis1 and hPBX1A individually and in combination. Co-transfection of these transcription factors individually and in pairwise combinations did not result in a significant change in the levels of luciferase activity compared with transfection with the pGL4.23-dmECR plasmid alone (Fig. 8A). However, the addition of all three expression vectors resulted in a ∼17-fold increase in the levels of luciferase activity over basal levels (Fig. 8A). Deletion of the 32 bp sequence encompassing the binding site consensus sequences for Hox, Pbx and Meis (Fig. 7C) from the 594 bp construct (pGL4.23-dmECRdel32) resulted in a severe reduction in enhancer activity (Fig. 8B). Most importantly, repeated transient transgenesis using the 594 bp dmECR construct conducted in parallel with the newly generated dmECRdel32 construct (Fig. 8C,D) demonstrated that the 32 bp sequence containing the Hox, Pbx and Meis binding sites is required for the enhancer to express in the dorsal aspect of BA1 and throughout BA2 (5/7 embryos versus 0/8 embryos; compare Fig. 8E-K with 8L-S). In addition, we noted that deletion of the 32 bp sequence also resulted in an expansion in two of eight embryos of the strong frontonasal expression domain rostrally into the medial and lateral nasal processes (Fig. S4H,I). A further four embryos showed weak rostral expansion with a diminution or loss of the frontonasal domain (Fig. S4J,K,M,N). Taken together, these data provide both in vivo and in vitro support for direct regulation of the Hmx1 gene in BA2 by the Hox-Pbx-Meis complex via the dmECR.
Fig. 8.
Hox-Pbx-Meis cooperatively bind to and regulate the dmECR via a 32 bp sequence. (A) Luciferase activity (relative firefly/Renilla levels) for the 594 bp dmECR sequence following transfection with the pcDNA3-mHoxa2, pcDNA3-mMeis1 or pCMX-PL1-hPBX1A expression vectors showed strong activation only in the presence of Hoxa2, Meis1 and PBX1A proteins (replicates n=3, Hoxa2 P=0.1386, Meis1 P=0.6913, PBX1A P=0.4998, Hoxa2+Meis1 P=0.1327, Hoxa2+PBX1A P=0.0862, Meis1+PBX1A P=0.9959, Hoxa2+Meis1+PBX1A ***P=0.0009). (B) Luciferase activity for the intact 594 bp dmECR sequence compared with the dmECRdel32 construct harboring the 32 bp deletion of the adjacent Hox-Pbx-Meis binding sites (see D). A significant reduction in enhancer activation from the dmECRdel32 construct compared with the intact dmECR construct was seen in the presence of Hoxa2, Meis1 and PBX1A proteins (replicates n=3, pGL4.23-dmECR+Hoxa2+Meis1+PBX1A P=0.0019, pGL4.23-dmECRdel32+Hoxa2+Meis1+PBX1A P=0.0031; pGL4.23-dmECR compared with pGL4.23-dmECRdel32 P=0.0152). (C-S) Transient transgenesis for the intact 594 bp dmECR construct (C) compared with the dmECRdel32 construct (D) showed almost complete loss of BA1/2 staining in E11.5 transient transgenic embryos generated from the dmECRdel32 construct (L-S, arrowheads) compared with the consistency of activity in these regions in transient transgenic embryos generated from the intact dmECR construct (E-K, arrowheads).
DISCUSSION
The distinctive lateral external ear placement that characterizes the dumbo phenotype is one of the most recognizable mutant presentations both in mice and in rats, and has made the dumbo rat a favorite amongst pet owners. However, Hmx1-related phenotypes have also been described in humans (OAS) and cattle (crop ear). In contrast to the loss-of-function mutations found in mice and humans, the mutations at the Hmx1 locus in rats and cattle implicate a putative regulatory region located ∼80 kb 3′ of the last exon (Quina et al., 2012a; Koch et al., 2013). At this site, we previously identified a highly conserved ∼600 bp segment (dmECR) that we hypothesized could represent a cis-regulatory enhancer of Hmx1 expression (Turner and Cox, 2014). In this article, we demonstrate that the homologous mouse ECR sequence functions as a bona fide distal enhancer containing information required to direct expression to defined structures of the developing lateral face, including the dorsal aspect of BA1, BA2, tissue caudal to the eye, and to a specific region of frontonasal mesenchyme. In the anterior branchial arches, the dmECR provides a regulatory link between patterning of the embryonic axis by classic homeotic factors and morphogenesis of the external ear.
Lateral cranioskeletal anomalies in dumbo mice are suggestive of a combination of primary and secondary defects
During our assessment of Hmx1dm/dm embryos, we identified previously unappreciated features of the dumbo phenotype. We noted an expansion of the central BA2-derived auricular hillock at E11.5 that leads to a bifurcated appearance of the caudal aspect of the developing pinnae at older embryonic ages. In the adult mutant, this highly penetrant trait is apparent as an ectopic cartilaginous flap on the back of the pinna. This may represent a simple duplication of a portion of the pinna structure or even a regionally restricted homeotic transformation. The additional postnatal cranioskeletal anomalies that we have described may reflect a combination of primary and secondary effects. It is unlikely that BA-derived muscles have a cell-autonomous requirement for Hmx1, as we have shown that dmECR activity and Hmx1 protein expression do not overlap the staining of Isl1, a marker for mesodermally derived muscle precursors in BA1 and BA2 (Nathan et al., 2008; Gopalakrishnan et al., 2015), which give rise to many muscles of the head and neck (Rinon et al., 2007). However, in dumbo animals, muscle attachment sites, such as the paired bony paraoccipital processes, are significantly hypoplastic. The paraoccipital processes, which are functionally analogous to the mastoid process in humans, are points of attachment of some of the major muscles of mastication and lateral head movement. Given that it is well established that mechanical strain forces from mastication can affect development of the bony mandibular angular process and patency of the cranial sutures (de Jong et al., 2011; Guerreiro et al., 2013; Rafferty et al., 2007), the altered shape of the mandibular angle and premature closure of the temporal sutures in dumbo mice are likely to be secondary to decreased biomechanical function. Additionally, we confirm here that dumbo mice exhibit nerve branching and organization defects of some lateral sensory facial nerves that innervate the pinnae and surrounding tissue, extending our prior findings (Quina et al., 2012b). Thus, the functional deficit in ear movement that we observed in dumbo mice could be the result of musculoskeletal defects, sensory defects, or a combination of both. Future fate-mapping experiments and more precise tissue-specific disruption of Hmx1 should help resolve the cellular contributions to the dumbo phenotype.
The 594 bp distal ECR functions as a facial mesenchyme-specific enhancer that is regulated in BA2 by the Hox-Pbx-Meis complex
The reproducible and restricted pattern of activity of the dmECR is strikingly similar to the activity of enhancers recently identified through a genome-wide analysis of Hox and Pbx transcriptional targets (Amin et al., 2015). Indeed, Hoxa2, Pbx1 and their binding partners, the Meis proteins, all share overlapping expression domains with Hmx1 in the craniofacial mesenchyme of BA1 and BA2 during development (Amin et al., 2015), and knockout of each results in a more severe ear malformation than that seen in the dumbo mice (Minoux et al., 2013; N.B. and T.C.C., unpublished). Consistent with this, we identified putative cis-binding sites for homeobox proteins and the Hox-Pbx complex within the dmECR (see Fig. 7C). These sites are found in the most conserved portion of the dmECR, shared not only among mammalian species but also in chick, frogs and zebrafish, albeit with one or two nucleotide differences in these latter species. The likely importance of these binding sites in mammals is emphasized by the crop ear cattle allele, which is a 76 bp duplication (Koch et al., 2013) involving this central core containing the Hox-Pbx-Meis binding motifs.
Consistent with the presence of predicted binding sites, genome-wide occupancy of Hoxa2 and Meis in vivo shows that both Hoxa2 and Meis bind to the dmECR in vivo. Moreover, our co-transfection analysis has shown that the dmECR enhancer is strongly activated in the presence of Hoxa2, Meis1 and PBX1A. Deletion of a 32 bp region containing the core Hox-Pbx-Meis binding sites within the 594 bp dmECR led to a significant reduction in enhancer activation. This was also confirmed using transient transgenesis where deletion of the 32 bp region containing the core Hox-Pbx-Meis binding sites resulted in the loss of enhancer expression in the dorsal aspect of BA1 and throughout BA2. These findings validate the earlier presumption that disruption of the ECR in both the dumbo rat and the crop ear cattle is the cause of the phenotypic abnormalities in these species. Together, these results place Hmx1 immediately downstream of the early branchial arch patterning genes, as one of the few validated targets of Hoxa2.
Hmx1 and the dmECR: conduits to elucidating lateral facial gene networks
The combined findings of additional lateral facial anomalies in dumbo mutants (mice and rats) and a critical distal enhancer of Hmx1 that is regulated by the Hox-Pbx-Meis complex, position Hmx1 as an early target of branchial arch patterning factors. In this regard, the dmECR is therefore likely to provide a unique conduit through which we will be able to elucidate additional upstream transcriptional regulators that help pattern the early arch and frontonasal mesenchyme. Candidates include members of the Six and Eya families (Cox et al., 2014), and both Tbx15 and Sall1, given the phenotypic presentation of auricular malformations in mice and humans when these genes are mutated (Cox et al., 2014). Furthermore, identification of downstream targets of Hmx1 itself, via gene expression studies in the dumbo mouse, should aid in the identification of specifiers of lateral facial tissues including the pinnae and their supporting structures as well as the facial sensory nerves innervating this region. Both approaches should ultimately identify new candidate genes for disorders involving lateral facial structures: upstream regulators may represent candidates for more severe malformations, while cases of ear malformation that occur without extensive changes in other branchial arch structures may lie immediately downstream of HMX1 (Cox et al., 2014). In fact, late-acting genes such as HMX1 are likely to be more relevant to pediatric practice as mild auricular and lateral facial anomalies, including facial palsies, collectively represent the third most commonly presenting group of conditions in craniofacial centers in the USA (Luquetti et al., 2012) and yet there is still little known about their underlying genetic basis. Given that mutations in the dmECR underlie isolated phenotypic presentations in multiple non-human mammalian species, this warrants inclusion of enhancers such as this in future mutational analysis of relevant patient cohorts.
MATERIALS AND METHODS
Animals and genotyping
Mice harboring the dumbo allele (Hmx1+/dm) were generated from the original B6:C3Fe-Hmx1dmbo/Rw/JcsJKjn mice, maintained on a C57Bl/6N background, and genotyped as described previously (Quina et al., 2012a,b). Hoxa2 mutant mice were described previously (Gendron-Maguire et al., 1993). We designated E0.5 to be noon on the day a plug was detected. Maintenance and genotyping of dumbo rats was also performed as previously described (Quina et al., 2012a). All animal use was approved by the appropriate Institutional Animal Care and Use Committee. All data were collected from at least three animals of a particular genotype at each gestational and postnatal time point described, unless otherwise stated. Body weights were measured postmortem.
Generation of transgenic mice
The 6094 bp fragment homologous to the sequence deleted in dumbo rats was PCR amplified from mouse genomic DNA using the NotI- and AscI-linkered oligonucleotides: dmbodelF, GGGATCGCGGCCGCGTGCACCATCTTTGAGGACTTAG; dmbodelR, GATCGGCGCGCCGTAGGGAAGCTGAGGCCAAG. The 594 bp ECR (hereafter called dmECR) was amplified using the NotI- and AscI-linkered oligonucleotides: dmECR-F, GGGATCGCGGCCGCGAATCCTGGCCAGTCAGTGTA; dmECR-R, GATCGGCGCGCCGGCTTGGGGGTGGCAAACTG. The underlined bases indicate the incorporated restriction endonuclease binding sites. The 6094 bp and 594 bp fragments were separately inserted into the Hsp68-lacZ-Gateway vector, and the resulting vectors linearized for injection using NotI. Transgenic mice were produced by pronuclear injection into C57Bl/6J or CD-1 single-cell embryos using standard techniques (Nagy et al., 2003). The 594 bp transgenic construct was also subsequently used to generate four stable lines (CH-5791, CH-5820, CH-5821 and CH-5831). Transgenic mice and embryos were genotyped by real-time PCR using transgene-specific oligonucleotides: LacZp1F, GCTGGATCAAATCTGTCGATCCTT and LacZp1R, CGCGTACATCGGGCAAATAATATC (95 bp product); LacZp2F, ATAGCGATAACGAGCTCCTGCACT and LacZp2R, ACTGTTTACCTTGTGGAGCGACATC (99 bp product).
X-gal and whole-mount neurofilament staining
Embryos with lacZ transgenes were stained with X-gal according to standard techniques (Nagy et al., 2003). Antibodies recognizing neurofilaments (2H3) were used to visualize nerves in E11.5-E13.5 embryos, as previously described (Vickerman et al., 2011). The 2H3 antibody was developed by T. Jessel and J. Dodd (Dodd et al., 1988) and was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of NICHD and maintained by the University of Iowa, Department of Biology, Iowa City, IA 52242, USA.
Immunohistochemistry
Embryos to be used for immunohistochemistry were prepared as previously described (Rosin et al., 2015). Cryosections (16-20 μm) were exposed to either anti-Hmx1 (1:2000; Quina et al., 2012b), anti-Isl1 (1:500; AB20670, Abcam), anti-cleaved caspase 3 (1:1200; 9664S, Cell Signaling) or anti-Ki67 (1:200; RM-9106 S0, Thermo Scientific) at 4°C overnight, then washed with PBS and exposed to secondary antibody (1:300; biotinylated donkey anti-rabbit; 711-065-152, Jackson ImmunoResearch) for 2 h at room temperature. This was followed by a 30 min incubation with Vectastain Elite ABC reagent (Vector Labs), and DAB Peroxidase (HRP) substrate color development for 5 min (Vector Labs). Images were captured on a Leica 4000B microscope. Brightness and/or contrast of the entire image were adjusted using Adobe Photoshop CS5.1 if deemed appropriate.
In situ hybridization and qPCR
Cryosections (16-20 μm) were exposed to a digoxigenin (DIG)-labeled Hmx1 riboprobe (1:300; Wang et al., 1998) at 68°C overnight. DIG-labeled riboprobes were detected with an alkaline phosphatase-conjugated anti-DIG antibody (11093274910, Roche) and BM Purple AP (Roche) was used as the color development substrate. BA2 of E11.5 embryos from Hoxa2+/− intercrosses were dissected out and snap-frozen in dry ice. After genotyping the embryos, pools were made with the wild-type and Hoxa2−/− BA2, and total RNA was extracted using Trizol. The sequences of the primers used in qPCR are: Hmx1 Fwd, 5′-CGGCTGCGGAGGTACAA-3′; Hmx1 Rev, 5′-AGTCCCGGTCGCTTGTG-3′; Hoxa2 Fwd, 5′-GCCTCGGCCACAAAGAA-3′; Hoxa2 Rev, 5′-CGGCGATTTCCACCCTGCG-3′.
OPT and microCT imaging
All tomographic imaging was conducted in the Small Animal Tomographic Analysis (SANTA) Facility at Seattle Children's Research Institute. Samples for OPT were prepared and imaged as described by Zovein et al. (2010). MicroCT imaging was performed at an isotropic resolution of 17.21 µm using a Skyscan 1076 scanner with the following settings: 55 kV, 180 µA, 1.0 mm Al filter, 360 ms exposure, 0.7° rotation step, and three-frame averaging. Raw OPT and microCT scan data were reconstructed using NRecon V1.6 software (Skyscan, Belgium) and rendered in 3D using the Drishti software V2.6 (Limaye, 2012). Reconstructed data were imported into Analyze 10.0 (Mayo Clinic) for mandible and skull segmentation and comprehensive shape analysis performed as previously described (Rolfe et al., 2013).
Luciferase assays
The dmECR and dmECRdel32 fragments were PCR amplified from genomic DNA using the KpnI and HindIII-linked oligonucleotides: ECR-F, GTGAGGTACCGAAGCCAGTCAGTGTA; ECR-R, CAGAAGCTTCTTGGGGGTGGCAAA (32 bp deletion primers: ECRdel32-F, CTGGAAACTCGGCTTCTGTTCACAAG and ECRdel32-R, CAGAAATTGATTCTCCAGAAAGGCAG) and separately ligated into the pGL4.23 firefly luciferase reporter plasmid (Promega) using standard techniques. The vectors were co-transfected into COS-1 cells (ATCC CRL-1650, passage 39) with either an empty pcDNA3.2 vector or in different combinations with the expression vectors pcDNA3-mHoxa2, pcDNA3-mMeis1 and/or pCMX-PL1-hPBX1A using Polyjet (SignaGen) according to the manufacturer's recommendations. Passive lysis buffer (PLB, Promega; 100 μl) was added to each well during collection. Firefly (LARII, Promega) and Renilla (Stop & Glo reagent, Promega) luciferase activities were assayed using a Monolight 2010 luminometer (Analytical Luminescence Laboratory, CA, USA). All assays were performed in triplicate.
Chromatin immunoprecipitation
ChIP for Hoxa2, Meis2 and Pbx were performed as described previously (Amin and Bobola, 2014). ChIP-qPCR analysis of Hoxa2 binding to the dmECR was accomplished using the oligonucleotides: Fwd, TCGAGCTCATAGCGCTTTT; Rev, GGGGGAGATAAAGTGAAACACAT. Enrichment was calculated as percent input, with IgG used as a negative control. Pou6f2 was used as a positive control and Itih4 as a negative control (Donaldson et al., 2012). The Hoxa2 antibody used for ChIP was described previously (Kutejova et al., 2008).
Behavioral analysis
Adult (P60+ mice) were recorded before, during and after they were exposed to a 10 s constant-pressure puff of air to determine if they could respond by (1) closing their eyes, and (2) tucking their ears back against their head. This was repeated three times for each animal.
Quantification methods and statistical analysis
Quantitative results for body weight, stained area, cell counts and luciferase activity are represented by mean scores±s.e.m. and were analyzed by two-tailed unpaired t-tests using Prism 3 (GraphPad Software). Genotype ratios were analyzed using a χ2 test.
Acknowledgements
The authors thank L. Quina, M. Deng and N. Kim for supplying antibodies; L. Quina and M. Deng for additional experimental assistance; I. Skuplik and J. Cobb for supplying luciferase vectors; V. Afzal for help with transgenic mouse experiments; and S. Vora and E. Camci for critical discussion. We are thankful to the National BioResource Project - Rat for providing dumbo rats (KFRS4/Kyo).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
The study was conceived and supervised by T.C.C. and E.E.T.; J.M.R. performed all experiments not otherwise mentioned; W.L. characterized the dumbo phenotype, and performed the initial transgenic work and behavioral analysis with academic and technical assistance from L.L.C.; S.M.R. did the shape analysis studies; V.L. and N.B. performed and contributed all ChIP data and analyses in the Hoxa2 embryos; J.A.A. and A.V. generated the transient transgenics; T.K. assisted with the rat studies. J.M.R., T.C.C. and E.E.T. wrote the manuscript with edits from all authors.
Funding
This work was supported in part by the Laurel Foundation Endowment for Craniofacial Research (T.C.C.); the National Institutes of Health (NIH) [R01-NS064993 to E.E.T.; R01-HG003988, U54-HG006997 and U01-DE024427 to A.V.]; a Seattle Children's Research Institute InterCenter Grant (T.C.C. and E.E.T.); and the Medical Research Council UK [MR/L009986/1 to N.B.]. Research conducted at the E.O. Lawrence Berkeley National Laboratory is performed under U.S. Department of Energy contract DE-AC02-05CH11231, University of California. J.M.R. and S.M.R. are supported by postdoctoral training fellowships from the Canadian Institutes of Health Research [MEF-140891] and the National Institute of Dental and Craniofacial Research [F32 DE025519], respectively. W.L. is supported by a University of Washington, School of Dentistry Institutional Trainee Award [R90 DE023059]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.133736.supplemental
References
- Alasti F., Sadeghi A., Sanati M. H., Farhadi M., Stollar E., Somers T. and Van Camp G. (2008). A mutation in HOXA2 is responsible for autosomal-recessive microtia in an Iranian family. Am. J. Hum. Genet. 82, 982-991. 10.1016/j.ajhg.2008.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S. and Bobola N. (2014). Chromatin immunoprecipitation and chromatin immunoprecipitation with massively parallel sequencing on mouse embryonic tissue. Methods Mol. Biol. 1196, 231-239. 10.1007/978-1-4939-1242-1_14 [DOI] [PubMed] [Google Scholar]
- Amin S., Donaldson I. J., Zannino D. A., Hensman J., Rattray M., Losa M., Spitz F., Ladam F., Sagerström C. and Bobola N. (2015). Hoxa2 selectively enhances Meis binding to change a branchial arch ground state. Dev. Cell 32, 265-277. 10.1016/j.devcel.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel B. R., Tvrdik P., Gaufo G. O. and Capecchi M. R. (2004). Hoxb1 functions in both motoneurons and in tissues of the periphery to establish and maintain the proper neuronal circuitry. Genes Dev. 18, 1539-1552. 10.1101/gad.1207204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox T. C., Camci E. D., Vora S., Luquetti D. V. and Turner E. E. (2014). The genetics of auricular development and malformation: new findings in model systems driving future directions for microtia research. Eur. J. Med. Genet. 57, 394-401. 10.1016/j.ejmg.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong W. C., Korfage J. A. M. and Langenbach G. E. J. (2011). The role of masticatory muscles in the continuous loading of the mandible. J. Anat. 218, 625-636. 10.1111/j.1469-7580.2011.01375.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd J., Morton S. B., Karagogeos D., Yamamoto M. and Jessell T. M. (1988). Spatial regulation of axonal glycoprotein expression on subsets of embryonic spinal neurons. Neuron 1, 105-116. 10.1016/0896-6273(88)90194-8 [DOI] [PubMed] [Google Scholar]
- Donaldson I. J., Amin S., Hensman J. J., Kutejova E., Rattray M., Lawrence N., Hayes A., Ward C. M. and Bobola N. (2012). Genome-wide occupancy links Hoxa2 to Wnt-beta-catenin signaling in mouse embryonic development. Nucleic Acids Res. 40, 3990-4001. 10.1093/nar/gkr1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron-Maguire M., Mallo M., Zhang M. and Gridley T. (1993). Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell 75, 1317-1331. 10.1016/0092-8674(93)90619-2 [DOI] [PubMed] [Google Scholar]
- Gillespie R. L., Urquhart J., Lovell S. C., Biswas S., Parry N. R. A., Schorderet D. F., Lloyd I. C., Clayton-Smith J. and Black G. C. (2015). Abrogation of HMX1 function causes rare oculoauricular syndrome associated with congenital cataract, anterior segment dysgenesis, and retinal dystrophy. Invest. Ophthalmol. Vis. Sci. 56, 883-891. 10.1167/iovs.14-15861 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S., Comai G., Sambasivan R., Francou A., Kelly R. G. and Tajbakhsh S. (2015). A cranial mesoderm origin for esophagus striated muscle. Dev. Cell 34, 694-704. 10.1016/j.devcel.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Guerreiro F. d. S., Diniz P., Carvalho P., Ferreira E., Avancini S. R. P. and Ferreira-Santos R. I. (2013). Effects of masticatory hypofunction on mandibular morphology, mineral density and basal bone area. Braz. J. Oral Sci. 12, 205-212. 10.1590/S1677-32252013000300010 [DOI] [Google Scholar]
- Koch C. T., Bruggmann R., Tetens J. and Drögemüller C. (2013). A non-coding genomic duplication at the HMX1 locus is associated with crop ears in highland cattle. PLoS ONE 8, e77841 10.1371/journal.pone.0077841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto T., Yokoe M., Yagasaki K., Kawaguchi T., Kumafuji K. and Serikawa T. (2010). Genetic analyses of fancy rat-derived mutations. Exp. Anim. 59, 147-155. 10.1538/expanim.59.147 [DOI] [PubMed] [Google Scholar]
- Kutejova E., Engist B., Self M., Oliver G., Kirilenko P. and Bobola N. (2008). Six2 functions redundantly immediately downstream of Hoxa2. Development 135, 1463-1470. 10.1242/dev.017624 [DOI] [PubMed] [Google Scholar]
- Limaye A. (2012). Drishti: a volume exploration and presentation tool. Proc. SPIE 8506, Developments in X-Ray Tomography VIII, 85060X 10.1117/12.935640 [DOI] [Google Scholar]
- Luquetti D. V., Heike C. L., Hing A. V., Cunningham M. L. and Cox T. C. (2012). Microtia: epidemiology and genetics. Am. J. Med. Genet. A 158A, 124-139. 10.1002/ajmg.a.34352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M., Kratochwil C. F., Ducret S., Amin S., Kitazawa T., Kurihara H., Bobola N., Vilain N. and Rijli F. M. (2013). Mouse Hoxa2 mutations provide a model for microtia and auricle duplication. Development 140, 4386-4397. 10.1242/dev.098046 [DOI] [PubMed] [Google Scholar]
- Munroe R. J., Prabhu V., Acland G. M., Johnson K. R., Harris B. S., O'Brien T. P., Welsh I. C., Noden D. M. and Schimenti J. C. (2009). Mouse H6 Homeobox 1 (Hmx1) mutations cause cranial abnormalities and reduced body mass. BMC Dev. Biol. 9, 27 10.1186/1471-213X-9-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K. and Behringer R. R. (2003). Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, New York, USA: Cold Spring Harbor Press. [Google Scholar]
- Nathan E., Monovich A., Tirosh-Finkel L., Harrelson Z., Rousso T., Rinon A., Harel I., Evans S. M. and Tzahor E. (2008). The contribution of Islet1-expressing splanchnic mesoderm cells to distinct branchiomeric muscles reveals significant heterogeneity in head muscle development. Development 135, 647-657. 10.1242/dev.007989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quina L. A., Kuramoto T., Luquetti D. V., Cox T. C., Serikawa T. and Turner E. E. (2012a). Deletion of a conserved regulatory element required for Hmx1 expression in craniofacial mesenchyme in the dumbo rat: a newly identified cause of congenital ear malformation. Dis. Model. Mech. 5, 812-822. 10.1242/dmm.009910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quina L. A., Tempest L., Hsu Y.-W. A., Cox T. C. and Turner E. E. (2012b). Hmx1 is required for the normal development of somatosensory neurons in the geniculate ganglion. Dev. Biol. 365, 152-163. 10.1016/j.ydbio.2012.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafferty K. L., Sun Z., Egbert M., Bakko D. W. and Herring S. W. (2007). Changes in growth and morphology of the condyle following mandibular distraction in minipigs: overloading or underloading? Arch. Oral Biol. 52, 967-976. 10.1016/j.archoralbio.2007.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijli F. M., Mark M., Lakkaraju S., Dierich A., Dollé P. and Chambon P. (1993). A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell 7, 1333-1349. 10.1016/0092-8674(93)90620-6 [DOI] [PubMed] [Google Scholar]
- Rinon A., Lazar S., Marshall H., Buchmann-Moller S., Neufeld A., Elhanany-Tamir H., Taketo M. M., Sommer L., Krumlauf R. and Tzahor E. (2007). Cranial neural crest cells regulate head muscle patterning and differentiation during vertebrate embryogenesis. Development 134, 3065-3075. 10.1242/dev.002501 [DOI] [PubMed] [Google Scholar]
- Rolfe S. M., Camci E. C., Mercan E., Shapiro L. G. and Cox T. C. (2013). A new tool for quantifying and characterizing asymmetry in bilaterally paired structures. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2013, 2364-2367. 10.1109/embc.2013.6610013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin J. M., Kurrasch D. M. and Cobb J. (2015). Shox2 is required for the proper development of the facial motor nucleus and the establishment of the facial nerves. BMC Neurosci. 16, 424 10.1186/s12868-015-0176-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet D. F., Nichini O., Boisset G., Polok B., Tiab L., Mayeur H., Raji B., de la Houssaye G., Abitbol M. M. and Munier F. L. (2008). Mutation in the human homeobox gene NKX5-3 causes an oculo-auricular syndrome. Am. J. Hum. Genet. 82, 1178-1184. 10.1016/j.ajhg.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleri L., Depew M. J., Jacobs Y., Chanda S. K., Tsang K. Y., Cheah K. S., Rubenstein J. L., O'Gorman S. and Cleary M. L. (2001). Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128, 3543-3557. [DOI] [PubMed] [Google Scholar]
- Turner E. E. and Cox T. C. (2014). Genetic evidence for conserved non-coding element function across species–the ears have it. Front. Physiol. 5, 7 10.3389/fphys.2014.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvrdik P. and Capecchi M. R. (2006). Reversal of Hox1 gene subfunctionalization in the mouse. Dev. Cell 11, 239-250. 10.1016/j.devcel.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Vaclavik V., Schorderet D. F., Borruat F.-X. and Munier F. L. (2011). Retinal dystrophy in the oculo-auricular syndrome due to HMX1 mutation. Ophthal. Genet. 32, 114-117. 10.3109/13816810.2011.562955 [DOI] [PubMed] [Google Scholar]
- Vickerman L., Neufeld S. and Cobb J. (2011). Shox2 function couples neural, muscular and skeletal development in the proximal forelimb. Dev. Biol. 350, 323-336. 10.1016/j.ydbio.2010.11.031 [DOI] [PubMed] [Google Scholar]
- Wang W., Van De Water T. and Lufkin T. (1998). Inner ear and maternal reproductive defects in mice lacking the Hmx3 homeobox gene. Development 125, 621-634. [DOI] [PubMed] [Google Scholar]
- Zovein A. C., Turlo K. A., Ponec R. M., Lynch M. R., Chen K. C., Hoffmann J. J., Cox T. C., Gasson J. C. and Iruela-Arispe M. L. (2010). Vascular remodeling of the vitelline artery initiates extravascular emergence of hematopoietic clusters. Blood 116, 3435-3444. 10.1182/blood-2010-04-279497 [DOI] [PMC free article] [PubMed] [Google Scholar]