Abstract
Vertebrate appendage patterning is programmed by Hox-TALE factor-bound regulatory elements. However, it remains unclear which cell lineages are commissioned by Hox-TALE factors to generate regional specific patterns and whether other Hox-TALE co-factors exist. In this study, we investigated the transcriptional mechanisms controlled by the Shox2 transcriptional regulator in limb patterning. Harnessing an osteogenic lineage-specific Shox2 inactivation approach we show that despite widespread Shox2 expression in multiple cell lineages, lack of the stylopod observed upon Shox2 deficiency is a specific result of Shox2 loss of function in the osteogenic lineage. ChIP-Seq revealed robust interaction of Shox2 with cis-regulatory enhancers clustering around skeletogenic genes that are also bound by Hox-TALE factors, supporting a lineage autonomous function of Shox2 in osteogenic lineage fate determination and skeleton patterning. Pbx ChIP-Seq further allowed the genome-wide identification of cis-regulatory modules exhibiting co-occupancy of Pbx, Meis and Shox2 transcriptional regulators. Integrative analysis of ChIP-Seq and RNA-Seq data and transgenic enhancer assays indicate that Shox2 patterns the stylopod as a repressor via interaction with enhancers active in the proximal limb mesenchyme and antagonizes the repressive function of TALE factors in osteogenesis.
KEY WORDS: Limb, Patterning, Shox2, Skeleton, Stylopod
Highlighted article: A Shox2-coordinated transcriptional program functions in the osteogenic lineage to regulate Hox-TALE factors and skeletogenesis, and to pattern the mouse limb.
INTRODUCTION
The body segments and appendages of vertebrates are patterned by clusters of Hox and paraHox genes that are expressed along the major body axes (Pearson et al., 2005). Mutations or even subtle changes in the expression of Hox genes cause homeotic transformation of the axial skeleton (Kondrashov et al., 2011; Pearson et al., 2005), and reduction or loss of skeletal elements in vertebrate limbs (Kondrashov et al., 2011; Zakany and Duboule, 2007). In contrast to the specific in vivo requirement of Hox gene expression for correct patterning, the in vitro binding specificity of Hox factors to DNA motifs remains controversial, raising the question whether additional machineries such as transcription co-factors exist to ensure the regional specific function of Hox genes (Mann et al., 2009). Indeed, a family of three amino acids loop extension (TALE) homeodomain proteins including Meis and Pbx subclasses has been extensively characterized as DNA binding co-factors for Hox proteins to achieve the DNA binding specificity and form a highly conserved Hox-TALE patterning system with its origin being traced back to ancestral species such as starlet sea anemone (Hudry et al., 2014; Mann et al., 2009; Parker et al., 2011; Slattery et al., 2011). However, it is still under debate whether TALE factors could fully satisfy the in vivo binding specificity of Hox proteins. The recently proposed low-affinity Hox-TALE binding motif clusters on Hox-TALE bound enhancers (Crocker et al., 2015) implies the existence of additional factors to confer sufficient in vivo binding specificity. Alternatively, instead of being the primary binding factor, Hox proteins are known to play an accessory role for the interaction of Meis transcription factors with specific enhancers. Moreover, Meis factors can even function without Hox on a large proportion of these enhancers in branchial arch (BA) patterning (Amin et al., 2015), suggesting that additional tissue-specific transcriptional mechanisms contribute to the in vivo binding specificity of enhancers with Hox and TALE factors. However, whether other co-factors exist for Hox-TALE system so far remains unknown.
In the developing vertebrate limb, bone elements form via endochondral ossification, whereas osteogenesis is preceded by the formation of cartilaginous template with Runx2+/Osx+ osteogenic precursors initially residing in the perichondium and later on invading into cartilaginous template (Long and Ornitz, 2013). Limb skeletal patterning by Hox-TALE transcriptional complexes was proposed to be essential for endochondral skeletogenesis by directly regulating cartilage template formation (Zakany and Duboule, 2007). Along the proximal to distal (PD) axis, the skeletal elements of tetrapod limbs are patterned by Hox9 to Hox13 located within the HoxA/D gene clusters. Additionally, the expression of TALE factors is also regulated by signaling pathways along the PD axis, in which context the proximal retinoic acid (RA) signaling and the distal FGF signaling antagonistically determine the proximal expression of Meis genes that marks the stylopodial segment and facilitates the nuclear localization of Pbx in the proximal limb (Cunningham and Duester, 2015; Mercader et al., 2000). Together with HoxA/D9 and HoxA/D10, Meis and Pbx provides patterning code for the stylopodial skeleton (Capellini et al., 2011; Cunningham and Duester, 2015; Penkov et al., 2013). Intriguingly, compound deletion of HoxA/D gene clusters produces considerably milder defects in the stylopodial skeleton than that in the distal zeugopodial and autopodial skeletons that are patterned by Hox10-Hox13 (Kmita et al., 2005; Raines et al., 2015), suggesting that the stylopod adopts a unique mechanism for patterning that is less dependent on HoxA/D factors.
We have shown previously that inactivation of Shox2, encoding a paired-like homeodomain transcription factor, causes developmental defects of multiple organs including the heart, palate and limb (Bobick and Cobb, 2012; Cobb et al., 2006; Espinoza-Lewis et al., 2009; Ye et al., 2015a; Yu et al., 2005, 2007). Strikingly, Shox2 mutation causes loss of the stylopod in both forelimbs and hindlimbs, which was thought to be attributed to the direct function of Shox2 in chondrogenesis (Bobick and Cobb, 2012; Yu et al., 2007). However, an epistatic additive interaction between HoxA/D genes and Shox2 was seen in limb development (Neufeld et al., 2014), suggesting an involvement of Shox2 in the Hox-TALE patterning system.
Here, using our unique Shox2 allelic toolsets, we undertook a comprehensive analysis of Shox2 expression and the fate of Shox2+ cells in the developing limb. Hereby, we reveal an unexpected direct role of Shox2 in osteogenesis for stylopodial skeletal patterning. Our ChIP-Seq and RNA-Seq analyses demonstrate that Shox2 functions by directly regulating enhancers that are co-occupied by Hox-TALE factors to specify the stylopod that emerges in the juxtaposition of the trunk with strong Meis and Pbx gene expression and the proximal limb where Shox2 is highly expressed. Moreover, by retrospective and de novo characterization of Shox2-occupied enhancers, we demonstrate that co-occupancy of Shox2 and TALE factors represents a key feature of the enhancer grammar for proximal limb-specific enhancer activity. Our results indicate that Shox2 acts as a repressor on these enhancers and is required for modulation of cell fate choices in limb development.
RESULTS
Shox2 is expressed in mesenchymal progenitors of multiple cell types in the proximal limb
We have shown previously that Shox2 is expressed in the developing proximal limb and Shox2 deficiency causes severely mispatterned stylopodial skeletal elements (Cobb et al., 2006; Yu et al., 2007). To unravel the functional mechanism of Shox2 in stylopod patterning, we first sought to comprehensively document and analyze Shox2 expression patterns and cell populations that are derived from Shox2+ cells. We took advantage of our recently generated Shox2HA/+ (Wang et al., 2014; Ye et al., 2015b) and Shox2Cre/+ (Sun et al., 2013) alleles for protein expression analysis and fate mapping.
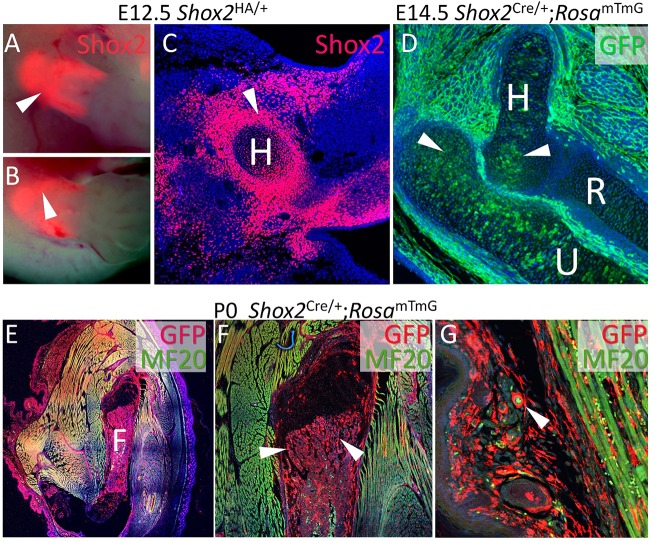
The Shox2HA/+ allele represents a knock-in of a Flag-2xHA-Shox2a-IRES-DsRed-pA coding cassette into the endogenous Shox2 locus (Wang et al., 2014; Ye et al., 2015a), which allows live imaging of Shox2 expression via DsRed and Shox2 protein localization using anti-HA antibodies. We chose to analyze Shox2 expression starting from embryonic day (E)12.5 at which stage the distinct anlagen of stylopodial and zeugopodial skeletal elements are formed. Live imaging revealed strong Shox2 expression in the proximal portion of the developing limb at E12.5 (Fig. 1A,B). An intensive Shox2 expression in the proximal limb, especially around the chondrogenic center of the stylopod, was observed by anti-HA staining at the same stage (Fig. 1C). Consistent with broad Shox2 expression at E12.5, fate mapping using Shox2Cre/+ allele revealed contribution of Shox2+ cells to multiple connective tissue cell types, including chondrocytes, osteoblasts, adipocytes, and dermal fibroblasts (Fig. 1D-G). In contrast to the extensive labeling of osteogenic cells in the bone-forming area (Fig. 1E), Shox2Cre/+ labeled only a portion of chondrocytes (Fig. 1D), consistent with relatively weak real-time Shox2 expression in the chondrogenic center (Fig. 1C).
Fig. 1.
Shox2 is expressed in mesenchymal progenitors of multiple cell types in the proximal limb. (A,B) Live imaging on Shox2HA/+ embryo reveals proximal expression of Shox2 (arrowhead) in embryonic forelimb (A) and hindlimb (B) at E12.5. (C) Immunofluorescence using anti-HA antibody on Shox2HA/+ embryo reveals strong perichondrial expression of Shox2 (arrowhead). (D-G) MF20 and GFP immunofluorescence on Shox2Cre/+;RosamTmG embryonic (D) and postnatal (E) mice reveals broad contribution of Shox2+ cells to connective tissue cell types such as chondrocyte (arrowheads in D), osteoblast/osteocyte (arrowheads in F) and dermal fibroblast (arrowhead in G). H, humerus; R, radius; U, ulna; F, femur.
Deletion of Shox2 results in specific loss of Shox2+/Runx2+ perichondrial cells
As the expression analysis and fate-mapping results did not indicate a lineage etiology for the phenotype observed in Shox2 mutants and Shox2-expressing cells contribute extensively to the proximal connective tissue cells, we speculated that virtual loss of the stylopod in Shox2 mutants results from absence of Shox2+ cells. We therefore compounded the RosamTmG allele to Shox2Cre/− mice (null mutant) to examine the contribution of Shox2+ cells in the absence of Shox2.
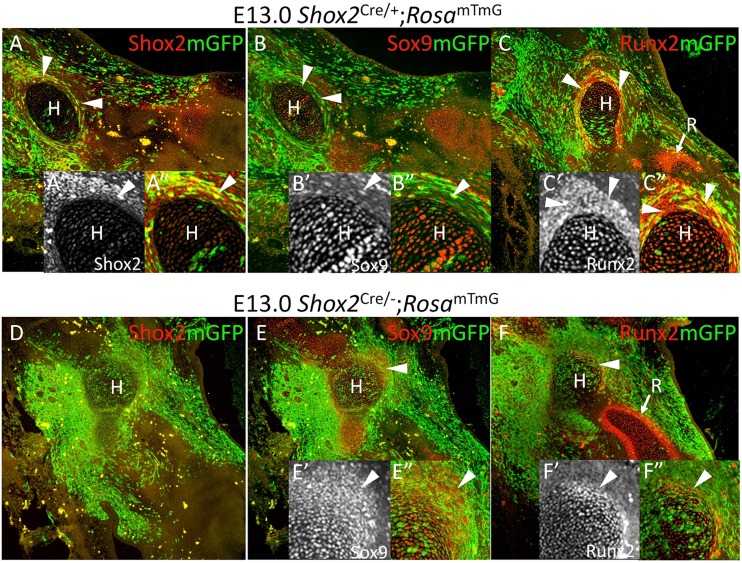
Shox2Cre/−;RosamTmG mice were analyzed at E13.0, the latest stage the Shox2 deficient embryos appear normal prior to severe complications caused by cardiac defects (Espinoza-Lewis et al., 2009; Ye et al., 2015b). In these embryos, the Shox2+ lineage is labeled by mGFP, and the expression pattern of mGFP reporter did not change significantly compared with controls (Fig. 2), indicating that Shox2 is not required for the proliferation and migration of the majority of Shox2+ cells in the stylopod. Consistent with the weak and transient expression of Shox2 in the chondrocytes of controls (Fig. 2A-C), the chondrogenic center of the stylopodial skeleton formed in Shox2-deficient Shox2Cre/−;RosamTmG embryos (Fig. 2D-F). Interestingly, intensive Shox2 expression in the perichondrium of the stylopod of control embryos was observed at this stage (Fig. 2A). Immunostaining on the adjacent sections showed that these cells are also positive for Runx2, a molecular marker for osteoblastic precursors (Fig. 2C). However, in Shox2Cre/−;RosamTmG embryos, these Shox2+/Runx2+ cells lost Runx2 expression completely and expressed Sox9 aberrantly, indicating defective fate decision (Fig. 2B,C,E,F).
Fig. 2.
Deletion of Shox2 results in specific loss of Shox2+/Runx2+ perichondrial cells. (A-F), Co-immunofluorescence of Shox2, mGFP, Sox9 and Runx2 in the forelimb of Shox2Cre/−;RosamTmG embryos at E13.0 (D-F) compared with littermate Shox2Cre/+;RosamTmG mice (A-C) shows loss of distinct stylopodial perichondrial structure (arrowheads) in the absence of Shox2 compared with that of zeugopod (arrow in F). Humerus regions are magnified in inserts A′,A″,B′,B″,C′,C″,E′,E″,F′,F″. H, humerus; R, radius.
Shox2 inactivation in osteogenic lineage precursors recapitulates limb defects in Shox2-null mutants
Given a potential role of the embryonic perichondrium as the osteogenic precursor pool (Colnot et al., 2004) and the extensive contribution of Shox2+ cells to osteoblasts/osteocytes (Fig. 1E), we asked whether Shox2+ osteogenic cells are derived from Shox2+/Runx2+ perichondrial cells and if the defective fate decision of early perichondrial Shox2+/Runx2+ cells could lead to aberrant osteogenic differentiation of Shox2+ cells at a later stage. To overcome the cardiac pacemaker defects that cause early embryonic lethality in Shox2 mutants, we made use of Nkx2-5F/F allele that, in combination with the Shox2Cre/−;RosamTmG alleles, conditionally rescues the cardiac defects and allows Shox2 mutants to survive to the birth without interfering defects in other organs irrelevant to Nkx2-5 (our unpublished results).
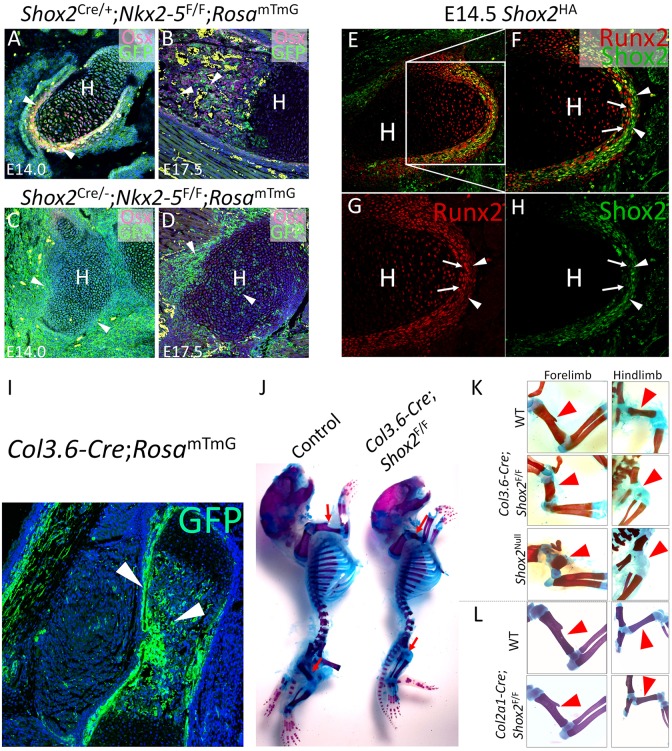
At E14.0, GFP+/Osterix+ cells could be clearly detected in the inner layer of perichondrium in the stylopod of Shox2Cre/+;Nkx2-5F/F;RosamTmG embryos (Fig. 3A). At E17.5, the GFP+/Osterix+ osteoblast/osteocytes were abundantly localized in the diaphysis of the stylopodial skeleton (Fig. 3B). Together with the restricted Shox2 expression in the perichondrium at E14.5 (Fig. 3E-H) and the complete lack of Shox2 expression in the stylopodial skeleton at E17.5 (data not shown), our observations indicate a direct contribution of the Shox2+ perichondrial cells to the osteogenic lineage. However, in Shox2-deficient embryos (Shox2Cre/−;Nkx2-5F/F;RosamTmG) at E14.0 and E17.5, the Osterix+ layer of perichondrial cells and the GFP+/Osterix+ osteogenic cells that were otherwise found abundantly in the diaphysis of control mice, were absent (Fig. 3C,D). The requirement of Shox2 for the formation of osteogenic cells appears to occur at the early fate determination phase. This is supported by the observations that at E14.5, Shox2 expression becomes predominantly restricted to the outer layer of the perichondrium that is characterized by relatively low levels of Runx2 expression (Fig. 3E-H) and was thought to have a less definitive role in osteogenesis (Swinehart et al., 2013).
Fig. 3.
Shox2 inactivation in osteogenic lineage recapitulates limb defects in Shox2−/− mice. (A-D) Co-immunofluorescence of Osx and GFP shows the absence of osteogenic cells (arrowheads) in the humerus of Shox2Cre/−;Nkx2-5F/F;RosamTmG mice at E14.0 (C) and E17.5 (D) in comparison to littermate control Shox2Cre/+;Nkx2-5F/F;RosamTmG mice (A,B). (E-H) Co-immunofluorescence on E14.5 Shox2HA/+ embryo shows restricted expression of Shox2 in the outer layer (arrowheads) of the perichondrium and strong Runx2 expression in the inner layer (arrows). (I) GFP staining shows specific osteogenic activation (arrowheads) by Col3.6-Cre in a Col3.6-Cre;RosamTmG embryo at E17.5. (J) Alcian Blue/Alizarin Red staining on a representative Col3.6-Cre;RosamTmG mouse in comparison to its littermate control at P0. Red arrows indicate stylopodial skeleton. (K,L) Alcian Blue/Alizarin Red staining shows specific stylopodial defect (arrowheads) in Col3.6-Cre;Shox2F/F mice similar to that of Shox2−/− mice, compared with the Col2a1-Cre;Shox2F/F mice (L) and WT mice at P0 (the littermate control for Col2a1-Cre;Shox2F/F mice at P0 is shown in L). H, humerus.
It was proposed that skeletal patterning in the limb by Hox genes is largely exerted by their expression in the perichondrium (Swinehart et al., 2013; Villavicencio-Lorini et al., 2010). Given that Shox2 deficiency causes a specific loss of perichondrial osteogenic cell type signature and leads to a depletion of osteogenic cells, we hypothesized that Shox2 functions to control stylopodial skeleton patterning through maintaining the fate of perichondrial osteogenic precursors that initially provide instructive cues for chondrocyte differentiation, and later differentiate to osteoblasts for bone formation. In supporting this hypothesis, we found that Shox2+ cells contribute specifically to the osteogenic population of the stylopodial skeleton but not that of the zeugopodial skeletal elements (Fig. S1), correlating well with the specific loss of the stylopod in Shox2 mutants.
To determine if Shox2 expression in the osteogenic cells is essential for stylopod patterning, we utilized Col3.6-Cre (Liu et al., 2004) to inactivate Shox2 specifically in osteogenic cells (Fig. 3I). In Col3.6-Cre;Shox2F/F mice, the stylopodial elements of both forelimb and hindlimb were severely shortened (Fig. 3J), mimicking the limb defects observed in Shox2-null mutants (Fig. 3K). Together with the fact that cartilage-specific deletion of Shox2 by Col2a1-Cre did not produce a similar limb phenotype (Fig. 3K; Fig. S1), our results demonstrate a pivotal role for Shox2 in the osteogenic lineage essential for stylopodial skeleton patterning.
Shox2 binds predominantly to limb-specific distal enhancers involved in skeletogenesis
To reveal the regulatory molecular framework behind this unique Shox2 function in osteogenic differentiation in the stylopod, we used the Shox2HA allele that allows efficient immunoprecipitation of endogenous HA-tagged Shox2 (Fig. 4A). We performed anti-HA ChIP-Seq using developing limbs from Shox2HA/+ and wild-type control embryos at E12.5 and E13.5. Resulting peaks with various levels of fold enrichment were successfully verified by ChIP-qPCR (Fig. S2).
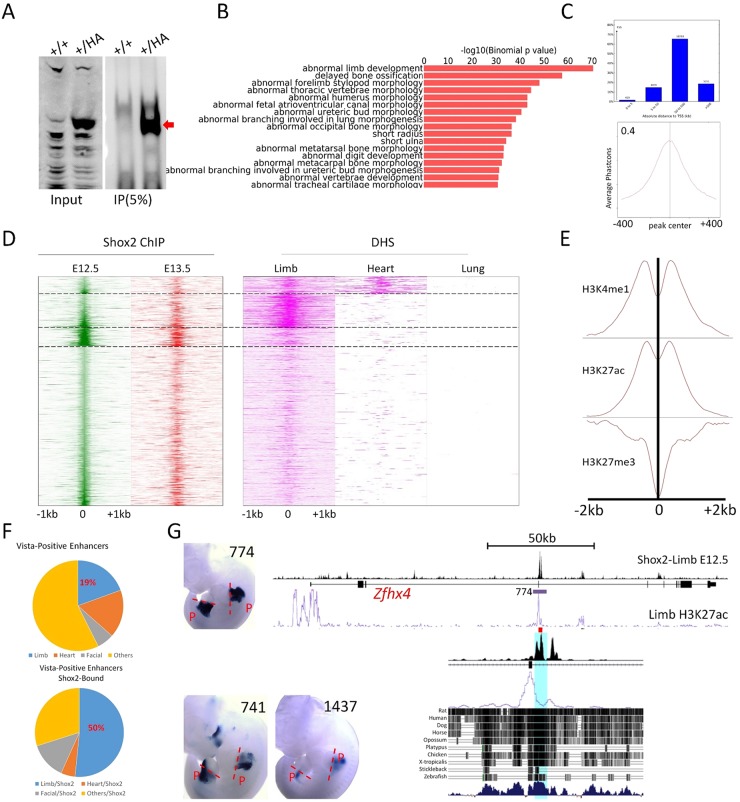
Fig. 4.
Shox2 predominantly binds to limb-specific distal-acting enhancers involved in skeletogenesis. (A) Immunoprecipitation after crosslinking shows efficiency and specific pull down of Shox2 from E12.5 Shox2HA/+ limbs after crosslinking. (B) GO analysis on gene list associated with Shox2 binding sites shows specific association of Shox2 occupancy with skeletogenic-related genes. (C) The majority of Shox2 binding peaks have great distance to its nearest transcription starting sites (TSS) (top panel), and aggregate plots based on average phastocons across vertebrates indicate that the majority of these sites are highly conserved (bottom panel). (D) Heat-map plot of Shox2 occupancy signals at E12.5 and E13.5 in relation to accessible chromatin landscape assessed by DNase I hypersensitivity sequencing (DHS) of E11.5 embryonic limb, and postnatal heart and lung. (E) Aggregate plots on binding signals of H3K4me1, H3K27ac and H3K27me3 in embryonic limb at E11.5 around summits of Shox2 binding sites. (F) Pie graph shows the percentage of enhancers with limb activities in total (top panel) and Shox2-occupied (bottom panel) enhancers verified by vista enhancer projects. (G) Representative transgenic embryos showing proximal (P) limb activity of enhancers harboring Shox2-occupied sites. In the right-hand panel, a genome browser view shows a prominent Shox2 binding site (orthologs to HS774) in the intron of Zfhx4 along with H3K27ac ChIP signal obtained in embryonic limb.
We first assigned the Shox2 binding peaks to the nearby coding genes and assessed the gene ontology (GO). The GO terms for skeletogenesis are highly enriched for nearby genes flanking Shox2 binding peaks, indicating a pivotal role of Shox2 in osteogenesis despite its broad expression in progenitor cells of multiple connective tissue cell types (Fig. 4B). Our analysis showed that Shox2 interacts predominantly with distal regulatory elements that are highly conserved among vertebrates as shown by plotting based on PhastCons (Fig. 4C). We next plotted Shox2 binding signals with the DNaseI hypersensitivity sequencing (DHS) data generated by the Encode project (Yue et al., 2014). Shox2 binding signals and DHS signals from the developing limb (E11.5), and adult heart and lung were categorized based on relative signal intensity using k-mean clustering into four groups. As shown in Fig. 4D, the genome-wide binding intensity of Shox2 at E12.5 and E13.5 is generally similar, suggesting a persistent function of Shox2. Moreover, the majority of Shox2 binding sites colocalize with DHS sites accessible specifically in the embryonic limb, which are enriched for skeletogenic-related GO terms (Fig. S3). Therefore, our identified limb-specific Shox2 target regions likely contain the unique set of Shox2-controlled cis-regulatory modules that is required for the transcriptional control of skeletal patterning genes in the stylopod.
To further characterize if these Shox2-bound elements represent active enhancers, we plotted the published histone modifications H3K4me1, H3K27ac and H3K27me3 ChIP-Seq data obtained from embryonic limbs (Attanasio et al., 2014) versus the Shox2 binding peak summits. Indeed, the active enhancer markers H3K4me1 and H3K27ac are generally enriched around Shox2 binding sites (Fig. 4E). In addition, H3K27me3, which is frequently associated with inactive enhancers, appears to be mildly enriched around summits of Shox2 binding sites, although it is clearly absent from the center of Shox2 binding peaks (Fig. 4E), suggesting that a set of Shox2 binding enhancers is in a poised state. Given the association of Shox2 occupancy with limb-specific DHS sites and enhancer marks, we sought to determine if the occupancy of Shox2 can be used as a feature to isolate limb-specific enhancers. We exploited the vista enhancer database (Visel et al., 2007) and found that only 19% of active enhancers validated through transgenic enhancer assays contain limb-specific activity (Fig. 4F). After filtering the active enhancers using Shox2 occupancy as a criterion, the percentage of the limb-specific active enhancers increased to 50%. Consistent with Shox2 expression in the developing proximal limb, many of the enhancers occupied by Shox2 displayed proximal specific activity in the limb, as exemplified in Fig. 4G. These observations indicate that the enhancers occupied by Shox2 and located near genes involved in skeletogenesis integrate the transcriptional program specifically required for stylopod formation.
Genome-wide co-occupancy of Shox2 and Hox-TALE factors on candidate enhancers of osteogenic genes
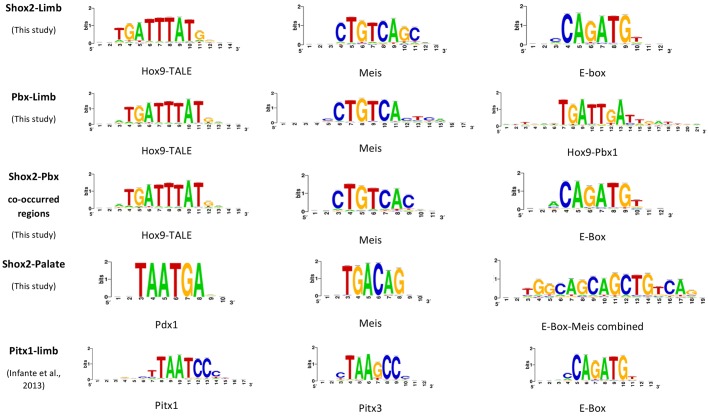
We next sought to search for transcription co-factors of Shox2 that co-operate in and underlie its unique function in stylopodial skeletogenesis. We first performed de novo motif discovery on Shox2 binding peaks in the embryonic limb. As shown in Fig. 5, in contrast to results discovered using the same methods in hindlimb ChIP-Seq top peaks of Pitx1 (Infante et al., 2013), which is also a paired-like homeodomain transcription factor, the top motifs discovered in the Shox2 binding sites are highly related to motifs for Hox-TALE factors, including those similar to the Hox9-TALE motif (Huang et al., 2012), Meis motif and Pbx-TALE motif. These analyses suggest an association of Shox2 with Hox-TALE factors.
Fig. 5.
TALEfactor-related motifs are enriched in Shox2-binding sites. Top motifs discovered in the binding sites of Shox2 (limb and palate), Pbx (limb), Shox2/Pbx co-occupied sites (limb) and Pitx1 binding sites (limb).
Given that Hox9, Pbx and Meis are all transcription factors that are highly abundant in the proximal limb (Capellini et al., 2011; Zakany and Duboule, 2007) and act potentially as determinants for accessible chromatin landscape, the outcome of the motif discovery on Shox2-occupied sites may be biased by the availability of transcription factor-accessible sites that are already established by Hox9, Pbx and Meis. To determine the specific association of Shox2 with Hox-TALE factors, we further conducted Shox2 ChIP-Seq on the developing palate where Shox2 is specifically expressed in the anterior domain overlapping with the future bony hard palate (Gu et al., 2008; Yu et al., 2005) and performed motif discovery similarly. The occupancy of Shox2 in the palate shows distal enhancer binding properties and high relevance to skeletogenesis similar to that in limb (Fig. 6A,B). TALE-related motifs were also discovered in the Shox2-bound peaks in the palate (Fig. 5). However, despite the similar association of Shox2 with TALE-Hox factors in the limb and palate, the top peak signals of the limb and palate do not overlap (Fig. 6C), suggesting that Shox2, along with TALE-Hox factors, is involved in different sets of machineries for skeletogenesis in the embryonic limb and palate, respectively.
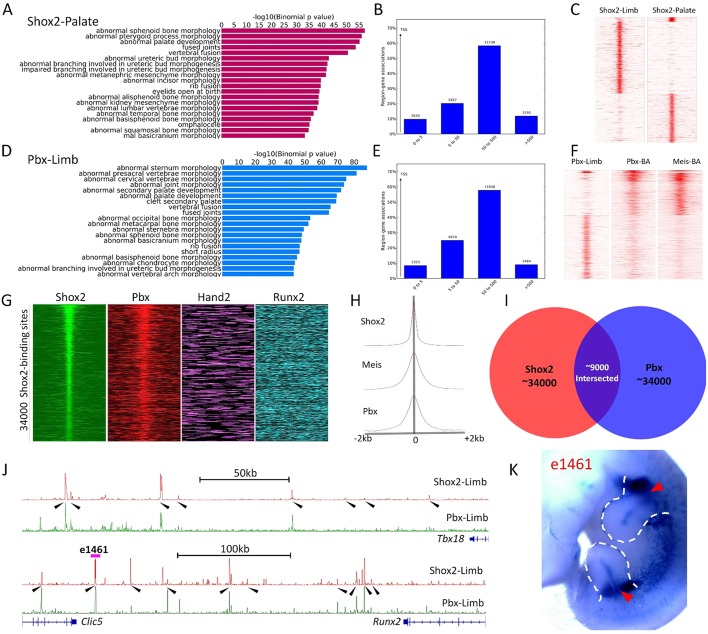
Fig. 6.
Genome-wide co-occupancy of Shox2 with Hox-TALE factors on osteogenic genes. (A-F), ChIP-Seq of Shox2 on the developing palate (A-C) and ChIP-Seq of Pbx on the developing limb (D-F) at E12.5. GO analysis and distance to TSSs histogram of Shox2-Palate (A,B) and Pbx-Limb (D,E) show similar functional and binding properties to Shox2-Limb ChIP-Seq data. K-mean cluster based on binding signal shows distinct binding preference of Shox2 occupancy in the limb compared with that in the palate (C). Similarly, Pbx preferentially binds to sites that are relatively weakly occupied by Pbx and Meis in the developing brachial arch (BA) (F). (G) Heat-map plot of binding signals of Shox2 (limb), Pbx (limb), Hand2 (limb) and Runx2 (osteogenic cell line) around Shox2 binding sites in E12.5 limb. (H) Aggregate plots on ChIP binding signal of Shox2 (limb), Pbx (limb) and Meis (BA) around Shox2 binding sites. (I) Venn diagram shows the ∼9000 co-binding sites of Shox2 and Pbx in proportion to the top ∼34,000 Shox2 and Pbx binding sites in E12.5 limb. (J) Shox2 and Pbx co-occupied enhancers cluster around Tbx18 and Runx2 (highlighted by arrowheads). (K) Representative transgenic embryo of e1461, a putative enhancer for Runx2 (indicated in J), shows proximal limb-specific activity (arrowheads).
To understand the collaborative regulatory manner of Shox2 and Hox-TALE, we performed ChIP-Seq of Pbx, a key TALE family homeodomain factor, using E12.5 limb tissue. The top motifs discovered from the Pbx-occupied sites (Fig. 5) are similar to that discovered from the Shox2-occupied sites, and GO analysis indicates a similar function for Pbx in controlling cis-regulatory modules near skeletogenic genes in the developing limb (Fig. 6D,E). Furthermore, the motif for Meis was abundant in the peak center of Pbx ChIP-Seq data, consistent with the fact that Pbx and Meis often exist in the same transcription complex and share the same binding sites (Choe et al., 2009; Moens and Selleri, 2006). Interestingly, a discrepant genome-wide binding pattern between Pbx in the limb and that of Pbx and Meis in the brachial arches (BAs) (Amin et al., 2015) was also discovered (Fig. 6F), indicating that Pbx, similar to Shox2, is involved in a set of transcription programs unique for the skeletal patterning of the limb. A unique genome-wide co-occupation between Shox2 and Pbx was further revealed by heat-map plotting (Fig. 6G) with the binding signals of Hand2 (Osterwalder et al., 2014) and Runx2 (Wu et al., 2014), which are crucial for skeletal formation. Moreover, aggregate plots (Fig. 6H) indicate that the binding peaks of Shox2 colocalize with those of Pbx in the embryonic limb (this study) and Meis (Amin et al., 2015), suggesting that Shox2 and TALE factors can function in the same transcription complex.
Interaction of Shox2 and Pbx binding peaks resulted in about 9000 co-occupied elements that cluster around skeletogenic-related genes (Fig. 6I). Moreover, multiple Shox2 and Pbx co-occupied sites were found around such genes, for example, flanking Tbx18 and Runx2 (Fig. 6J), suggesting that these putative regulatory elements function additively on their putative target gene as a ‘regulatory archipelago’ – a phenomenon that was found for the regulation of Hox genes (Montavon et al., 2011). Remarkably, within the cluster of Shox2-Pbx co-occupied enhancers downstream of Runx2, an enhancer that is located within the same topologically associated domain (TAD) as Runx2 [inferred from published Hi-C data (Dixon et al., 2012)] showed proximal limb specific activity in the transgenic enhancer assay (Fig. 6K). This observation suggests that the regional control of Runx2 expression by a Shox2-TALE-Hox mechanism contributes to stylopodial skeleton patterning.
Shox2 exhibits a complementary expression gradient with and represses the expression of TALE factors in the stylopod
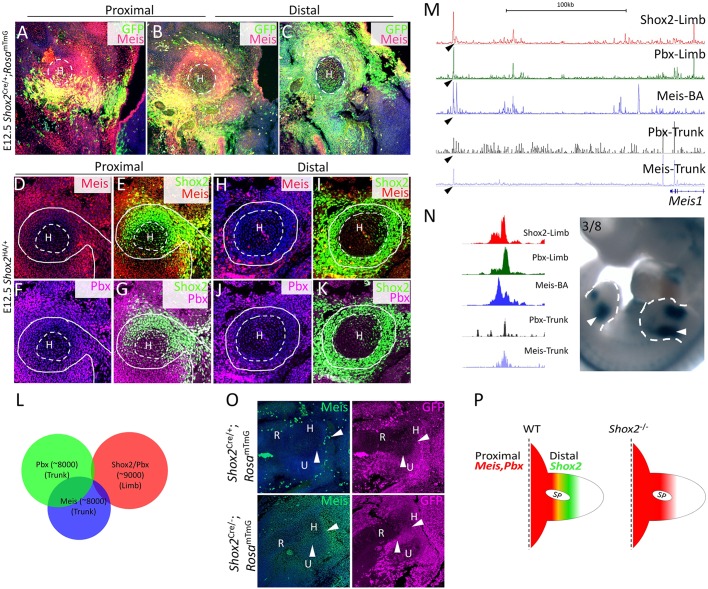
Given a strict requirement of Shox2 for Runx2 expression in the stylopod and the co-occupancy of Shox2 and Pbx on the regulatory elements of Runx2, we initially hypothesized that Shox2 and TALE-Hox factors would be collectively required for the expression of key genes for stylopod patterning. This notion seemed to be supported by the hypothesis that Meis and Pbx genes are essential for stylopodial skeleton patterning based on their strong proximal limb expression (Capellini et al., 2011; Tabin and Wolpert, 2007). To determine if an epistatic additive output of Shox2 and TALE factors contributes to stylopod patterning, we first analyzed the Shox2+ domain in relation to that of Meis gene expression. As shown in Fig. 7A-C, similar to previous reports (Mercader et al., 2000), Meis1/2 is expressed most strongly in the proximal limb adjacent to the trunk, and the expression intensity decreases distally. To our surprise, the expression domain of Meis1/2 appears partially overlapped with, but complementary to the Shox2+ domain that increases distally in the stylopod (Fig. 7A-C). A closer examination on the expression of Meis and Pbx proteins and Shox2 in the developing stylopod also identified a partial exclusive expression of both Meis and Pbx proteins from Shox2-expressing cells surrounding the cartilage primordium (Fig. 7D-K). These observations together suggest that the Shox2+ perichondrial mesenchymal cells acquire a distinct distal identity within the stylopod, and Shox2 may enforce this identity by repressing the expression of TALE transcription factors. Consistently, Shox2 colocalizes with both Pbx and Meis transcriptional regulators on putative cis-regulatory elements near Meis and Pbx genes (Fig. S4), suggesting that Shox2 exerts its repressive function on Meis and Pbx via a self-regulatory machinery.
Fig. 7.
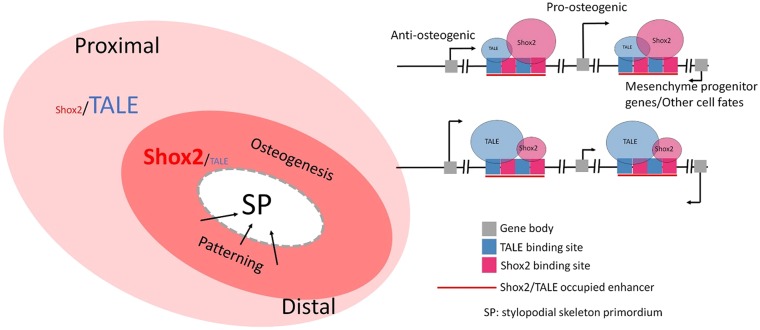
Shox2 exhibits an opposite expression gradient with and represses the expression of TALE factors in the stylopod. (A-C) Co-immunofluorescence of E12.5 Shox2Cre/+;RosamTmG limb shows opposite gradients of Meis expression and Shox2+ domains along the proximal (A) to distal (C) axis in the stylopodi. The cartilaginous domains are circled by dashed lines. (D-K) Co-immunofluorescence on cross sections of E12.5 Shox2HA/+ forelimb shows complementary expression of Shox2 to that of Meis (D,E,H,I) and Pbx (F,G,J,K) in the stylopod. Note that the signals for both Meis and Pbx staining are stronger in the proximal domain (D-G) compared with that in distal domain (H-K). The cartilaginous domains are circled by dashed lines and the Shox2-expressing domain is highlighted by solid lines. (L) Venn diagram shows very little overlap of Pbx and Meis binding sites to the Shox2/Pbx co-occupied sites. (M,N) Binding signal of Shox2, Pbx (limb and trunk), Meis (BA and trunk) downstream of Meis1, the most prominent binding site with co-binding of Shox2, Pbx and Meis is indicated by arrowheads. The limb-specific enhancer activity (arrowheads) of this site, along with short flanking sequence (1013-bp) (chr11:18612148-18613160) was confirmed by transgenic enhancer assay (N). (O) Immunofluorescence on sections of E12.5 Shox2Cre/−;RosamTmG limb at the distal portion of the stylopod shows elevated expression of Meis (arrowheads) compared with that in littermate control Shox2Cre/+;RosamTmG embryos. (P) Schematic model shows that stylopodial skeleton primordium (SP) is developed in the juxtaposition of the most proximal trunk-limb transition domain, where TALE factors are present, and the relative distal Shox2 dominant domain. Such identity is lost in the absence of Shox2. H, humerus; R, radius; U, ulna.
To further understand the epigenomic change underlying the unique identity of Shox2+ perichondrial mesenchymal cells, we compared the Pbx/Meis ChIP-Seq data obtained from embryonic trunk (Penkov et al., 2013) with our Shox2/Pbx ChIP-Seq data. As summarized in the Venn diagram (Fig. 7L), the Shox2/Pbx binding sites in the limb overlap very little with the Pbx/Meis binding sites in the trunk, consistent with the notion that the Shox2+ perichondrial cells have a distinct chromatin landscape accessible to TALE factors.
The cis-regulatory elements for Meis genes have not been characterized in vivo. Although Meis1/2 are expressed in both the trunk and proximal limb, we reasoned that if the occupancy of Shox2/TALE transcription factors underlies the trunk to limb epigenomic change and that Shox2 represses the expression of Meis and Pbx genes directly by acting on their putative enhancers, then the strong co-occupancy of Shox2 and Pbx in the limb and the weak occupancy of Meis and Pbx in the trunk would serve as a principle to identify limb-specific enhancers for Meis. As shown in Fig. 7M, a potential enhancer for Meis1 showed prominent co-occupancy of Shox2 and Pbx in the limb, but mild occupancy by Meis and Pbx in the trunk. Transgenic enhancer assay confirmed its enhancer activity in the developing limb (Fig. 7N). Interestingly, this site does not show identifiable H3K27ac ChIP-Seq signal in the limb (Fig. S4) (Shen et al., 2012), suggesting that co-occupancy by context-specific transcription factors could serve as a more sensitive approach for identifying tissue-specific enhancers. In addition, this enhancer showed H3K27ac enrichment in differentiated osteoblast-like cell line (Fig. S4) (St John et al., 2014), suggesting that it is an osteogenic enhancer for Meis1 and that Shox2 represses this enhancer to prevent Meis1 expression in perichondrial osteogenic lineage cells. In support of this notion, in the absence of Shox2, an elevated expression of Meis was observed in the distal region of the humerus (Fig. 7O), and elevated Meis and Pbx expression was also detected in the Shox2+ perichondrial mesenchymal cells (Fig. S5). Shox2 appears to establish a trunk to limb transition status (Fig. 7P). It was reported previously that excessive expression of Pbx (Gordon et al., 2010) or Meis (Mercader et al., 2009) inhibited osteogenesis. Shox2 thus probably functions to antagonize the inhibitory effect of Meis and Pbx for stylopodial skeletogenesis. This hypothesis is also in line with the fact that excessive proximal RA signaling that is upstream of Meis could also cause failure of proper skeletogenesis in the developing limb, including the stylopod (Cunningham and Duester, 2015).
Shox2 functions as a dominant repressor in the mesenchyme of embryonic limb
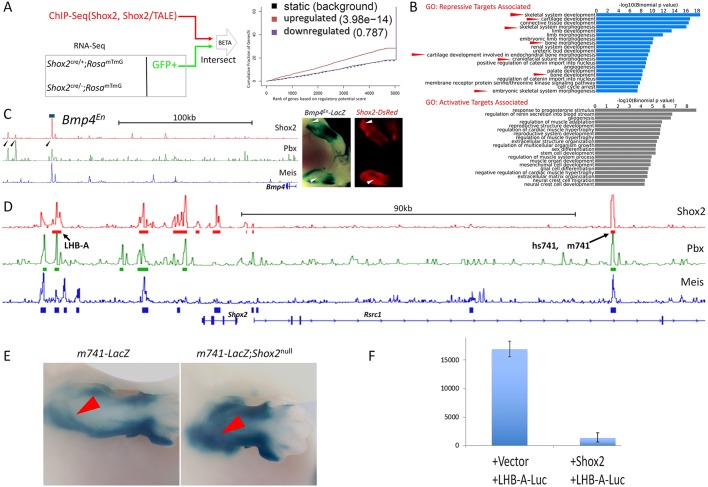
The likely repressive function of Shox2 on the expression of Meis and Pbx genes prompted us to further understand the general function of Shox2 in transcriptional regulation of its putative target genes. We accordingly performed RNA-Seq to profile the transcriptome of GFP+ cells sorted from E12.5 Shox2Cre/−;RosamTmG limb and compared results with those from Shox2Cre/+;RosamTmG mice. The list of differentially expressed genes (DEGs) was intersected to the list of genes with Shox2-bound peaks in their putative regulatory elements to identify Shox2-targeted genes. Subsequent analysis of the intersected DEGs and gene-peak association by BETA, a program designed for generalizing the function of transcription factors (Wang et al., 2013), showed that Shox2 functions primarily as a repressor (Fig. 8A). Additionally, the same analysis using Shox2/Pbx co-bound peaks as input also revealed a repressive function for Shox2 (data not shown), suggesting that TALE-Hox machinery is also involved in the repressive action of Shox2. GO analysis further demonstrated that the putative Shox2 repressed targets are more relevant to limb development compared with targets activated by Shox2 (Fig. 8B). Indeed, many putative direct repressive targets of Shox2, such as Prrx1, Osr2, Gria2, Epha7, Rsrc1 and Bmp4 (Tables S1 and S2), were found to show elevated/ectopic expression in the absence of Shox2 in our previous studies by microarray and in situ hybridization on E10.5 and E11.5 limbs (Vickerman et al., 2011; Yu et al., 2007). Similar to Runx2 and Tbx18 (Fig. 6J), Shox2-TALE co-occupied sites were also identified as clusters around their associated genes (Fig. S4), supporting the notion of a convergent function of Shox2-Hox-TALE machinery toward their target genes.
Fig. 8.
Shox2 functions as a dominant repressor in the mesenchyme of embryonic limb. (A) Schematic and representative results for integrative interpretation of the general function of Shox2. (B) GO analysis for genes associated with Shox2 occupancy that are upregulated (putative repressive targets) and that are downregulated (putative activate targets) in the absence of Shox2. (C) Binding signal of Shox2, Pbx (limb), Meis (BA) downstream of Bmp4, the co-binding sites of Shox2, Pbx and Meis are indicated by arrowheads. The 2237 bp fragment (chr14:46840958-46843194) containing this binding sites was characterized by transgenic enhancer assay and a representative transgenic embryo is shown next to real-time Shox2 expression at the same stage. (D) Binding signal of Shox2, Pbx (limb), Meis (BA) around the Shox2-Rsrc1 locus with two characterized enhancers indicated (LHB-A, m741 and its human ortholog hs741). (E) In the absence of Shox2 in the embryonic limb (mm741-lacZ; Prrx1-Cre;Shox2F/F), the β-galactosidase activity driven by the Rsrc1 enhancer (m741) is elevated. (F) Normalized luciferase activity of co-transfection luciferase assay in C3h10T1/2 cells shows direct repressive activity of Shox2 on the LHB-A enhancer.
We next investigated whether Shox2 represses target enhancers directly by using a transgenic enhancer assay. First, we used an enhancer containing a Shox2-Pbx-Meis co-occupied element downstream of Bmp4, which revealed complementary activity of the transgenic enhancer compared with the expression domains of Shox2 in developing limbs (Fig. 8C), suggesting a repressive role of Shox2 on this enhancer. We next created a stable transgenic line (mm741-lacZ) from a Shox2/Pbx1/Meis co-bound enhancer in intron 3 of the Rsrc1 in the Rsrc1-Shox2 locus (Fig. 8D), given that Rsrc1 is a robust repressive target of Shox2 and that Shox2 also shows self-repressive activity (Fig. S6). Similar to its human ortholog hs741 (Fig. 5G), this enhancer displayed strong limb-specific activity (Fig. 8E). Compounding this transgenic enhancer-reporter allele to Shox2−/− background caused significant elevation of the enhancer activity in the proximal limb (Fig. 8E; N=3). A direct repressive activity of Shox2 toward the Shox2-Rsrc1 locus is further supported by the co-occupancy of Shox2, Pbx and Meis on an enhancer of Shox2 itself (Rosin et al., 2013) and a strong repressive activity of Shox2 on this enhancer in luciferase reporter assay in vitro (Fig. 8F). Interestingly, many genes we identified as putative repressive targets of Shox2 were shown or known previously to be broadly expressed in mesenchymal progenitor cells of the early developing limb (Vickerman et al., 2011; Yu et al., 2007) and less widely expressed in the osteogenic lineage, raising the possibility that the repressive action of Shox2 helps to steer the progenitor cells away from a non-osteogenic cell fate.
DISCUSSION
A Shox2-coordinated transcription program for stylopod patterning
In this study, we identified a Shox2+ cell population specifically required for the patterning of the stylopodial skeleton, as summarized in Fig. 9. This population of Shox2+ perichondrial mesenchymal cells appears to instruct the maturation and hypertrophic differentiation of chondrocytes in the cartilaginous mode and contributes to the osteogenic lineage at later stage, because in the Shox2−/− limb, the stylopodial cartilage mode fails to undergo hypertrophy and lacks bone formation (Cobb et al., 2006; Yu et al., 2007), in contrast to the milder limb phenotype in mice carrying cartilage-specific inactivation of Shox2 (Bobick and Cobb, 2012; and this study). In the perichondrial mesenchymal cells, Shox2 acts to determine the fate of osteogenic lineage, as it is required for the expression of key osteogenic genes including Runx2. On the other hand, Shox2 represses a panel of genes, as demonstrated by integrated analysis of our RNA-Seq and ChIP-Seq data. Among the repressed genes are Meis and Pbx, both of which contribute to correct proximal skeleton patterning. Together with the fact that Shox2, Meis and Pbx show broad genome-wide co-occupation on osteogenic genes, the repressive activity of Shox2 toward Meis and Pbx assists to establish a Shox2 dominant form of Shox2/TALE occupancy on Shox2/TALE co-occupied regulatory elements and favors pro-osteogenic transcription output essential for the instructive role of the perichondrial cells towards cartilaginous mode patterning as well as osteogenic cell fate (Fig. 9).
Fig. 9.
A Shox2-coordinated transcription program for stylopod patterning. In the developing proximal limb, juxtaposition of the proximal TALE-dominant domain and the relative distal Shox2-dominant domain (where Shox2 is the dominant identity and the dominant form of Shox2/TALE co-occupancy on their co-occupied enhancers) results in transcription that is favorable for osteogenesis of the perichondial Shox2+ progenitor cells. These cells simultaneously instruct the chondrogenic differentiation of stylopodial skeleton primordia (SP) and undergo osteogenic differentiation to provide osteoblasts to form bone.
Enhancer grammar for proximal limb patterning and epigenomic specification of the proximal limb
Understanding the grammar for tissue-specific enhancers is essential for deciphering the subregion-specific transcription output. In addition to the limb enhancers characterized by approaches utilizing whole-limb tissue (Visel et al., 2009), the enhancers for the zone of polarizing activity (ZPA) and apical ectodermal ridge (AER) were also characterized by microdissection with H3K27ac ChIP-Seq (VanderMeer et al., 2014). However, the enhancer grammar underlying the PD patterning of the limb and whether the PD specificity can be recapitulated by enhancer elements are yet to be addressed. We took the advantage of the specific expression domain of Shox2 in the proximal limb and its essential function for proximal limb patterning to characterize putative functional enhancers with proximal limb-specific activities. By computational and comparative analyses on our Shox2 ChIP-Seq data obtained from the limb and palate as well as Pbx ChIP-Seq on the limb with existing ChIP-Seq data, we found that the regulatory elements for Shox2/TALE co-occupation probably represent the key features for the proximal limb-specific enhancers. This notion is further supported by a de novo discovered proximal limb enhancer for Meis1 (Meis1En) based on Shox2/TALE co-occupancy.
Although Meis and Pbx are strongly expressed in the trunk region (Mercader et al., 2000; Penkov et al., 2013) and our study also revealed an expression gradient of Meis from the trunk to the distal end of the stylopod, the Meis1En identified in this study is weakly occupied by Meis and Pbx in the trunk region (Penkov et al., 2013), suggesting that the alternative use of the limb enhancer that is repressed by Shox2 underlies the trunk to limb (proximal to distal) expression gradient of Meis. Notably, a large population of Shox2/Pbx co-occupied sites is not (or is only weakly) accessible to Pbx and Meis in the trunk region (Fig. 7L), suggesting that the Shox2/TALE co-occupied elements that are specifically accessible in the limb represent the epigenomic change required for the specification of the proximal limb mesenchyme. This highlights the importance of Shox2 in specification of the stylopod that emerges in juxtaposition of the trunk, with its strong Meis/Pbx expression and the proximal limb, where Shox2 is highly expressed. However, because of the closely located binding centers of Shox2 and Pbx genome wide (Fig. S7), and the lack of a Pbx de novo motif – a phenomenon that is typical for cooperative binding of transcription factors (Jolma et al., 2015) – Shox2-TALE co-occupancy appears to be competitive. Moreover, since Hox9/Hox10 proteins were thought to interact with Meis/Pbx to pattern the proximal limb (Capellini et al., 2011; Zakany and Duboule, 2007), the fact that Shox2 interacted directly with Hoxa9 in co-IP assay (Fig. S8) further suggests that on the Shox2/TALE co-occupied enhancers, Shox2 may also act as an alternative co-factor for Hox9/Hox10 to exert a different transcription output. Interestingly, in the palate that is derived from the Hox-free first BA (Amin et al., 2015), Shox2 displays a distinct pattern of genome-wide occupancy, implying that, in the absence of Hox factors, Shox2-TALE factors regulate a unique set of palate-specific modules.
Interestingly, a large population of Shox2/TALE co-occupied sites is also occupied by Gli3 (Vokes et al., 2008), an early anterior-posterior limb patterning factor and an effector for Shh signaling, in the developing limb bud (Fig. S9). In light of the fact that Hox and Shh show substantial epistatic cross talks and that Gli3 is essential for proximal limb skeleton development (Barna et al., 2005), the co-occupation of Gli3 and Shox2/TALE suggests that such cross talk is realized by converged transcription output on the same enhancers.
Cell lineages that are commissioned for patterning
The fact that Shox2 mutation results in a specific ablation of the perichondrial Shox2+/Runx2+ osteogenic precursors and subsequently mispatterned cartilaginous mode in the stylopod highlight a crucial role for osteogenic precursors in chondrogenic differentiation. This conclusion is supported by phenotypic reminiscence in mice carrying Shox2 inactivation in the osteogenic lineage. Notably, the expression of Shox2 in many connective tissue cell types and its later expression in perichondrial outer layer cells are reminiscent of that of Hox11 (Swinehart et al., 2013), which patterns the zeugopodial skeleton elements. Together with the fact that the perichondrial expression of Hox13 was also proposed to pattern the autopodial skeleton (Villavicencio-Lorini et al., 2010), our study indicates that the patterning can be exerted by controlling the specification and differentiation of the perichondrial osteogenic precursor cells to simultaneously modulate the patterning of cartilaginous mode and bone formation. Interestingly, in the HoxA/D compound mutant, the expression of Shox2 in the embryonic perichondrium, but not chondrocytes, is severely affected (Neufeld et al., 2014), indicating that Shox2 itself is a key effector for Hox-TALE patterning system in the stylopod and further supports the idea that Hox-TALE patterns skeletal elements by commissioning perichondrial cells. Given the instructive role of perichondrial cells on chondrocyte differentiation and maturation (Ohbayashi et al., 2002), further dissection of Shox2-specific targets in the osteogenic lineage would further highlight key factors involved in such patterning function.
MATERIALS AND METHODS
Mouse models
The Shox2HA, Shox2+/−, Shox2LacZ, Shox2Cre, Shox2Flox, RosamTmG, Nkx2-5Flox, Col2a1-Cre and Col3.6-Cre alleles used in this study were described previously (Cobb et al., 2006; Liu et al., 2004; Ovchinnikov et al., 2000; Sun et al., 2013; Wang et al., 2014; Ye et al., 2015b; Yu et al., 2005). The animal experiments in this study were approved by The Tulane University Institutional Animal Care and Use Committee. Transgenic animal work performed at the Lawrence Berkeley National Laboratory (LBL) was reviewed and approved by the LBL Animal Welfare and Research Committee. Transgenic mouse assays were performed in Mus musculus FVB strain mice. Sample sizes were selected empirically based on our previous experience of performing transgenic mouse assays for >2000 total putative enhancers. Mouse embryos were only excluded from further analysis if they did not carry the reporter transgene or if they were not at the correct developmental stage. As all transgenic mice were treated with identical experimental conditions, and as there were no groups of animals directly compared in this section of the study, randomization and experimenter blinding were unnecessary and not performed.
Collection of embryos, histology, immunohistochemistry and skeletal staining
Embryos from timed pregnant females were fixed in ice-cold 4% paraformaldehyde (PFA), embedded in paraffin, and sectioned at 8 µm for immunohistochemistry analyses. Antibodies used in the study are described in supplementary Materials and Methods. Alcian Blue/Alizarin Red skeletal staining was performed using established protocol, as described previously (Bobick and Cobb, 2012; Yu et al., 2007).
Immunoprecipitations and western blotting
Immunoprecipitation (IP), co-immunoprecipitation (Co-IP), chromatin-immunoprecipitation (ChIP) and western blotting were performed as described previously (Ye et al., 2015b). ChIP-Seq and associated analysis are described in Supplemental Information. Antibodies used for ChIP and ChIP-Seq are described in supplementary Materials and Methods.
FACS, RNA extraction and RNA sequencing
The GFP+ proximal embryonic limbs were dissected out from E12.5 Shox2Cre/+;RosamTmG and Shox2Cre/−;RosamTmG embryos under a fluorescence dissecting scope and subjected to digestion by a cocktail of collagenase I, II and IV, followed by a brief trypsin treatment. Suspended cells were sorted based on GFP signal and subsequently subjected to RNA extraction and RNA sequencing. The associated analysis is described in supplementary Materials and Methods.
Statistical analysis
All experiments were repeated at least three times. Quantification results are presented as mean±s.d., and statistical analysis was conducted using Student's t-test. For qPCR, reactions for each sample were also performed in triplicate. P<0.05 was considered significant.
Acknowledgements
The LHB-A-Luc plasmid used in this study was kindly provided by Jessica Rosin currently at Seattle Children's hospital.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
W.Y. and Y.C. conceived the project. W.Y. and Y.S. performed most experiments, collected and analyzed data. W.Y. and Z.H. performed and analyzed ChIP-Seq results. W.Y. and Z.H. generated constructs for transgenic enhancer assay. Z.H. performed luciferase assay. J.X., A.L., B.B., N.R., and R.S. helped with x-gal staining, histological and skeletal preparation. S.A.-O., D.Y., L.Z., B.B., C.-L.C, J.C., and Y.Z. provided necessary reagents. M.O. and A.V. performed transgenic enhancer assay described in the manuscript. W.Y. prepared figures and wrote the manuscript. J.C., M.O., A.V. and Y.Z. provided insights and helped in manuscript editing. Y.C. conducted final revision and editing of the manuscript.
Funding
We acknowledge financial support by grants from the National Institutes of Health [R01DE017792 and R01DE024152 to Y.C.; R24HL123879, U01DE024427, R01HG003988, U54HG006997, and UM1HL098166 to A.V.], an American Heart Association Predoctoral Fellowship [13PRE1375003 to W.Y.], a grant [WKJ-FJ-24] from the National Health and Family Planning Commission of the People's Republic of China, and a grant [201510011] from the International Collaboration Program of Fujian Province, China. M.O. was supported by a Swiss National Science Foundation (Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung) Fellowship. Research conducted at the E.O. Lawrence Berkeley National Laboratory was performed under U.S. Department of Energy Contract DE-AC02-05CH11231, University of California. Deposited in PMC for release after 12 months.
Data availability
ChIP-Seq and RNA-Seq data have been deposited in Gene Expression Omnibus (GEO) with accession numbers GSE81897 and GSE82300, respectively. Available at: ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81897 and ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82300.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.138750.supplemental
References
- Amin S., Donaldson I. J., Zannino D. A., Hensman J., Rattray M., Losa M., Spitz F., Ladam F., Sagerström C. and Bobola N. (2015). Hoxa2 selectively enhances Meis binding to change a branchial arch ground state. Dev. Cell 32, 265-277. 10.1016/j.devcel.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C., Nord A. S., Zhu Y., Blow M. J., Biddie S. C., Mendenhall E. M., Dixon J., Wright C., Hosseini R., Akiyama J. A. et al. (2014). Tissue-specific SMARCA4 binding at active and repressed regulatory elements during embryogenesis. Genome Res. 24, 920-929. 10.1101/gr.168930.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M., Pandolfi P. P. and Niswander L. (2005). Gli3 and Plzf cooperate in proximal limb patterning at early stages of limb development. Nature 436, 277-281. 10.1038/nature03801 [DOI] [PubMed] [Google Scholar]
- Bobick B. E. and Cobb J. (2012). Shox2 regulates progression through chondrogenesis in the mouse proximal limb. J. Cell Sci. 125, 6071-6083. 10.1242/jcs.111997 [DOI] [PubMed] [Google Scholar]
- Capellini T. D., Zappavigna V. and Selleri L. (2011). Pbx homeodomain proteins: TALEnted regulators of limb patterning and outgrowth. Dev. Dyn. 240, 1063-1086. 10.1002/dvdy.22605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S.-K., Lu P., Nakamura M., Lee J. and Sagerström C. G. (2009). Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev. Cell 17, 561-567. 10.1016/j.devcel.2009.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J., Dierich A., Huss-Garcia Y. and Duboule D. (2006). A mouse model for human short-stature syndromes identifies Shox2 as an upstream regulator of Runx2 during long-bone development. Proc. Natl. Acad. Sci. USA 103, 4511-4515. 10.1073/pnas.0510544103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnot C., Lu C., Hu D. and Helms J. A. (2004). Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev. Biol. 269, 55-69. 10.1016/j.ydbio.2004.01.011 [DOI] [PubMed] [Google Scholar]
- Crocker J., Abe N., Rinaldi L., McGregor A. P., Frankel N., Wang S., Alsawadi A., Valenti P., Plaza S., Payre F. et al. (2015). Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160, 191-203. 10.1016/j.cell.2014.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham T. J. and Duester G. (2015). Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 16, 110-123. 10.1038/nrm3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S. and Ren B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376-380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis R. A., Yu L., He F., Liu H., Tang R., Shi J., Sun X., Martin J. F., Wang D., Yang J. et al. (2009). Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 327, 376-385. 10.1016/j.ydbio.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J. A. R., Hassan M. Q., Saini S., Montecino M., van Wijnen A. J., Stein G. S., Stein J. L. and Lian J. B. (2010). Pbx1 represses osteoblastogenesis by blocking Hoxa10-mediated recruitment of chromatin remodeling factors. Mol. Cell. Biol. 30, 3531-3541. 10.1128/MCB.00889-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S., Wei N., Yu X., Jiang Y., Fei J. and Chen Y. (2008). Mice with an anterior cleft of the palate survive neonatal lethality. Dev. Dyn. 237, 1509-1516. 10.1002/dvdy.21534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Sitwala K., Bronstein J., Sanders D., Dandekar M., Collins C., Robertson G., MacDonald J., Cezard T., Bilenky M. et al. (2012). Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood 119, 388-398. 10.1182/blood-2011-03-341081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudry B., Thomas-Chollier M., Volovik Y., Duffraisse M., Dard A., Frank D., Technau U. and Merabet S. (2014). Molecular insights into the origin of the Hox-TALE patterning system. Elife 3, e01939 10.7554/eLife.01939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante C. R., Park S., Mihala A. G., Kingsley D. M. and Menke D. B. (2013). Pitx1 broadly associates with limb enhancers and is enriched on hindlimb cis-regulatory elements. Dev. Biol. 374, 234-244. 10.1016/j.ydbio.2012.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A., Yin Y., Nitta K. R., Dave K., Popov A., Taipale M., Enge M., Kivioja T., Morgunova E. and Taipale J. (2015). DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527, 384-388. 10.1038/nature15518 [DOI] [PubMed] [Google Scholar]
- Kmita M., Tarchini B., Zakany J., Logan M., Tabin C. J. and Duboule D. (2005). Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature 435, 1113-1116. 10.1038/nature03648 [DOI] [PubMed] [Google Scholar]
- Kondrashov N., Pusic A., Stumpf C. R., Shimizu K., Hsieh A. C., Xue S., Ishijima J., Shiroishi T. and Barna M. (2011). Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383-397. 10.1016/j.cell.2011.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Woitge H. W., Braut A., Kronenberg M. S., Lichtler A. C., Mina M. and Kream B. E. (2004). Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int. J. Dev. Biol. 48, 645-653. 10.1387/ijdb.041816fl [DOI] [PubMed] [Google Scholar]
- Long F. and Ornitz D. M. (2013). Development of the endochondral skeleton. Cold Spring Harb. Perspect. Biol. 5, a008334 10.1101/cshperspect.a008334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R. S., Lelli K. M. and Joshi R. (2009). Hox specificity: unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 88, 63-101. 10.1016/S0070-2153(09)88003-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N., Leonardo E., Piedra M. E., Martinez A. C., Ros M. A. and Torres M. (2000). Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development 127, 3961-3970. [DOI] [PubMed] [Google Scholar]
- Mercader N., Selleri L., Criado L. M., Pallares P., Parras C., Cleary M. L. and Torres M. (2009). Ectopic Meis1 expression in the mouse limb bud alters P-D patterning in a Pbx1-independent manner. Int. J. Dev. Biol. 53, 1483-1494. 10.1387/ijdb.072430nm [DOI] [PubMed] [Google Scholar]
- Moens C. B. and Selleri L. (2006). Hox cofactors in vertebrate development. Dev. Biol. 291, 193-206. 10.1016/j.ydbio.2005.10.032 [DOI] [PubMed] [Google Scholar]
- Montavon T., Soshnikova N., Mascrez B., Joye E., Thevenet L., Splinter E., de Laat W., Spitz F. and Duboule D. (2011). A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132-1145. 10.1016/j.cell.2011.10.023 [DOI] [PubMed] [Google Scholar]
- Neufeld S. J., Wang F. and Cobb J. (2014). Genetic interactions between Shox2 and Hox genes during the regional growth and development of the mouse limb. Genetics 198, 1117-1126. 10.1534/genetics.114.167460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi N., Shibayama M., Kurotaki Y., Imanishi M., Fujimori T., Itoh N. and Takada S. (2002). FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 16, 870-879. 10.1101/gad.965702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M., Speziale D., Shoukry M., Mohan R., Ivanek R., Kohler M., Beisel C., Wen X., Scales S. J., Christoffels V. M. et al. (2014). HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme. Dev. Cell 31, 345-357. 10.1016/j.devcel.2014.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov D. A., Deng J. M., Ogunrinu G. and Behringer R. R. (2000). Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 26, 145-146. [DOI] [PubMed] [Google Scholar]
- Parker H. J., Piccinelli P., Sauka-Spengler T., Bronner M. and Elgar G. (2011). Ancient Pbx-Hox signatures define hundreds of vertebrate developmental enhancers. BMC Genomics 12, 637 10.1186/1471-2164-12-637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. C., Lemons D. and McGinnis W. (2005). Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6, 893-904. 10.1038/nrg1726 [DOI] [PubMed] [Google Scholar]
- Penkov D., Mateos San Martin D., Fernandez-Diaz L. C., Rossello C. A., Torroja C., Sanchez-Cabo F., Warnatz H. J., Sultan M., Yaspo M. L., Gabrieli A. et al. (2013). Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Rep. 3, 1321-1333. 10.1016/j.celrep.2013.03.029 [DOI] [PubMed] [Google Scholar]
- Raines A. M., Magella B., Adam M. and Potter S. S. (2015). Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development. BMC Dev. Biol. 15, 28 10.1186/s12861-015-0078-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin J. M., Abassah-Oppong S. and Cobb J. (2013). Comparative transgenic analysis of enhancers from the human SHOX and mouse Shox2 genomic regions. Hum. Mol. Genet. 22, 3063-3076. 10.1093/hmg/ddt163 [DOI] [PubMed] [Google Scholar]
- Shen Y., Yue F., McCleary D. F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V. V. et al. (2012). A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116-120. 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M., Riley T., Liu P., Abe N., Gomez-Alcala P., Dror I., Zhou T., Rohs R., Honig B., Bussemaker H. J. et al. (2011). Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147, 1270-1282. 10.1016/j.cell.2011.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John H. C., Bishop K. A., Meyer M. B., Benkusky N. A., Leng N., Kendziorski C., Bonewald L. F. and Pike J. W. (2014). The osteoblast to osteocyte transition: epigenetic changes and response to the vitamin D3 hormone. Mol. Endocrinol. 28, 1150-1165. 10.1210/me.2014-1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Zhang T., Liu C., Gu S. and Chen Y. (2013). Generation of Shox2-Cre allele for tissue specific manipulation of genes in the developing heart, palate, and limb. Genesis 51, 515-522. 10.1002/dvg.22397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinehart I. T., Schlientz A. J., Quintanilla C. A., Mortlock D. P. and Wellik D. M. (2013). Hox11 genes are required for regional patterning and integration of muscle, tendon and bone. Development 140, 4574-4582. 10.1242/dev.096693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabin C. and Wolpert L. (2007). Rethinking the proximodistal axis of the vertebrate limb in the molecular era. Genes Dev. 21, 1433-1442. 10.1101/gad.1547407 [DOI] [PubMed] [Google Scholar]
- VanderMeer J. E., Smith R. P., Jones S. L. and Ahituv N. (2014). Genome-wide identification of signaling center enhancers in the developing limb. Development 141, 4194-4198. 10.1242/dev.110965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman L., Neufeld S. and Cobb J. (2011). Shox2 function couples neural, muscular and skeletal development in the proximal forelimb. Dev. Biol. 350, 323-336. 10.1016/j.ydbio.2010.11.031 [DOI] [PubMed] [Google Scholar]
- Villavicencio-Lorini P., Kuss P., Friedrich J., Haupt J., Farooq M., Turkmen S., Duboule D., Hecht J. and Mundlos S. (2010). Homeobox genes d11-d13 and a13 control mouse autopod cortical bone and joint formation. J. Clin. Invest. 120, 1994-2004. 10.1172/JCI41554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Minovitsky S., Dubchak I. and Pennacchio L. A. (2007). VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88-D92. 10.1093/nar/gkl822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Blow M. J., Li Z., Zhang T., Akiyama J. A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F. et al. (2009). ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854-858. 10.1038/nature07730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes S. A., Ji H., Wong W. H. and McMahon A. P. (2008). A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 22, 2651-2663. 10.1101/gad.1693008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., Tang Q., Meyer C. A., Zhang Y. and Liu X. S. (2013). Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 8, 2502-2515. 10.1038/nprot.2013.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Bai Y., Li N., Ye W., Zhang M., Greene S. B., Tao Y., Chen Y., Wehrens X. H. T. and Martin J. F. (2014). Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc. Natl. Acad. Sci. USA 111, 9181-9186. 10.1073/pnas.1405411111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Whitfield T. W., Gordon J. A. R., Dobson J. R., Tai P. W. L., van Wijnen A. J., Stein J. L., Stein G. S. and Lian J. B. (2014). Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol. 15, R52 10.1186/gb-2014-15-3-r52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W., Song Y., Huang Z., Zhang Y. and Chen Y. (2015a). Genetic regulation of sinoatrial node development and pacemaker program in the venous pole. J. Cardiovasc. Dev. Dis. 2, 282-298. 10.3390/jcdd2040282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W., Wang J., Song Y., Yu D., Sun C., Liu C., Chen F., Zhang Y., Wang F., Harvey R. P. et al. (2015b). A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 142, 2521-2532. 10.1242/dev.120220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Gu S., Alappat S., Song Y., Yan M., Zhang X., Zhang G., Jiang Y., Zhang Z., Zhang Y. et al. (2005). Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development 132, 4397-4406. 10.1242/dev.02013 [DOI] [PubMed] [Google Scholar]
- Yu L., Liu H., Yan M., Yang J., Long F., Muneoka K. and Chen Y. (2007). Shox2 is required for chondrocyte proliferation and maturation in proximal limb skeleton. Dev. Biol. 306, 549-559. 10.1016/j.ydbio.2007.03.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B. D. et al. (2014). A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355-364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany J. and Duboule D. (2007). The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 17, 359-366. 10.1016/j.gde.2007.05.011 [DOI] [PubMed] [Google Scholar]