Abstract
Solid-state NMR spectroscopy allows the characterization of structure, interactions and dynamics of insoluble and/or very large proteins. Sensitivity and resolution are often major challenges for obtaining atomic-resolution information, in particular for very large protein complexes. Here we show that the use of deuterated, specifically CH3-labelled proteins result in significant sensitivity gains compared to previously employed CHD2 labelling, while line widths only marginally increase. We apply this labelling strategy to a 468 kDa-large dodecameric aminopeptidase, TET2, and the 1.6 MDa-large 50S ribosome subunit of Thermus thermophilus.
Biomolecular magic-angle spinning (MAS) solid-state NMR (ssNMR) has made substantial progress over the last decade in terms of sensitivity, resolution and information content, and established as an important technique in structural biology, addressing structure and dynamics in systems with increasing complexity.1–3 The progress was enabled by improved NMR instrumentation, in particular probes allowing fast MAS (up to 100 kHz), improved pulse sequences, as well as better sample preparation and isotope labelling techniques. Experiments based on proton-detection are particularly powerful for the study of protein structure, interactions and dynamics. The large gyromagnetic ratio of protons translates to high intrinsic detection sensitivity, and ability to measure long-range distances. However, the large 1H gyromagnetic ratio and the corresponding strong 1H-1H dipolar couplings also result in large 1H line widths. Proton-detected experiments are, therefore, particularly powerful when protons are introduced only at selected sites of interest in an otherwise deuterated environment, and when high MAS frequencies are employed, leading to efficient averaging of 1H-1H interactions. Methyl groups are particularly interesting reporters of protein structure and dynamics in proteins, due to their widespread abundance in proteins, often located in important hydrophobic core regions4. In solution-state NMR, methyl-directed studies have provided a wealth of information about dynamics, interactions and structure in particular for large complexes (up to 1 MDa), because in this case methyls are the only observable NMR reporters.5,6 Protocols for introducing methyls of all iso-topomers (CH3, CH2D, CHD2) into otherwise deuterated proteins4 have been established. The three isotopomers differ in the number and nature of interactions, and thus in the spin relaxation properties, sensitivity and resolution. In solution-state NMR, the most commonly employed isotopomer is CH3, which generally provides the highest sensitivity due to the three-fold spin multiplicity. As long as optimized pulse schemes are used7, the spectra of all isotopomers exhibit similar line widths.8 In ssNMR, only few studies have reported the use of specifically methyl-labelled samples for studying dynamics and structure.3,6–8 All these studies have employed CHD2 labelling, with the exception of a brief note of CH3 labelling.9 The primary argument for using the CHD2 isotopomer is related to the fact that there are no intra-methyl 1H-1H dipolar couplings in CHD2, and that these interactions are expected to lead to strong line broadening; this assumption was supported by pioneering experiments on stochastically labelled proteins at low (20 kHz) MAS frequency,10 but not systematically investigated. Here we show that, in experiments performed at high MAS frequency (50-60 kHz) the line widths in CHD2 and CH3 methyl are comparable, and that CH3 labelling results in improved sensitivity compared to CHD2. The enhanced sensitivity opens possibilities to study even very large objects, as demonstrated with the 1.6 MDa-large 50S ribosome, where sensitivity is a great challenge.
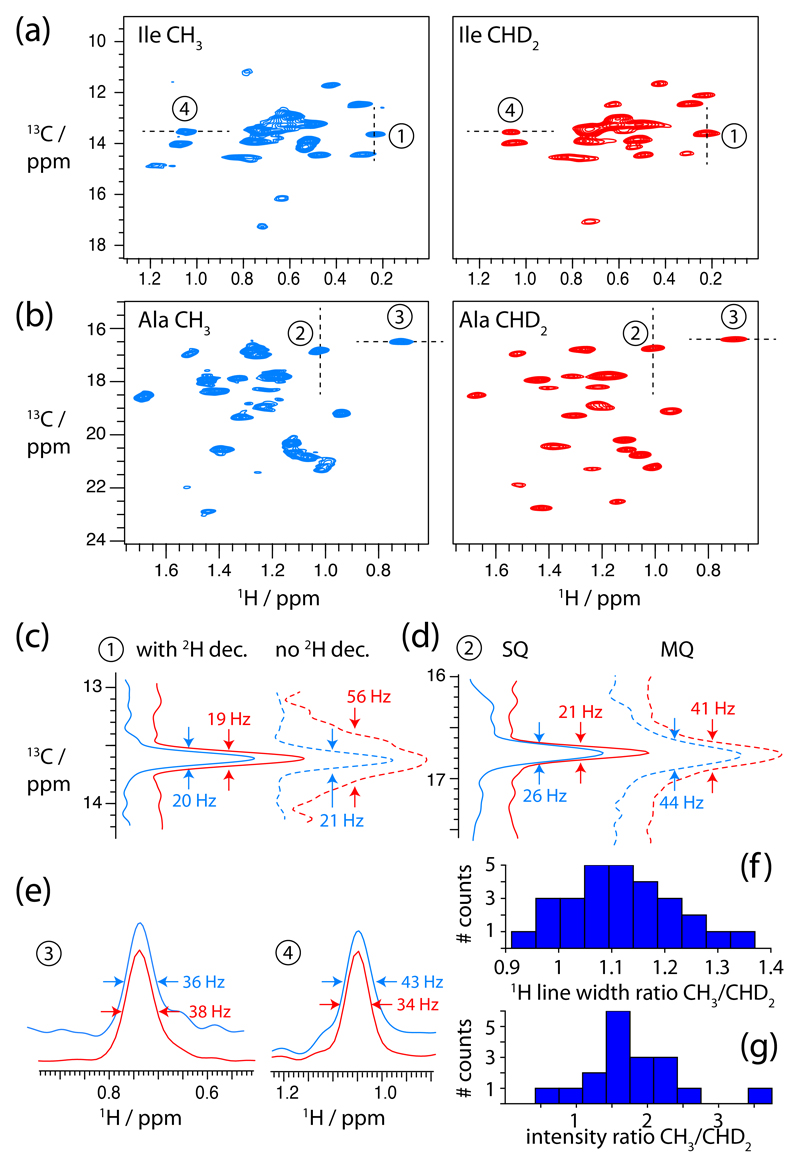
In order to investigate the properties of the different methyl isotopomers in ssNMR, we compared four samples of the enzyme TET2 from Pyrococcus horikoshii, a dodecameric (12x39 kDa) aminopeptidase involved in the cellular protein quality control, with different methyl labelling schemes. (Details about sample preparation can be found in the ESI.) With 353 residues per monomer and a total oligomer mass of 468 kDa, TET2 is among the largest proteins studied so far by ssNMR, making it thus a considerable challenge in terms of resolution and sensitivity. In the samples we incorporated either CH3 or CHD2 methyl groups on isoleucine residues or on alanines, respectively, in an otherwise deuterated background. The different lengths and flexibility of Ala and Ile side chains generally lead to different dynamics, reflecting the range of methyl cases encountered in proteins. Figure 1a,b shows comparisons of 1H-13C correlation spectra of these four samples, obtained with cross-polarization transfer steps at a MAS frequency of 57 kHz. High-resolution spectra are obtained for this very large protein assembly for both isotopomers and both residue types. The spectra are similar to solution-state spectra of TET, showing that structure and dynamics in our solid-state preparations are conserved in the solid state, and allowing us to transfer previously known assignments from solution-state NMR11 (see Supporting Information Figure S2). In CHD2 groups, the observed line widths are of the order of 30 – 45 Hz (1H) and 20-30 Hz (13C), respectively (Figure 1c-e). These values are in good agreement with previously reported line widths obtained in fast-MAS solid-state NMR experiments on smaller model proteins with CHD2 labelling9,12,13. For the CH3 isotopomer, the line widths along the 13C dimension are similar to the ones observed with CHD2 groups (20-30 Hz), suggesting that the intra-methyl heteronuclear 1H-13C dipolar coupling is not the dominant factor for line width under the chosen conditions (heteronuclear decoupling, fast MAS). This is expected, given that fast-MAS, the high deuteration level of the surroundings, and the 3-fold reduced dipolar 1H-13C coupling that results from the rapid methyl rotation, make decoupling very efficient. Remarkably, the 1H line widths in the CH3 sample are only slightly larger than the ones in CHD2 samples (Figure 1e,f). The fact that reducing the number of methyl proton spins from three (CH3) to one (CHD2) has only a modest effect on line widths suggests that the intra-methyl dipolar couplings are not determining the 1H line widths. Sample inhomogeneity, dipolar inter-actions to remote protons as well as dynamics may be important contributions to line widths, such that intra-methyl 1H-1H dipolar interactions do not dominate. Of note, given that the line widths observed here are similar to microcrystalline samples,9,12 our conclusions hold also for those systems. In more heterogeneous systems (fibrils, membrane proteins) the differences in line widths between the two isotopomers are expected to be similar or smaller than here, because line widths due to sample heterogeneity is independent of the labelling scheme. Importantly, in terms of sensitivity the CH3 labelling scheme outcompetes CHD2 labelling, on average by a factor of about 1.9 (Figure 1g). This is due to the proton spin multiplicity (expected gain of 3), reduced by the somewhat faster relaxation of CH3 methyls (see below).
Fig. 1.
Proton-detected 2D methyl 1H-13C spectra of CH3 (blue) and CHD2 (red) methyls of (a) isoleucines and (b) alanines in TET2, obtained at 14.1 T (600 MHz 1H Larmor frequency). These spectra were obtained with the CP-based pulse sequence shown in ESI Figure S1a. Traces along the positions indicated with dashed lines and numbers are shown in (c)-(e). Dashed 1D traces were obtained at the indicated positions in the spectra, but different pulse sequences, either omitting 2H decoupling, or using MQ evolution (see Figure S1). Line-widths (full-width at half maximum) are indicated for these selected peaks. (f) shows the ratio of line widths observed for CH3 and CHD2 correlations, merged for both Ala and Ile samples. (g) Intensity ratios for cross-peaks observed in the Ile spectrum, taking into account the protein amounts in CH3/CHD2 samples. For all these spectra, resolution enhancement was applied using a 70-degrees shifted sine-bell square function (90 degrees would correspond to a cosine, i.e. no resolution enhancement), see ESI.
The experiments shown in Figure 1a,b were recorded with 2H decoupling during chemical-shift evolution periods, which requires a probehead able to tune to 2H frequency. Such equipment is not standard in contemporary MAS probes. As expected, while omission of 2H decoupling has virtually no effect on line widths and sensitivity in CH3-labelled methyls (Figure 1c), due to the absence of 1JCD couplings, in CHD2 groups the undecoupled 13C line widths are dramatically increased due to the scalar coupling (˜20 Hz) multiplet structure.12 Therefore, if 2H decoupling is impossible with the available hardware, the CH3 isotopomer is clearly favoured in terms of resolution and sensitivity. The effects of 2H decoupling on 1H line widths are negligible, as the 2JHD coupling is small (˜1-3 Hz).12 We have also investigated two different types of 13C evolution, single-quantum (SQ), and multiple-quantum (MQ) evolution. In solution-state NMR, in the case of slow overall molecular tumbling, MQ coherence life times are significantly longer than SQ life times14. In contrast, Figure 1d establishes that MQ lines in our ssNMR experiments are almost two-fold larger than their SQ counterparts. This can be understood by considering that due to the absence of overall tumbling, cross-correlated relaxation is small (reducing the potential benefit of MQ evolution), and dephasing due to remote protons outweighs the potential gains from cross-correlated relaxation. We also investigated the relative sensitivity obtained with INEPT-transfer and CP-transfer. INEPT-based transfer is favoured by large-scale dynamics and extensive deuteration, because both factors increase coherence life times and thus INEPT transfer efficiency. We find indeed that for the more dynamic Ile residues INEPT outcompetes CP-transfer, while for Ala we find similar sensitivity (Figure S4).
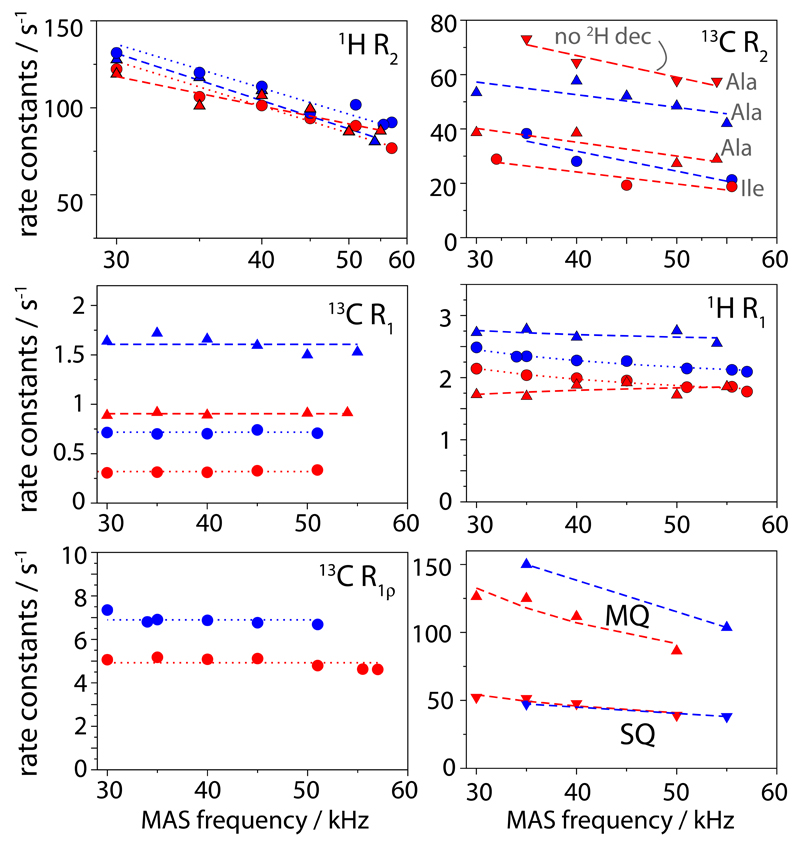
Line widths are related to the life times of spin coherences, which are determined by spin relaxation (i.e., dynamics) and dephasing that is due to incompletely averaged dipolar interactions; the latter contribution depends on the MAS frequency. To further gain insight into the properties of the different labellings, we performed systematic analyses of apparent relaxation rate constants. These measurements, shown in Figure 2, corroborate the trends shown in Figure 1: (i) 1H R2’ relaxation rates are similar in CH3 and CHD2 samples, with values of ˜100 s-1 and ˜80 s-1, respectively, at high MAS frequency (55-57 kHz). The expected line widths from these numbers (R2’/π, ˜25-30 Hz) are smaller than the experimentally observed values (˜35-45 Hz), showing that part of the line width originates from sample and/or magnetic-field heterogeneity. (ii) 13C R2’ of ˜20-40 s-1 are found at high MAS frequencies (˜55 kHz). The line widths predicted from these numbers (6-12 Hz) are significantly smaller than the experimentally observed line widths in this study and crystalline ubiquitin before12 (˜18-25 Hz), showing that heterogeneity limits the resolution of 13C lines. (iii) Omission of 2H decoupling leads to significantly shorter 13C T2’, in CHD2 (but not CH3), in agreement with the larger line widths (Figure 1c) (iv) MQ coherences decay faster than their SQ counterpart, again in line with data shown in Figure 1d. (v) Furthermore, proton R1 rate constants are ˜15% higher in CH3 compared to CHD2. This allows higher repetition rate of experiments, and thus slightly increased sensitivity for CH3 samples (see Figure S4). (vi) We also measured 13C R1 and 13C R1ρ rate constants, and find that they are not MAS-frequency dependent. This finding establishes that coherent dephasing (i.e. contribution to the decay that is not due to dynamics, and which would be MAS dependent) is negligible, allowing quantitative interpretation of methyl dynamics. While 13C R1 has been used for this purpose before,13 the possibility to quantitatively interpret R1ρ enables the study of slow (ns-ms) motion.15
Fig. 2.
MAS-frequency dependence of various spin relaxation parameters in Ile-labelled (circles) and Ala-labelled (triangles) TET2, with CH3 (blue) and CHD2 (red) isotopomers. Bulk methyl relaxation rate constants were obtained from a series of 1D spectra, using pulse sequences shown in ESI Figure S1 .
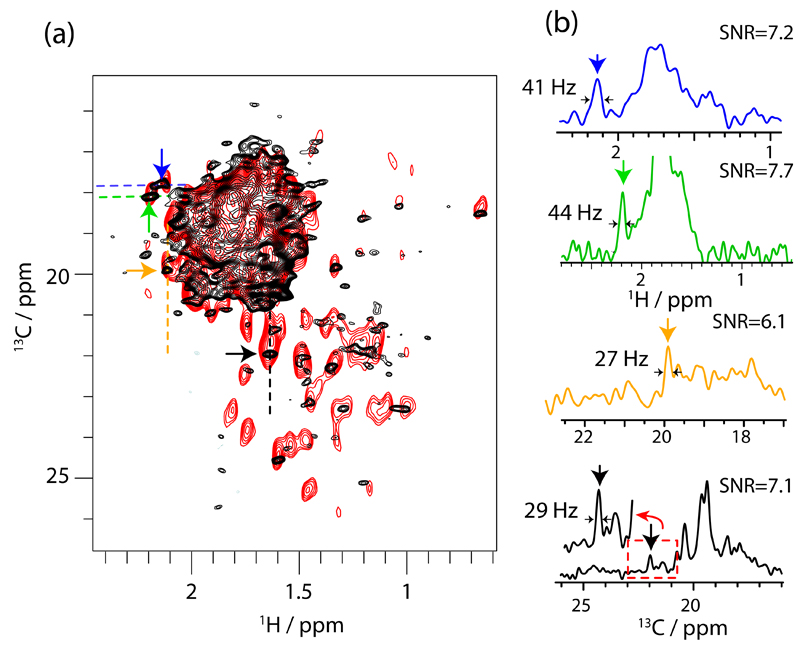
Having thus established CH3 labelling provides enhanced sensitivity compared to CHD2, with similar resolution, we turned to a very challenging application, the 50S ribosome particle from Thermus thermophilus. Ribosomes are among the most complex biological machineries, composed of RNAs and proteins, with a total molecular mass of 1.6 MDa (for the 50S subunit). Unlike TET2, where the 12-fold symmetry improves sensitivity and reduces spectral complexity, ribosomes are not symmetric. Sensitivity and resolution are thus a great challenge. Earlier studies by solution-state NMR have observed only highly mobile isotope-labelled proteins attached to ribosomes16, or flexible nascent polypeptides exiting the ribo-some,17,18 while less flexible proteins are invisible due to unfavourable relaxation properties. Recently, ssNMR spectra have been reported for tightly ribosome-bound proteins, which were isotope labelled and bound to unlabelled 50S and 30S subunits, respectively.19,20 This selective labelling of a single protein overcomes the severe resonance overlap problem but requires reconstitution of ribosomes from labelled/unlabelled subunits, which is not generally possible. We investigate here the possibility to study uniformly labelled ribosome (50S subunit, 1.6 MDa), obtained from bacterial cells grown in uniformly deuter-ated, Ala-CH3-labeled media, and sedimentation into a 1.3 mm ssNMR rotor21. The HSQC-type 1H-13C correlation spectrum obtained in a total experimental time of 14 hours is shown in Figure 3 (black). As expected, the large number of non-equivalent Ala methyl sites – 337 in total – results in significant resonance overlap. Nonetheless, for the resolved cross-peaks the line widths are similar to TET2 (˜25-30 Hz for 13C and 40-50 Hz for 1H). Signal-to-noise ratios of these cross-peaks are of the order of 3 to 8. Given the small amount of sample (1.2 nmoles) these results are encouraging, and demonstrate that methyl labelling is a route to gain residue-specific information of very large complexes. We also performed an additional experiment, using a shorter 13C evolution time and larger number of scans to favour sensitivity and confirm that the detected cross peaks are indeed reproducible (red spectrum in Figure 3). Resonance assignment and the spectral complexity are of course challenging. Solutions may involve unlabelling of protein components that can be reconstituted, or chemically methyl-labelled amino acid with low abundance, such as methylated cysteines22 or lysines. The data shown here – to our knowledge the first reported ssNMR spectra of uniformly methyl-labelled ribosomes – establish that in terms of sensitivity and line widths, studies of ribosomes are feasible.
Fig. 3.
Alanine methyl ssNMR spectra of Thermus thermophilus ribosome, obtained with 1.5 mg of deuterated, Ala-13CH3 labelled material. (a) 2D spectra obtained with INEPT-transfer (black, pulse sequence Figure S1E), obtained in a total time of 14 hours. The spectrum in red was obtained in 9 hours, using CP-transfers (sequence Figure S1A) with increased number of scans at the expense of resolution (see ESI for details). (b) 1D traces of the black spectrum along the 1H and 13C dimensions were extracted at the positions indicated with dashed lines, and the colours of the traces correspond to the colours of the arrows.
In conclusion, we have shown here that CH3-methyl-labelling allows for high-resolution proton-detected ssNMR at high MAS frequencies. In terms of sensitivity, CH3 labelling significantly outcompetes CHD2 labelling. We believe that CH3 methyl labelling will be very useful for further ssNMR studies of structure, dynamics and interactions, in particular for large systems, as exemplified here.
Supplementary Material
Electronic Supplementary Information (ESI) available: comprehensive experimental details. See DOI: 10.1039/C6CC04484K
Acknowledgments
This work was supported by the European Research Council (ERC-StG-2012-311318 and ERC-StG-2010-260887) and used the platforms of the Grenoble Instruct Center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB).
References
- 1.Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Griesinger C, Kolbe M, Baker D, Becker S, Lange A. Nature. 2012;486:276–9. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Science. 2008;319:1523–6. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 3.Andreas LB, Reese M, Eddy MT, Gelev V, Ni QZ, Miller EA, Emsley L, Pintacuda G, Chou JJ, Griffin RG. J Am Chem Soc. 2015;137:14877–14886. doi: 10.1021/jacs.5b04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sprangers R, Velyvis A, Kay LE. Nat Methods. 2007;4:697–703. doi: 10.1038/nmeth1080. [DOI] [PubMed] [Google Scholar]
- 5.Ruschak AM, Kay LE. J Biomol NMR. 2010;46:75–87. doi: 10.1007/s10858-009-9376-1. [DOI] [PubMed] [Google Scholar]
- 6.Sheppard D, Sprangers R, Tugarinov V. Prog Nucl Magn Reson Spectrosc. 2010;56:1–45. doi: 10.1016/j.pnmrs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. J Am Chem Soc. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- 8.Ollerenshaw JE, Tugarinov V, Skrynnikov NR, Kay LE. J Biomol NMR. 2005;33:25–41. doi: 10.1007/s10858-005-2614-2. [DOI] [PubMed] [Google Scholar]
- 9.Andreas LB, Le Marchand T, Jaudzems K, Pintacuda G. J Magn Reson. 2015;253:36–49. doi: 10.1016/j.jmr.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Agarwal V, Diehl A, Skrynnikov N, Reif B. J Am Chem Soc. 2006;128:12620–12621. doi: 10.1021/ja064379m. [DOI] [PubMed] [Google Scholar]
- 11.Amero C, Asunción Durá M, Noirclerc-Savoye M, Perollier A, Gallet B, Plevin MJ, Vernet T, Franzetti B, Boisbouvier J. J Biomol NMR. 2011;50:229–236. doi: 10.1007/s10858-011-9513-5. [DOI] [PubMed] [Google Scholar]
- 12.Huber M, With O, Schanda P, Verel R, Ernst M, Meier BH. J Magn Reson. 2012;214:76–80. doi: 10.1016/j.jmr.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal V, Xue Y, Reif B, Skrynnikov NR. J Am Chem Soc. 2008;130:16611–16621. doi: 10.1021/ja804275p. [DOI] [PubMed] [Google Scholar]
- 14.Ollerenshaw JE, Tugarinov V, Kay LE. Magn Reson Chem. 2003;41:843–852. doi: 10.1002/mrc.1819. [DOI] [PubMed] [Google Scholar]
- 15.Schanda P, Ernst M. Prog Nucl Magn Reson Spectrosc. 2016;96:1–46. doi: 10.1016/j.pnmrs.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulder FAA, Bouakaz L, Lundell A, Venkataramana M, Liljas A, Akke M, Sanyal S. Biochemistry. 2004;43:5930–5936. doi: 10.1021/bi0495331. [DOI] [PubMed] [Google Scholar]
- 17.Cabrita LD, et al. Nat Struct Mol Biol. 23:278–285. doi: 10.1038/nsmb.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu S-TD, Fucini P, Cabrita LD, Launay H, Dobson CM, Christodoulou J. Proc Natl Acad Sci U S A. 2007;104:16516–16521. doi: 10.1073/pnas.0704664104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbet-Massin E, Huang CT, Daebel V, Hsu STD, Reif B. Angew Chemie - Int Ed. 2015;54:4367–4369. doi: 10.1002/anie.201409393. [DOI] [PubMed] [Google Scholar]
- 20.Gelis I, Vitzthum V, Dhimole N, Caporini MA, Schedlbauer A, Carnevale D, Connell SR, Fucini P, Bodenhausen G. J Biomol NMR. 2013;56:85–93. doi: 10.1007/s10858-013-9721-2. [DOI] [PubMed] [Google Scholar]
- 21.Bertini I, Luchinat C, Parigi G, Ravera E, Reif B, Turano P. Proc Natl Acad Sci U S A. 2011;108:10396–10399. doi: 10.1073/pnas.1103854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Religa TL, Ruschak AM, Rosenzweig R, Kay LE. J Am Chem Soc. 2011;133:9063–9068. doi: 10.1021/ja202259a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.