Abstract
Gastric contractions are governed by a bioelectrical event known as slow waves. High-resolution electrical mapping has recently been applied to study complex gastric slow wave spatiotemporal propagations in detail. As these methods are translated to clinical and experimental applications, it is evident that efficient and automated methods are a necessity for analysis. Despite automated methods to detect slow wave events, manual review and correction remains necessary due to the presence of experimental noise in the recordings. Manual deletion of invalid slow wave events is time consuming and inefficient. We have therefore developed an algorithm to eliminate invalid markers of slow waves, via the use of frequency and morphological analysis. The techniques were validated with experimental data using serosal gastric slow wave recordings from animals and humans with a sensitivity of 90% and specificity of 85%. It is anticipated these methods will facilitate analyzing high-resolution slow wave mapping data and accelerate clinical translation of electrical mapping to clinical and diagnostic gastroentrology.
I. INTRODUCTION
Gastric contractions are governed by rhythmic bio-electric slow wave activity, which occurs around 3 cycles per minutes (cpm) in humans [1]. Slow waves are generated and propagated through the interstitial cells of Cajal present in the gastrointestinal (GI) musculature. Abnormalities of slow waves are being linked to major GI functional motility disorders [2], [3]. Recent advances in recording technology, especially with high-resolution (HR) mapping (100s of electrodes) have been able to define and quantify propagation characteristics of slow wave dysrhythmias [4], [5], [6]. Gastroparesis is a motility disorder involving unremitting nausea and vomiting. HR mapping were undertaken in gastropatetic patients who showed a reduced ICC count, has revealed dysrhythmic patterns of slow wave propagation [3].
When HR mapping techniques were first introduced into the GI field, most of the analysis were performed manually, which was inefficient and subject to observer bias. In recent years, automated techniques, along with software platforms and frameworks have been introduced which has aided in progressing the GI mapping field [7], [8]. Briefly, the overall analysis procedures is as follows; raw data is pre-processed to remove baseline wander and high-frequency noise with validated filtering techniques [9], [10]. Events of interest, such as slow wave activation times, are then marked using fiducial markers and clustered into their propagating wavefronts using automated techniques [11], [12]. Quantification of the slow wave amplitude and velocity profile, followed by identification of slow wave activation patterns can be performed using automated methods, followed by visualization and statistical analysis [13], [6].
One of the time consuming tasks that remains is the manual correction of any automatically detected invalid fiducial markers for slow waves, prior to clustering of events. This occurs due to potential noise artifact in the recordings, and current automated detection techniques have assumed signals are relatively noise free. Some examples of common experimental artifacts for analysis are (i) non-contact or semi-contact of electrode array with electrically active regions of the gut, (ii) placement of electrodes in regions of the gut which are electrically silent (e.g., the fundus region of the stomach), (iii) deteriorated electrodes tips recording bio-signals with low signal to noise ratio and (iv) motion or electrical artifacts, due to electrode repositioning during an experiment, handling of the subject, subject ventilation, or interference from surrounding electrical devices. Some of the noise artifacts are unavoidable during experimental studies, and they manifest as various morphologies. In this study, a novel approach has been developed to eliminate invalid markers to improve the signal processing and analysis pipeline for multi-electrode GI recordings.
II. METHODS
A. Detection methods
The algorithm uses a two stage process to eliminate invalid fiducial markers representing the activation time for each slow wave event (Fig. 1). The first step is based on frequency which does not take into account the fiducial marker positions. This step is primarily aimed at eliminating signals that do not resemble any physiological gastric slow waves. The second step is a localized approach to assess whether the detected marker is a valid gastric slow wave event. Descriptions of each of the steps are detailed below. The first stage is based on frequency analysis for each electrode. The time domain signals X(t) was transformed to the frequency domain via a fast Fourier Transform (FFT): Y = FFT(X(t)), where a fraction (FFTfrac) was calculated for each electrode signal, where the subscript of Y are the frequencies in cycles per minute (cpm).
| (1) |
Figure 1.
Algorithm to eliminate invalid slow wave events. A two-step process is used where the first step is a global check and uses frequency, while the second stage uses a local check which uses the signal amplitude, correlations, and kurtosis.
For robustness, a short term Fourier transform (STFT) was calculated with a moving window size of 2 minutes, and overlapped by 50%. STFT captures the change in frequency over time and here it is primarily used to acquire multiple estimates of the frequency for a given signal. If the average of the FFTfrac for any of the electrode was greater than 1, it denotes that the dominant frequency of the signal lies in the frequency range of 9 to 20 cpm, which is outside the expected frequency range of gastric slow wave events [14], [15], [16]. Thus, if FFTfrac ≥ 1, the signal was not included for analysis. The FFTfrac measure was designed to eliminate the fiducial markers in signals which has dominant ventilator frequency (normally around 9–12 cpm), and maintain fiducial markers in signals which have a dominant slow wave frequency (normally around 3 cpm) [14].
The second stage assesses the signal around the fiducial marker to quantify if the morphology of the signal segment resembles a gastric slow wave event. Three metrics were used to quantify each of the signal segments around a fiducial marker; (i) amplitude of slow wave event, (ii) cross correlation of slow wave events and (iii) kurtosis of the gradient of the slow wave event.
If maximum and minimum voltage limits of the signal segment did not fall within physiologically observed amplitude limits for a gastric slow wave event (0–2 mV) [14], [15], then that fiducial marker was removed. Secondly, cross correlations were computed to assess the shape of the signal segment. It is based on the fact that slow waves in a single channel (or a particular segment) typically have similar morphologies. For each electrode signal, a window was placed around the fiducial marker, and stacked according to the activation time of the slow wave event (Fig. 2). Pearson correlation coefficients (PCC) (Equation 2) were computed against each of the windows. For each fiducial marker, an average correlation coefficient value was obtained across all windows. If it was below a certain threshold (empirically set as 0.4) it was considered an invalid slow wave fiducial marker. To take into account variations of slow wave morphologies in time (which can occur during dysrhythmia), cross correlations were computed on a rolling time segment or for a defined number of slow waves (set at 6 for this study). The PCC measure was chosen for morphological analysis because it is an amplitude insensitive measure, as variations occur physiologically. Fig. 2 shows a set of valid and invalid fiducial markers along with average cross correlation values.
| (2) |
In Equation 2, cov(X, Y) is the covariance of variables X and Y , which are the window segments, while σX and σY are the standard deviation of variables X and Y.
Figure 2.
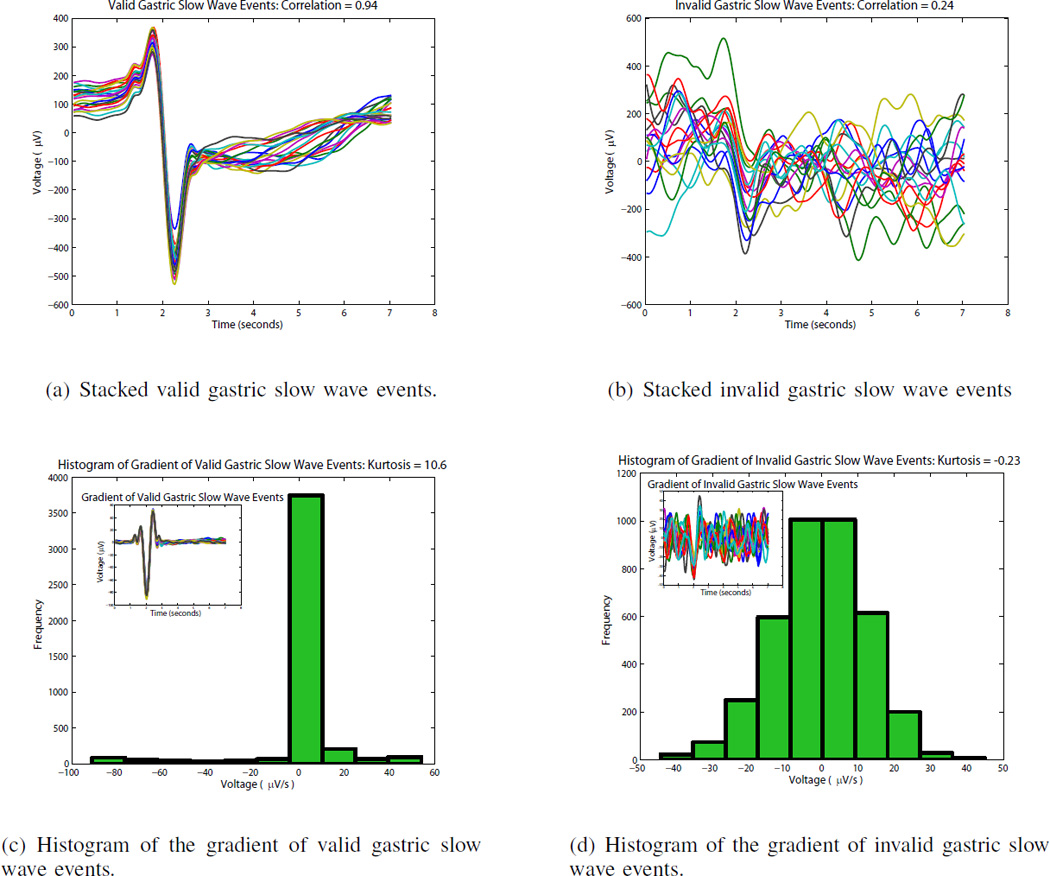
Stacking of valid and invalid gastric slow wave events. (a) shows valid detected slow wave events stacked on top of each other and are characterized by a consistent morphology. (b) shows invalid detected slow wave events stacked on top of each other and is characterized by varying amplitudes and an inconsistent morphology. (c) show the histogram of the gradient of the stacked slow wave events of (a) with the gradient signal shown in the inset. The valid slow wave events have a sharp deflection which can be quantified via the histogram using kurtosis. (d) shows the histogram of the gradient of invalid detected slow wave events in (b) with the gradient signal shown in the inset. The invalid slow wave signals do not have a sharp deflection and this can be quantified via the histogram using kurtosis.
Kurtosis was the third local metric used to assess whether the fiducial marker represents a slow wave event. This metric measured the peakedness of the histogram distribution and was estimated using Equation 3. The estimation of kurtosis was used to assess the gradient of the negative deflection of the slow wave signal, which represents the activation time [12]. As seen in Fig. 2, a slow wave event with a high signal to noise ratio will yield a high kurtosis value (leptokurtic or platykurtic distribution), while a noisy signal would yield a low kurtosis value (mesokurtic distribution). One of the drawbacks of estimating kurtosis is that it can be sensitive to outliers making the kurtosis estimate misleading [17].
| (3) |
B. Experimental mapping methods
Ethical approval for these experimental studies were granted by University of Auckland Ethics Committee for the animal studies, and by the University of Mississippi Medical Center Institutional Review Board for human studies along with patient consent. Six experimental gastric multielectrode mapping datasets (3 pig and 3 human) ranging from 4.7–8.3 minutes were used for experimental validation of the algorithm. The experimental method used to acquire the datasets are described previously [14], [15]. The data was recorded at 512 Hz using an ActiveTwo Biosemi data acquisition system modified for passive recordings, with up to 256 simultaneous recordings.
Filtering was applied to remove baseline wander and high frequency noise on the datasets [10], after which an automated detection algorithm was used to detect the slow wave activation time [12]. To assess the usefulness of the automated ‘clean up’ algorithm, it was tested against the gold standard, which was manual deletion of fiducial markers. Two experts in gastric slow wave mapping manually cleaned up the data sets. Two metrics, sensitivity and specificity were defined as in Equation 4, to determine the effectiveness of the automated algorithm at eliminating invalid fiducial markers for slow waves. Sensitivity estimates the true positive rate of discarding invalid slow wave markers, while specificity estimates the true negative rate of not discarding valid slow wave markers.
| (4) |
True positive (TP): invalid markers detected as invalid
True negative (TN): valid markers detected as valid
False negative (FN): invalid markers detected as valid
False positive (FP): valid marker detected as invalid
III. RESULTS
Overall there were on average 3475±1755 (median: 3024) marks per data set, as defined by the automated detection method. Manual review undertaken by two researchers eliminated on average 44±7% (median: 46%) of those marks as invalid. The automated clean up algorithm resulted in an average of 90±5% (median: 91%) in sensitivity and 85±5% (median: 85%) in specificity to eliminate incorrect fiducial marker for gastric slow wave events (Fig. 3). A default set of parameters has been used for testing of this algorithm. Varying the parameters based on a priori knowledge of the recorded data may increase the accuracy by removing incorrect slow wave events. For example, if the only noise present in the recorded data was the respiration artifact, the FFTfrac measure in the global check can be adjusted accordingly to increase the specificity of the algorithm.
Figure 3.
Quantification of the accuracy of the automated data clean up algorithm by comparison with the manual analysis. With the default parameters chosen for the algorithm to remove incorrect slow wave fiducial markers, the average sensitivity was 90% and specificity was 85%.
IV. CONCLUSIONS
HR gastric slow wave mapping is now being utilized in clinical research because it provides a unique view of spatiotemporal propagation, which was not possible with sparse recording techniques. Its usage in clinical research is being driven by the development of automated and real time techniques for analysis, allowing researchers to traverse large amounts of data in a short period of time [18], [6], [11]. Noise is omnipresent and cannot be avoided or predicted accurately. The automated slow wave detection algorithm assumes relatively noise free signals and the presence of slow wave events [12]. The automated clustering wavefront is capable of handling some noise in slow wave markers to group a propagating wavefront [11], but eliminating incorrect markers will allow for efficient and reliable automated clustering of wavefront. An algorithm has been developed to eliminate incorrect slow wave markers which were detected using automated techniques, and is shown to be highly effective.
The described techniques are applicable for certain noise characteristics, and further novel algorithms would be required to deal with other noise types. For example, presence of pacing artifacts during experimental slow wave recordings will not allow for accurate slow wave detection, and will require signal preprocessing to suppress the artifact prior to automated detection. The techniques herein have been applied for gastric slow wave recordings and maybe applicable to intestinal slow wave recordings with the modification of parameters along with validation on experimental datasets. It is anticipated these additional automated methods will allow for ease of analyzing HR slow wave mapping data and further facilitate clinical translation of electrical mapping to clinical and diagnostic gastroentrology.
Acknowledgments
FUNDING
This work is funded through the NIH (R01 DK64775) and the Health Research Council of New Zealand
REFERENCES
- 1.Cheng L, Du P, O’Grady G. Mapping and modeling gastrointestinal bioelectricity: From engineering bench to bedside. Physiology. 2013;28(5):310–317. doi: 10.1152/physiol.00022.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin Z, Eaker EY, Sarosiek I, McCallum RW. Gastric myoelectrical activity and gastric emptying in patients with functional dyspepsia. Am. J. Gastroenterol. 1999;94(9):2384–2389. doi: 10.1111/j.1572-0241.1999.01362.x. [DOI] [PubMed] [Google Scholar]
- 3.O’Grady G, Angeli T, Du P, Lahr C, Lammers W, Windsor J, Abell T, Farrugia G, Pullan A, Cheng L. Abnormal initiation and conduction of slow-wave activity in gastroparesis, defined by high resolution electrical mapping. Gastroenterology. 2012;143:589–598. doi: 10.1053/j.gastro.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lammers W, al Kais A, Singh S, Arafat K, el Sharkawy TY. Multielectrode mapping of slow-wave activity in the isolated rabbit duodenum. J. Appl. Physiol. 1993;74(3):1454–1461. doi: 10.1152/jappl.1993.74.3.1454. [DOI] [PubMed] [Google Scholar]
- 5.Du P, O’Grady G, Egbuji J, Lammers W, Budgett D, Nielsen P, Windsor J, Pullan A, Cheng L. High-resolution mapping of in vivo gastrointestinal slow wave activity using flexible printed circuit board electrodes: methodology and validation. Ann. Biomed. Eng. 2009;37(4):839–846. doi: 10.1007/s10439-009-9654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paskaranandavadivel N, Gao J, Du P, O'Grady G, Cheng L. Automated classification and identification of slow wave propagation patterns in gastric dysrhythmia. Ann Biomed Eng. 2014;42(1):177–192. doi: 10.1007/s10439-013-0906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yassi R, O’Grady G, Paskaranandavadivel N, Du P, Angeli T, Pullan A, Cheng L, Erickson J. The gastrointestinal electrical mapping suite (GEMS): software for analyzing and visualizing high resolution (multi-electrode) recordings in spatiotemporal detail. BMC Gastroenterol. 2012;12(1):60. doi: 10.1186/1471-230X-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lammers W. Smoothmap v3.05. Al Ain, United Arab Emirates. Al Ain, United Arab Emirates: UAE University; 2009. [Accessed on 10 April 2013]. Available: http://www.fmhs.uaeu.ac.ae/smoothmap/. [Online]. Available: http://www.fmhs.uaeu.ac.ae/smoothmap/ [Google Scholar]
- 9.Paskaranandavadivel N, O’Grady G, Du P, Cheng L. Comparison of filtering methods for extracellular gastric slow wave recordings. Neurogastroenterol Motil. 2013;25(1):79–83. doi: 10.1111/nmo.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paskaranandavadivel N, Cheng LK, Du P, O’Grady G, Pullan A. Improved signal processing techniques for the analysis of high resolution serosal slow wave activity in the stomach. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:1737–1740. doi: 10.1109/IEMBS.2011.6090497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson J, O’Grady G, Du P, Egbuji J, Pullan A, Cheng L. Automated gastric slow wave cycle partitioning and visualization for high-resolution activation time maps. Ann. Biomed. Eng. 2011;39(1):469–483. doi: 10.1007/s10439-010-0170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson J, O’Grady G, Du P, Obioha C, Qiao W, Richards W, Bradshaw L, Pullan A, Cheng L. Falling-edge, variable threshold (FEVT) method for the automated detection of gastric slow wave events in high-resolution serosal electrode recordings. Ann. Biomed. Eng. 2010;38(4):1511–1529. doi: 10.1007/s10439-009-9870-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paskaranandavadivel N, O’Grady G, Du P, Pullan A, Cheng L. An improved method for the estimation and visualization of velocity fields from gastric high-resolution electrical mapping. IEEE Trans. Biomed. Eng. 2012;59(3):882–889. doi: 10.1109/TBME.2011.2181845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Grady G, Du P, Cheng L, Egbuji J, Lammers W, Windsor J, Pullan A. Origin and propagation of human gastric slow wave activity defined by high-resolution mapping. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;299(3):G585–G592. doi: 10.1152/ajpgi.00125.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egbuji J, O’Grady G, Du P, Cheng L, Lammers W, Windsor J, Pullan A. Origin, propagation and regional characteristics of porcine gastric slow wave activity determined by high-resolution mapping. Neurogastroenterol Motil. 2010;22(10):e292–e300. doi: 10.1111/j.1365-2982.2010.01538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lammers W, Ver Donck L, Stephen B, Smets D, Schuurkes JA. Focal activities and re-entrant propagations as mechanisms of gastric tachyarrhythmias. Gastroenterology. 2008;135(5):1601–1611. doi: 10.1053/j.gastro.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 17.Hyvärinen A. New approximations of differential entropy for independent component analysis and projection pursuit. Proceedings of the 1997 Conference on Advances in Neural Information Processing Systems. 1998:273–279. [Google Scholar]
- 18.Bull S, O’Grady G, Du P, Cheng L. A system and method for online high-resolution mapping of gastric slow-wave activity. IEEE Trans Biomed Eng. 2014 Nov;61(11):2679–2687. doi: 10.1109/TBME.2014.2325829. [DOI] [PMC free article] [PubMed] [Google Scholar]




