Abstract
The T box riboswitch is a cis-acting regulatory RNA that controls expression of amino acid-related genes in response to the aminoacylation state of a specific tRNA. Multiple genes in the same organism can utilize this mechanism, with each gene responding independently to its cognate tRNA. The uncharged tRNA interacts directly with the regulatory RNA element, and this interaction promotes readthrough of an intrinsic transcriptional termination site upstream of the regulated coding sequence. A second class of T box elements uses a similar tRNA-dependent response to regulate translation initiation. This review will describe the current state of our knowledge about this regulatory system.
Keywords: tRNA, riboswitch, transcription attenuation, gene regulation, RNA structure
1. Introduction
The T box regulatory system was one of the first regulatory mechanisms identified on the basis of the presence of conserved elements in the leader region of a gene or operon, between the promoter and the start of the first regulated coding sequence. Comparative sequence analysis revealed a complex set of conserved features, including the presence of a triplet sequence that was postulated to specify binding of the cognate tRNA [1]. The demonstration that a specific uncharged tRNA stimulates antitermination, that the tRNA acts in the absence of translation in vivo and in vitro, and that no other cellular factors are required for the tRNA-leader RNA interaction [1–4], indicated that this system can be considered a member of the riboswitch family, albeit one that utilizes a ligand other than a cellular metabolite [5]. This review will describe the identification and characterization of this unique class of regulatory elements, and will discuss the interesting variations on a theme that are emerging.
2. Identification of the T box system
The T box system was first identified by characterization of a single gene, the Bacillus subtilis tyrS gene, which encodes tyrosyl-tRNA synthetase (TyrRS) [6]. The levels of TyrRS activity in B. subtilis had been shown to increase when cells were grown under tyrosine depletion conditions [7], and this effect was shown to occur at the level of readthrough of an intrinsic transcriptional terminator located in the tyrS leader region [6]. Conservation of the leader region terminator in multiple tyrS genes in Bacillus species, coupled with the identification of a short conserved sequence upstream of the terminator, designated the T box sequence, suggested a conserved regulatory mechanism. Mutation of the conserved sequence resulted in loss of readthrough of the terminator, indicating an important role for this sequence [6].
Identification of the T box sequence in additional amino acid-related genes in B. subtilis, including multiple aminoacyl-tRNA synthetase genes and the ilv-leu branched chain amino acid biosynthesis operon, suggested that this regulatory mechanism extends beyond tyrS genes [6]. This was confirmed when a complex pattern of conserved primary sequence elements (in addition to the T box sequence) and structural elements (in addition to the intrinsic terminator) was identified in the leader regions of all of these genes (Fig. 1) [1,8].
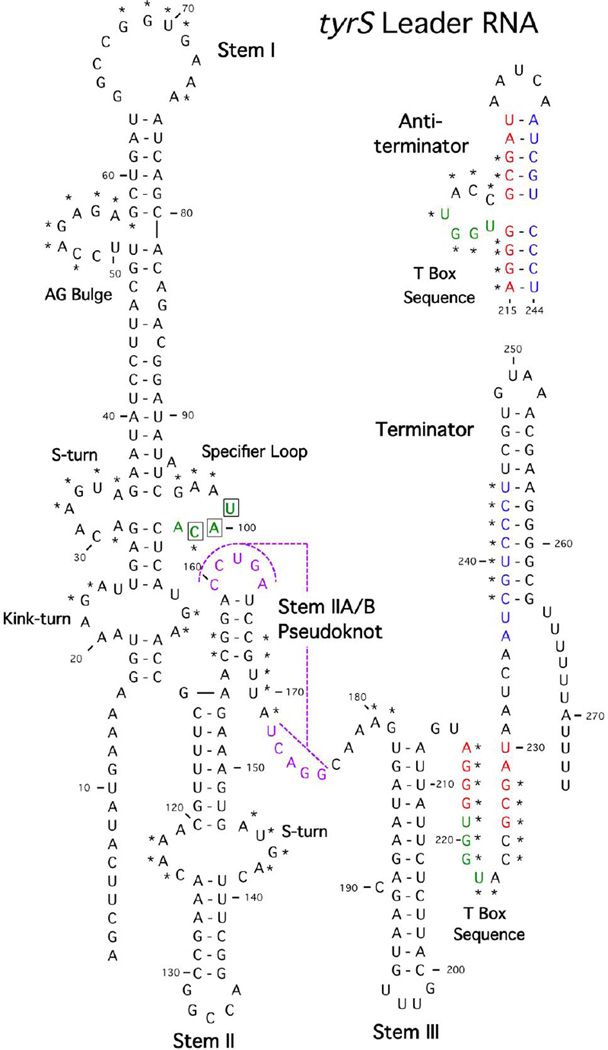
Figure 1.
Structural model of the B. subtilis tyrS leader RNA. The sequence is shown from the transcription initiation site through the termination site. The terminator form is shown, with the antiterminator form above the terminator helix. Structural domains and conserved sequence elements are labeled. Highly conserved residues are marked with asterisks. The pseudoknot pairing is shown in purple. Sequences on the 5’ side of the terminator (blue) pair with a portion of the T box sequence (red) to form the antiterminator. The residues that interact with tRNATyr (UAC Specifier Sequence and UGGU in antiterminator bulge) are shown in green. Modified from [5].
Since it was likely that genes of different amino acid classes responded specifically to limitation for the cognate amino acid, the conservation of this overall pattern led to the question of how individual genes in this group could respond individually to the availability of the appropriate amino acid. This was resolved by identification of a triplet sequence embedded within the structural pattern that corresponded to a codon matching the amino acid specificity of the downstream gene [1]. Mutation of this triplet, termed the “Specifier Sequence,” in tyrS from a UAC tyrosine codon to a UUC phenylalanine codon was sufficient to block induction of the expression of a tyrS-lacZ reporter gene in response to tyrosine limitation, and to promote induction in response to limitation for phenylalanine.
The positive role of tRNA in this response was demonstrated by introduction of nonsense mutations into the position of the Specifier Sequence, which resulted in loss of expression (because a normal cell lacks tRNAs with the matching anticodon); suppression of the nonsense mutations by the corresponding nonsense suppressor tRNAs provided positive proof that tRNA is the effector [1,2]. tRNA mutants that are unchargeable in vivo conferred expression during growth in rich medium, which demonstrated that uncharged tRNA is the key effector.
The basic properties of the B. subtilis tyrS regulatory system were shown to apply to a number of other genes in this family, including the B. subtilis thrS, thrZ, ilv-leu and valS genes [9–11], as well as genes from related Gram-positive organisms [12–15]. Each gene that was analysed was shown to be induced in response to limitation for the cognate amino acid, and in the case of the B. subtilis ilv-leu operon, a mutation predicted to result in reduced aminoacylation in vivo resulted in increased expression in the absence of amino acid limitation [16], adding further support to the model that uncharged tRNA is the signal to which the system responds.
3. Structural features of T box leader RNAs
Leader RNAs in the T box family were identified initially by simple searches for conservation of the highly conserved T box element [1,2], and subsequently by bioinformatics analyses that relied on a combination of conserved features [17–19]. These studies revealed that the majority of T box family RNAs contained a full complement of the conserved features that were initially described [1], as shown in Fig. 1. These include a complex Stem I structure with a predicted kink-turn motif at the base, an S turn in the Specifier Loop region, an extended domain above the Specifier Loop that included conserved elements in the terminal loop and AG bulge [20,21]. Stem I is immediately followed by a second helix that most commonly contains an internal bulge with another predicted S turn motif, followed by a complex Stem IIA/B pseudoknot element [21]. A linker region is then followed by another helix (Stem III) that precedes the competing antiterminator and terminator elements. Mutational analysis revealed the importance of these conserved features, as even single nucleotide substitutions of conserved elements were often sufficient to disrupt tRNA-dependent antitermination in vivo [20–22].
Analysis of additional genomic sequences revealed subsets in which individual features exhibited specific patterns of variability; in many cases, the variants correlate with amino acid class, suggesting a basis either in evolutionary history or in tRNA recognition specificity [19]. These variants include the absence of the Stem II and Stem IIA/B pseudoknot domains (glycyl genes and some alanyl genes), absence of the S turn element in the Specifier Loop (threonyl genes), and absence of the region above the Specifier Loop (some isoleucyl genes). In each case, examples of these variant classes have been shown to be functional in vivo or in vitro (see below; [3, 23, Sherwood, Grundy and Henkin, unpublished; Liu, Grundy and Henkin, unpublished]). The ability of natural variants to function in the absence of elements that are highly sensitive to mutation in T box RNAs that contain these elements indicates that these variants must have evolved to compensate for the absence of these elements, through other structural changes or through other interactions with their cognate tRNAs.
4. tRNA features required for recognition in vivo
Initial studies of tRNA requirements for antitermination in vivo focused on expression of Specifier Sequence mutants of T box leader RNAs in response to limitation for the amino acid specified by the codon that was introduced. As described above, the initial switch of the UAC tyrosine codon in the B. subtilis tyrS gene to a UUC phenylalanine codon resulted in induction in response to limitation for phenylalanine [1]; however, the level of expression was much lower than that observed for the wild-type construct in response to limitation for tyrosine. This pattern was observed for a number of Specifier Sequence mutations, even in conjunction with mutations in the antiterminator that allowed pairing with the tRNA discriminator base (the residue upstream of the terminal CCA) [24]. Similar results were observed for other T box family genes [25]. These results suggested that tRNA features in addition to the anticodon and discriminator base are important for proper recognition of the tRNA. A detailed mutational analysis further supported the importance of the entire tRNA structure [26]; however, this study was limited by the requirement for in vivo expression of tRNA variants.
5. Biochemical analyses of the T box riboswitch
While in vivo studies could show that the cognate uncharged tRNA is necessary for antitermination, demonstration that tRNA is sufficient to induce antitermination required in vitro analyses with purified components. Initial in vitro transcription experiments using tyrS as a model failed to reproduce tRNATyr-dependent antitermination [22]. The B. subtilis glyQS gene was therefore tested as an alternative, because the leader RNA of this gene lacks the highly conserved Stem II and Stem IIA/B pseudoknot elements [3]. Transcription of a DNA fragment that included the region extending from upstream of the glyQS promoter to the start of the coding region downstream of the terminator resulted in efficient termination in the absence of tRNA. Addition of tRNAGly, but not noncognate tRNA, promoted antitermination; this effect was observed using either B. subtilis or Escherichia coli RNAP, despite the fact that no T box-regulated genes are present in E. coli. As was observed in vivo, the tRNA response required pairing at both the Specifier Sequence-anticodon and antiterminator-tRNA acceptor end domains. The ability of the tRNA to act alone to promote the regulatory response provided a first clear indication of riboswitch-like behavior of the leader RNAs in this family [3].
Analysis of the B. subtilis thrS gene, which encodes threonyl-tRNA synthetase, also provided evidence for tRNAThr-dependent antitermination in vitro [23]. This leader RNA, unlike glyQS, includes the Stem II and IIA/B pseudoknot domains, and has a different arrangement in the Specifier Loop that lacks the S turn motif. tRNAThr-dependent antitermination required the addition of either spermidine or a cell extract, suggesting that there are additional constraints for this leader RNA that are absent for the simpler glyQS gene. Furthermore, efficient antitermination required modified tRNA, whereas unmodified tRNAGly was sufficient for glyQS. This may relate to the fact that tRNAGly lacks anticodon loop modifications in vivo, while tRNAThr is more heavily modified.
The efficiency of the glyQS in vitro antitermination assay allowed more detailed characterization of effects of the kinetics of transcription [27]. These studies showed that antitermination was not sensitive to the speed of transcription, and that specific pause sites observed during transcription in vitro were dispensable for antitermination. A charged tRNA mimic (in which an extra residue was present at the 3’ end of the tRNA) was used to demonstrate that binding of uncharged tRNA could be blocked by this mimic; this result suggests that the system monitors not only uncharged tRNA but also charged tRNA, so that the key parameter is the tRNA charging ratio rather than the absolute amount of uncharged tRNA [28]. This observation is consistent with in vivo experiments in which induction of expression with an unchargeable tRNA variant is more efficient if there is no chargeable tRNA present in the cell the anticodon of which is capable of binding to the Specifier Sequence [2].
A more detailed analysis of the competence of transcription complexes to interact with both uncharged and charged tRNA was carried out by using a transcriptional roadblock to generate complexes in which varying amounts of the leader RNA were available for interaction with the tRNA [29]. These studies indicated that transcription complexes were fully capable of binding either charged or uncharged tRNA until the antiterminator domain was fully present in the nascent transcript; at this point, binding of uncharged tRNA is stable, and cannot be reversed by addition of excess charged tRNA mimic. This result suggests that the interaction of the antiterminator bulge with the tRNA acceptor end, which occurs only with uncharged tRNA, locks the complex into a more stable form that is resistant to challenge by charged tRNA.
Binding assays between the leader RNA and tRNA were also carried out, and showed dependence upon Specifier Sequence-tRNA anticodon matching consistent with that observed both in vivo and in the in vitro antitermination assay [4]. Unlike antitermination, binding was not completely dependent on the antiterminator-tRNA acceptor end interaction, suggesting that the Stem I interaction dominates the binding affinity. Fluorescence assays also demonstrated that RNAs containing only the bottom portion of Stem I (including the kink-turn motif and the Specifier Loop) exhibit specific binding to both full-length tRNA and an anticodon helix mimic [30]. Binding studies also were carried out between a small RNA mimic of the antiterminator domain and either full-length tRNA of an acceptor end minihelix; in this case, the major specificity determinant appears to be the tRNA discriminator base, which pairs with the variable position of the antiterminator bulge (UGGN; [31]). These studies showed that conservation in other parts of the antiterminator bulge (notably the first C in the conserved UGGNACC sequence) plays a key role in tRNA affinity, despite the lack of specific base-pairing to tRNA [31, 32].
Structural mapping studies of the glyQS leader RNA in the presence or absence of tRNA using a variety of RNases provided additional evidence for the interactions proposed on the basis of genetic experiments [4]. Protection of the Specifier Loop occurred with both uncharged tRNA and the charged tRNA mimic, whereas protection of the antiterminator bulge was observed only in the presence of uncharged tRNA. Protection of residues in other regions of the leader RNA also was observed only with uncharged tRNA, supporting the proposal that the tRNA acceptor end/antiterminator interaction promotes formation of a more stable complex with changes throughout the leader RNA. Parallel experiments examining changes in the tRNA revealed protection of the anticodon loop, consistent with pairing of this domain with the Specifier Sequence. Protection of residues in the D loop of the tRNA was also observed (see below).
The availability of the in vitro assays permitted testing of a variety of variants in both the glyQS leader RNA and tRNAGly for effects on tRNA binding and antitermination. These studies indicated that most of the residues and motifs recognized as conserved in T box leader sequences, and sensitive to mutation in vivo, were also sensitive to mutation in vitro, validating their functional importance as well as the physiological relevance of the in vitro assays (Grundy, Rollins, Green, Henkin, unpublished). Analysis of tRNA variants indicated that the entire tertiary structure was required [28]. One interesting observation was that although the anticodon stem was very sensitive to alterations in length, the acceptor stem could be extended by 11 bp (a full turn of the RNA helix) without any deleterious effect on antitermination; addition of a half-turn of the helix resulted in loss of antitermination activity, suggesting that presentation of the acceptor end of the tRNA to the antiterminator bulge exhibits face-of-the-helix dependence, and that there is linear flexibility in the relative positions of the Specifier Loop and antierminator, but limitations on rotational flexibility [28]. It is not clear whether this flexibility is unique to glyQS, which lacks Stem II and Stem IIA/B, or is general to all T box leader RNAs, as these elements could make additional unknown tRNA contacts that place additional constraints on tRNA positioning.
6. Structural analysis of T box leader RNAs
The first detailed structural information about T box RNAs was derived from NMR analysis of the antiterminator domain [31, 32]. These studies revealed that the RNA folded into the predicted helix-bulge-helix arrangement, with an arrangement of the bulge in which the UGGN residues that pair with the tRNA acceptor end are splayed out to make those contacts. The bulge residues were somewhat flexible in solution, presumably to facilitate interaction with the tRNA. However, introduction of a C to U substitution in the penultimate position of the bulge increase flexibility, and reduced tRNA affinity, which suggested that too much flexibility in this region interfered with tRNA binding [32]. Binding of the tRNA to the bulge stabilized the bulge residues, consistent with the physiological model, in which the terminator dominates in the absence of tRNA, and binding of tRNA allows the antiterminator to form.
The next domain of T box leader RNAs to be explored structurally was the Stem I region that includes the Specifier Loop, using the B subtilis tyrS leader as a model. NMR analysis of this region demonstrated that the predicted kink-turn domain at the base of Stem I, and the predicted S turn in the Specifier Loop adjacent to the Specifier Sequence, both formed in solution [33, 34]. Furthermore, the residues of the Specifier Sequence were shown to be splayed out for binding to the tRNA anticodon. Similar results were obtained for the B. subtilis glyQS Stem I domain [35]; this study also demonstrated that the conserved purine downstream of the Specifier Sequence assists in stabilization of the interaction with the tRNAGly anticodon but does not base-pair with the conserved U adjacent to the anticodon in the tRNA.
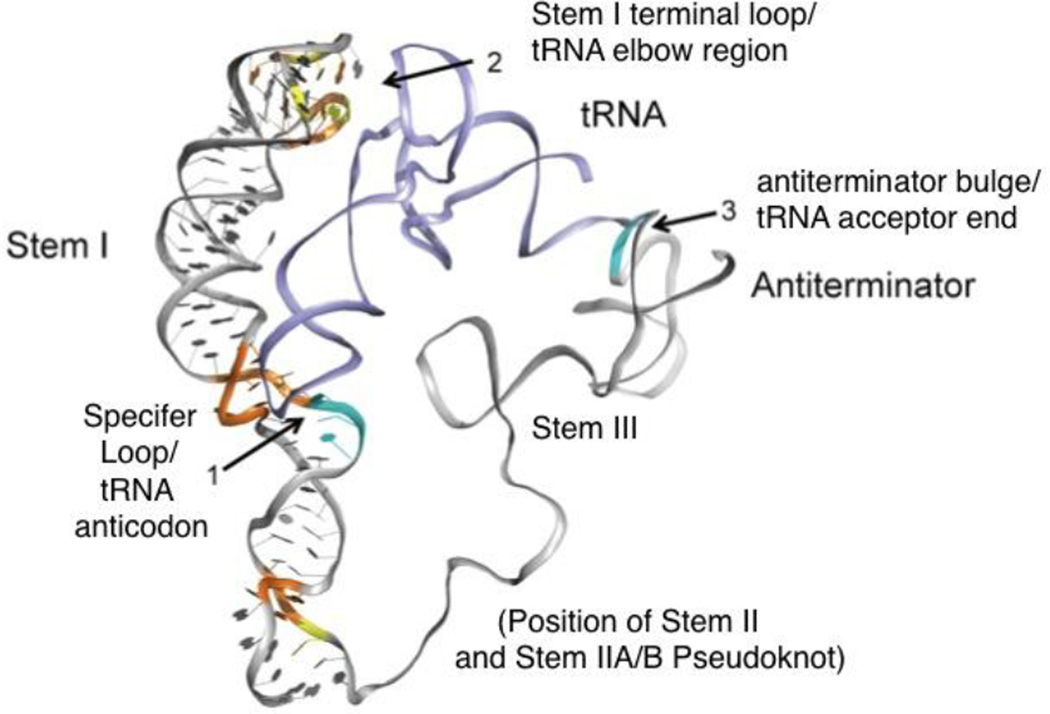
The crystal structure of the terminal region of Stem I, which contains conserved residues within both the AG bulge and terminal loop, was recently reported for leader RNAs of glyQS genes [36, 37]. Both of these structures revealed an interaction between residues in the AG bulge, Stem I terminal loop, and the elbow region of the tRNA (Fig. 2). These results, in conjunction with phylogenetic and genetic analyses (K. Kreuzer, N. Green, F. Grundy, T. Henkin, unpublished), suggest that this domain of Stem I may provide a third interaction that assists in maintaining the specificity of tRNA recognition. It should be noted that residues that participate in this interaction in glyQS are arranged differently in most other T box leader sequences, suggesting that alternate arrangements may be functional in other contexts. Furthermore, a subset of isoleucyl genes lack the entire upper portion of Stem I, yet are functional in vitro (Sherwood, Grundy, Henkin, unpublished; see below).
Figure 2.
Model of the interaction between the glyQS leader RNA and tRNAGly. Direct interactions between the Specifier Loop and the tRNA anticodon (1) and between the antiterminator bulge and tRNA acceptor end (3) have been demonstrated by genetic, biochemical and structural approaches. The interaction between the Stem I terminal loop and the tRNA elbow region (2) is indicated by structural evidence, and the residues that participate in this interaction in glyQS are not conserved in all T box leaders, suggesting that this interaction may vary in different genes. No structural information is available for Stem III, or the Stem II and IIA/B pseudoknot domains that are absent from glyQS leader RNAs (modified from [36]).
No structural information is available for the Stem II and Stem IIA/B pseudoknot domains (which are absent from glyQS leader RNAs) or for the highly variable Stem III domain. It is interesting to note that while Stem II and IIA/B are absent in certain T box leader sequences, such as all glycyl genes, these elements are highly sensitive to mutation in RNAs in which they are found (e.g., tyrS; [21]). This variability in leader RNA structure is likely to correlate with tRNA recognition in unknown ways. Structural analysis of other classes of T box leader RNAs, and of complete leader RNA-tRNA complexes, will be crucial for a full understanding of how the tRNA is recognized and the complex is stabilized.
7. Regulatory mechanism
As described above, the T box riboswitch was first described as a regulatory mechanism that functions at the level of premature transcription termination (or transcription attenuation). Bioinformatics analyses identified a large number of sequences with the features of the characterized T box RNAs in genomic data, and most of the genes identified were also predicted to operate at this level, based on the identification of canonical transcriptional terminator elements [17–19]. A small subclass of these sequences lack obvious terminator elements, and instead contain a final helix that is predicted to sequester the Shine-Dalgarno (SD) sequence of the downstream coding sequence. As with the terminators, the sequences that pair with the SD region (i.e., the anti-SD sequence, or ASD) can alternatively base-pair with a portion of the T box sequence to form a structure similar to the canonical antiterminator that acts to sequester the ASD, allowing the SD to be available for ribosome binding and translation initiation.
Genes predicted to be regulated at the level of translation are found predominantly in high G+C organisms in the Actinobacteria group, and are predominantly ileS genes, which encode isoleucyl-tRNA synthetase. Functional studies of representative leader RNAs in this class demonstrated specific tRNAIle binding (Sherwood, Grundy & Henkin, unpublished). Binding of the tRNA also was shown by ribosomal toeprint assays to allow binding of 30S ribosomal subunits, consistent with the model that tRNA binding stabilizes the anti-ASD structural element, in parallel to tRNA-dependent stabilization of the antiterminator in transcription attenuation systems. It therefore appears that these predicted translational control systems function as expected. This subclass of T box leaders includes the class in which the terminal domain of Stem I is absent, further indicating the variability in structure and function of leader RNAs of this type. It remains to be determined if the regulatory mechanism employed provides additional constraints on the leader RNA-tRNA interaction.
8. Targeting of the T box riboswitch with novel antimicrobial compounds
Three major factors suggested that the T box system might be an appropriate target for development of antimicrobial agents. First, many T box-regulated genes encode essential aminoacyl-tRNA synthetases. Second, most Gram-positive organisms have multiple genes that use this mechanism, reducing the probability that a single target site mutation could yield resistance. Third, the the default state of the riboswitch is off (i.e., gene expression is off in the absence of ligand binding); as a result, most mutations in T box family leader RNAs result in disruption of ligand binding, and loss of expression, and therefore would not allow survival of the organism.
The antiterminator element was identified as the most highly conserved element in T box RNAs, and its arrangement (interrupted helix with conserved 7 nt bulge) is retained even in leader RNAs predicted to regulate at the translational level. This element must be stabilized by interaction with the acceptor end of the tRNA. The structure of this domain [31–32] was therefore used as the basis for design of families of compounds that were predicted to bind to the antiterminator, and the resulting compounds were tested for effects on tRNA binding, tRNA-dependent stabilization of the antiterminator and antitermination [38–43]. Further development of these compounds holds promise for development of useful new antimicrobial compounds that are predicted to be specific for Gram-positive organisms, avoiding more general effects on the commensal flora.
9. Conclusions and perspectives
The T box system represents an interesting variant on the riboswitch theme. As in metabolite binding riboswitches, binding of a specific ligand mediates a structural rearrangement of the leader RNA that affects gene expression. The majority of metabolite-binding riboswitches operate as feedback repression systems, where the endproduct of a biosynthetic pathway acts to repress expression of genes involved in acquisition or synthesis of that compound. In contrast, the T box system monitors the ratio between the substrate of the pathway (uncharged tRNA) and its product (charged tRNA) to regulate genes involved in conversion of substrate to product (i.e., genes encoding aminacyl-tRNA synthetases, amino acid biosynthesis, amino acid transport, and regulators of amino acid biosynthesis [45]. Another interesting difference is that while the overall shape of the tRNA ligand, including the elbor domain, is clearly crucial for its recognition, the primary interactions with the leader RNA are by Watson-Crick basepairing between the Specifier Sequence and tRNA anticodon, and between the antiterminator bulge and tRNA acceptor end. This allows small changes in leader RNA sequence to shift the specificity of the regulatory response, and therefore has allowed this system to confer differential regulation genes of many different amino acid classes within the same organism. The physiological utility of this system is apparent from its widespread use throughout the Gram-positive branch of bacteria. This in turn justifies the attempt to target this system for developent of novel antimicrobial agents specific to Gram-positive pathogens.
Highlights.
The T-box riboswitch responds to tRNA aminoacylation
Multiple genes in the same organism respond individually to the cognate tRNA
Uncharged tRNA stabilizes an antiterminator element to allow downstream gene transcription
Some T-box RNAs regulate at the level of translation initiation
tRNA-dependent gene regulation can be reproduced in vitro
Acknowledgments
Work in the Henkin lab is supported by National Institutes of Health Institute of General Medical Sciences Grant R01-GM47823.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 2.Grundy FJ, Rollins SM, Henkin TM. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: A new role for the discriminator base. J. Bacteriol. 1994;176:4518–4526. doi: 10.1128/jb.176.15.4518-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grundy FJ, Winkler WC, Henkin TM. tRNA-mediated transcription antitermination in vitro: Codon-anticodon pairing independent of the ribosome. Proc. Natl. Acad. Sci. USA. 2002;99:11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T box leader RNA. J. Mol. Biol. 2005;349:273–287. doi: 10.1016/j.jmb.2005.03.061. [DOI] [PubMed] [Google Scholar]
- 5.Green NR, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584:318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henkin TM, Glass BL, Grundy FJ. Analysis of the Bacillus subtilis tyrS gene: Conservation of a regulatory sequence in multiple tRNA synthease genes. J. Bacteriol. 1992;174:1299–1306. doi: 10.1128/jb.174.4.1299-1306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale BA, Nester EW. Regulation of tyrosyl-transfer ribonucleic acid synthetase in Bacillus subtilis. J. Bacteriol. 1971;108:586–588. doi: 10.1128/jb.108.1.586-588.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundy FJ, Henkin TM. Conservation of a transcription antitermination mechanism in aminoacyl-tRNA synthetase and amino acid biosynthesis genes in gram-positive bacteria. J. Mol. Biol. 1994;235:798–804. doi: 10.1006/jmbi.1994.1038. [DOI] [PubMed] [Google Scholar]
- 9.Putzer H, Laalami S, Brakhage AA, Condon C, Grunberg-Manago M. Aminoacyl-tRNA synthetase gene regulation in Bacillus subtilis: Induction, repression and growth-rate regulation. Mol. Microbiol. 1995;16:709–718. doi: 10.1111/j.1365-2958.1995.tb02432.x. [DOI] [PubMed] [Google Scholar]
- 10.Marta PT, Ladner RD, Grandoni JA. A CUC triplet confers leucine-dependent regulation of the Bacillus subtilis ilv-leu operon. J. Bacteriol. 1996;178:2150–2153. doi: 10.1128/jb.178.7.2150-2153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo D, Leautey J, Grunberg-Manago M, Putzer H. Structure and regulation of the Bacillus subtilis valyl-tRNA synthetase gene. J. Bacteriol. 1997;179:2472–2478. doi: 10.1128/jb.179.8.2472-2478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frenkiel H, Bardowski J, Ehrlich SD, Chopin A. Transcription of the trp operon in Lactococcus lactis is controlled by antitermination in the leader region. Microbiology. 1998;144:2103–2111. doi: 10.1099/00221287-144-8-2103. [DOI] [PubMed] [Google Scholar]
- 13.Delorme C, Ehrlich SD, Renault P. Regulation of expression of the Lactococcus lactis histidine operon. J. Bacteriol. 1999;181:2026–2037. doi: 10.1128/jb.181.7.2026-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Guchte M, Ehrlich SD, Chopin A. tRNATrp as a key element of antitermination in the Lactococcus lactis trp operon. Mol. Microbiol. 1998;29:61–74. doi: 10.1046/j.1365-2958.1998.00903.x. [DOI] [PubMed] [Google Scholar]
- 15.Grundy FJ, Haldeman MT, Hornblow GM, Ward JM, Chalker AF, Henkin TM. The Staphylococcus aureus ileS gene, encoding isoleucyl-tRNA synthetase, is a member of the T-box family. J. Bacteriol. 1997;179:3767–3772. doi: 10.1128/jb.179.11.3767-3772.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrity DB, Zahler SA. Mutations in the gene for a tRNA that functions as a regulator of a transcriptional attenuator in Bacillus subtilis. Genetics. 1994;137:627–636. doi: 10.1093/genetics/137.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitreschak AG, Mironov AM, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wels M, Kormelink TG, Kleerebezem M, Siezen RJ, Francke C. An in silico analysis of T-box regulated genes and T-box evolution in prokaryotes, with emphasis on prediction of substrate specificity of transporters. BMC Genomics. 2008;9:330. doi: 10.1186/1471-2164-9-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez-Preciado A, Henkin TM, Yanofsky C, Merino E. RNA-based T box regulation: New insights revealed by comparative genomics. Microbiol. Mol. Biol. Rev. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkler WC, Grundy FJ, Murphy BA, Henkin TM. The GA motif: An RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001;7:1165–1172. doi: 10.1017/s1355838201002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rollins SM, Grundy FJ, Henkin TM. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol. 1997;25:411–421. doi: 10.1046/j.1365-2958.1997.4851839.x. [DOI] [PubMed] [Google Scholar]
- 22.Grundy FJ, Moir TR, Haldeman MT, Henkin TM. Sequence requirements for terminators and antiterminators in the T-box transcription antitermination system: disparity between conservation and functional requirements. Nucl. Acids Res. 2002;30:1646–1655. doi: 10.1093/nar/30.7.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putzer H, Condon C, Brechemier-Baey D, Brito R, Grunberg-Manago M. Transfer RNA-mediated antitermination in vitro. Nucl. Acids Res. 2002;30:3026–3033. doi: 10.1093/nar/gkf415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grundy FJ, Hodil SE, Rollins SM, Henkin TM. Specificity of tRNA-mRNA interactions in Bacillus subtilis tyrS antitermination. J. Bacteriol. 1997;179:2587–2594. doi: 10.1128/jb.179.8.2587-2594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van de Guchte M, Ehrlich SD, Chopin A. Identity elements in tRNA-mediated transcription antitermination: implication of tRNA D- and T-arms in mRNA recognition. Microbiology. 2001;147:1223–1233. doi: 10.1099/00221287-147-5-1223. [DOI] [PubMed] [Google Scholar]
- 26.Grundy FJ, Collins JC, Rollins SM, Henkin TM. tRNA determinants for transcription antitermination of the Bacillus subtilis tyrS gene. RNA. 2000;6:1131–1141. doi: 10.1017/s1355838200992100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grundy FJ, Henkin TM. Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro. J. Bacteriol. 2004;186:5392–5399. doi: 10.1128/JB.186.16.5392-5399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yousef MR, Grundy FJ, Henkin TM. tRNA requirements for glyQS antitermination: A new twist on tRNA. RNA. 2003;9:1148–1156. doi: 10.1261/rna.5540203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grundy FJ, Yousef MR, Henkin TM. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J. Mol. Biol. 2005;346:73–81. doi: 10.1016/j.jmb.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 30.Nelson AR, Henkin TM, Agris PF. tRNA regulation of gene expression: Interactions of an mRNA 5’-UTR with a regulatory tRNA. RNA. 2006;12:1–8. doi: 10.1261/rna.29906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerdeman MS, Henkin TM, Hines JV. In vitro structure-function studies of the Bacillus subtilis tyrS antiterminator: Evidence for factor-independent tRNA acceptor stem binding specificity. Nucl. Acids Res. 2002;30:1065–1072. doi: 10.1093/nar/30.4.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerdeman MS, Henkin TM, Hines JV. Solution structure of the B. subtilis T box antiterminator RNA: Seven-nucleotide bulge characterized by stacking and flexibility. J. Mol. Biol. 2003;326:189–201. doi: 10.1016/s0022-2836(02)01339-6. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Henkin TM, Nikonowicz EP. NMR structure and dynamics of the Specifier Loop domain from the Bacillus subtilis tyrS T box leader RNA. Nucl. Acids Res. 2010;38:3388–3398. doi: 10.1093/nar/gkq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Nikonowicz EP. Solution structure of the K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T box leader RNA. J. Mol. Biol. 2011;408:99–117. doi: 10.1016/j.jmb.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang AT, Nikonowicz EP. Solution NMR determination of hydrogen bonding and base pairing between the glyQS T box riboswitch Specifier domain and the anticodon loop of tRNAGly . FEBS Lett. 2013;587:3495–3499. doi: 10.1016/j.febslet.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grigg JC, Chen Y, Grundy FJ, Henkin TM, Pollack L, Ke A. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc. Natl. Acad. Sci. USA. 2013;110:7240–7245. doi: 10.1073/pnas.1222214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Ferre-D’Amare AR. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature. 2013;500:363–366. doi: 10.1038/nature12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Means J, Katz S, Nayek A, Anupam R, Hines JV, Bergmeier SC. Structure-activity studies of oxazolidinone analogs as RNA-binding agents. Bioorg. Med. Chem. Lett. 2006;16:3600–3604. doi: 10.1016/j.bmcl.2006.03.068. [DOI] [PubMed] [Google Scholar]
- 39.Means JA, Wolf S, Agyeman A, Burton JS, Simson CS, Hines JV. T box riboswitch antiterminator affinity modulated by tRNA structural elements. Chem. Biol. Drug Des. 2007;69:139–145. doi: 10.1111/j.1747-0285.2007.00476.x. [DOI] [PubMed] [Google Scholar]
- 40.Anupan R, Nayek A, Green NJ, Grundy FJ, Henkin TM, Means JA, Bergmeier SC, Hines JV. 4,5-Disubstituted oxazolidinones: high affinity molecular effectors of RNA function. Bioorg. Med. Chem. Lett. 2008;18:3541–3544. doi: 10.1016/j.bmcl.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orac CM, Zhou S, Means JA, Boehm D, Bergmeier SC, Hines JV. Synthesis and stereospecificity of 4,5-disubstituted oxazolidinone ligands binding to T-box riboswitch RNA. J Med Chem. 2011;54:6786–6795. doi: 10.1021/jm2006904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou S, Means JA, Acquaah-Harrison G, Bergmeier SC, Hines JV. Characterization of a 1,4-disubstituted 1,2,3-triazole binding to T box antiterminator RNA. Biorg. Med. Chem. 2012;20:1298–1302. doi: 10.1016/j.bmc.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 43.Jentzsch F, Hines JV. Interfacing medicinal chemistry with structural bioinformatics: implications for T box riboswitch RNA drug discovery. BMC Bioinformatics. 2012;13(Suppl 2):S5. doi: 10.1186/1471-2105-13-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith AM, Fuchs RT, Henkin TM. Riboswitch RNAs: Regulation of gene expression by direct monitoring of a physiological signal. RNA Biol. 2010;7:104–110. doi: 10.4161/rna.7.1.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarsero JP, Merino E, Yanofsky C. A Bacillus subtilis operon containing genes of unknown function senses tRNATrp charging and regulates expression of the genes of tryptophan biosynthesis. Proc. Natl. Acad. Sci. USA. 2000;97:2656–2661. doi: 10.1073/pnas.050578997. [DOI] [PMC free article] [PubMed] [Google Scholar]