Abstract
Inherited leukodystrophies are a group of neurological disorders with significant morbidity and mortality. Children and their families can experience lengthy diagnostic odysseys; however, there is no data on the charges related to testing for diagnosis in leukodystrophy patients, compared to approaches using next-generation sequencing (NGS). Our objective was to determine charges related to the determination of diagnosis, and overall yield of diagnostic testing, for leukodystrophy patients. We determined and quantified all inpatient and outpatient lab testing, including brain MRIs, obtained for the purpose of diagnosis, in a retrospective population cohort of children with inherited leukodystrophies. Each patient had average charges of $8,231 (range $543–26,437) for diagnostic testing. Overall charges related to diagnosis for the entire cohort was $526,794. A final etiological diagnosis was determined in 34% of patients. In those in whom a specific diagnosis was determined, average time to diagnosis was 1.4 years. If NGS on the entire cohort had been performed instead, charges would have been ~$359,600 (at $5,800/patient). Alternatively, a two-tier approach consisting of first, biochemical testing (serum very-long chain fatty acids and leukocyte lysosomal enzyme testing), and then with NGS for remaining undiagnosed patients, would have resulted in total cohort charges of $361,309. We have determined the charges directly associated with diagnostic testing in a population cohort of children with leukodystrophy. We conclude that appropriately incorporating NGS into diagnostic algorithms could lower charges; reduce time to diagnosis; and reduce amount of testing.
Keywords: leukodystrophy, charges, diagnostic odyssey, testing
INTRODUCTION
Inherited leukodystrophies are a group of genetically diverse diseases with more than 30% mortality by age 8 years [Bonkowsky et al., 2010; Raymond et al., 2011]. Clinical trials and therapy development are inhibited by low rates of diagnosis [Schiffmann and Van Der Knaap, 2009; Bonkowsky et al., 2010]. Next-generation sequencing (NGS) approaches, including whole exome, whole genome, and gene panels, offer the potential to improve diagnostic yield and reduce the time and cost to diagnosis [Yang et al., 2013]. There is no comparative data, however, on the charges related to testing for diagnosis in leukodystrophy patients using current standards of practice.
Our objective was to determine charges related to the determination of diagnosis and overall yield of diagnostic testing for leukodystrophy patients. This information would guide options for diagnostic algorithms incorporating NGS in the leukodystrophies, and more broadly can inform processes to reduce the diagnostic odyssey in patients with rare diseases.
METHODS
The Institutional Review Boards of the University of Utah and Intermountain Healthcare (IH) approved this study. The study was a retrospective population cohort collected from a 9-year period at Primary Children’s Hospital (PCH) and the University of Utah Pediatric Neurology Clinic, with a minimum additional 4 years of follow-up. All charges related to testing for the purpose of diagnosis were collected, including both in- and out-patient testing and brain MRIs. We determined, both manually and by computer search, all lab and radiology testing related to diagnosis, including brain MRIs performed after the initial MRI performed at presentation. Tests included general and disease screening labs; as well as disease-specific testing (for example, respectively: blood chemistry, hemoglobin; leukocyte lysosomal enzymes, chromosome karyotype; very long chain fatty acids, Pelizaeus–Merzbacher gene testing). Tests related to clinical patient care (such as monitoring of drug levels) and professional fees were not included. The initial MRI was not included since it formed the basis of the inclusion criteria we used. Charges for each test were only included for the first instance of that test being obtained. However, we did include charges for repeat MRIs. PCH is part of IH, and complete charge data was extracted for each patient from the electronic data warehouse and were standardized to 2013 constant U.S. dollars [Nelson et al., 2013].
RESULTS
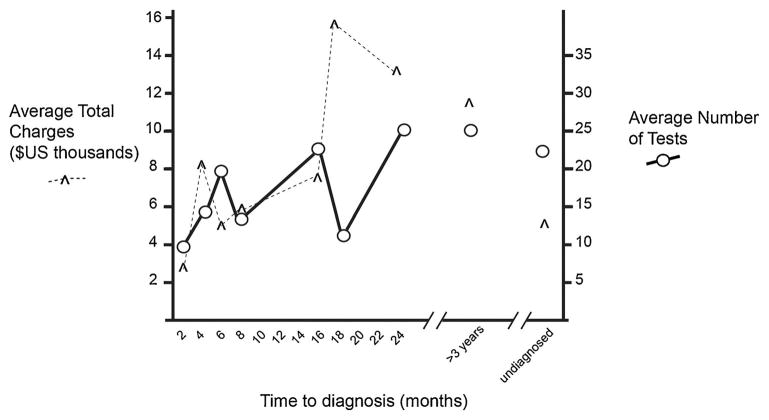
From 2002–2010, all patients presenting with an apparent inherited leukodystrophy were identified and followed through June 2014 using our previous criteria [Bonkowsky et al., 2010]. The cohort consisted of 62 leukodystrophy patients (Table I); siblings or relatives of known leukodystrophy patients were excluded. Each patient had average charges of $8,231 (range $543–26,437) for diagnostic testing. Overall charges related to diagnosis for the entire cohort was $526,794. A final etiological diagnosis was determined in 34% of patients. In those in whom a specific diagnosis was determined, average elapsed time to diagnosis was 1.4 years; higher charges and an increased number of tests were associated with increasing time to diagnosis (Fig. 1).
TABLE I.
Demographics of the Cohort and Charges Associated With Diagnostic Testing
| Characteristic | Number (Percentage) |
|---|---|
| Gender | |
| Male | 35 (56%) |
| Female | 27 (44%) |
| Race | |
| Caucasian | 53 (85%) |
| Hispanic | 7 (11%) |
| African-American | 1 (2%) |
| Native American | 1 (2%) |
| Diagnosis | |
| Unknown | 41 (66%) |
| Krabbe disease | 2 (3%) |
| Mitochondrial | 2 (3%) |
| Pelizaeus–Merzbacher disease | 2 (3%) |
| Othera | 15 (24%) |
| Patient Charges | |
| Total | $526,794 |
| Average (range) | $8,231 ($543–26,437) |
| Average per test cost (range) | $531 ($7–7,930) |
Other diagnoses: 2q24.3 syndrome, 4H syndrome, del17p13.3, 18q-syndrome, 46, XY dup(9) (q21.1q22.31), Aicardi–Goutieres, Batten, FG syndrome, Gaucher type II, Metachromatic leukodystrophy, Sandhoff, Tay-Sachs, TUBB4a, X-linked Adrenoleukodystrophy, Vanishing White Matter.
FIG. 1.
Average charges per patient (y-axis, ^marks) and average number of tests (circles), based on time to diagnosis (x-axis).
DISCUSSION
We have determined the charges directly associated with diagnostic testing in a population cohort of children with leukodystrophy. Charges reflect the end-costs experienced by patients and families. While charges vary significantly from institution to institution, and are dependent on a variety of financial circumstances that are often not directly related to the cost of providing the test, we decided to use charges because it is most reflective of what most families and clinicians are familiar with; and what insurance companies commonly deal with. Further, information about diagnosis charges can be most easily compared to the charges for NGS.
Conservatively assuming a similar rate of diagnostic yield using NGS, charges for NGS on the entire cohort would have been ~$359,600 (at $5,800/patient) [Ambry Genetics]. Alternatively, a two-tier approach could consist of first, biochemical testing (serum very-long chain fatty acids and leukocyte lysosomal enzyme testing) for treatable leukodystrophies (for X-linked adrenoleukodystrophy and for lysosomal disorders), and then with NGS for remaining undiagnosed patients. In this two-tier approach, the first-tier would have charges of $42,309, and the second tier charges would be $319,000; for total cohort charges of $361,309. Turnaround time for NGS can be as few as 12 weeks, which is substantially shorter than the average time to diagnosis we found, but is longer than biochemical testing for adrenoleukodystrophy and lysosomal disorders.
Limitations of our work are that it was a single center, and consisted of retrospectively collected data. In addition, since the cohort was relatively small, ordering preferences of a few physicians could affect the costs and limit generalizability. However, the group of pediatric neurologists caring for these children consists of over 12 physicians; and the ability to longitudinally track all diagnostic charges are unique strengths of the cohort and database used.
We conclude that the advantages of appropriately incorporating NGS into diagnostic algorithms may include lower charges; reduced time to diagnosis; and reduced amount of testing including reducing repeat MRIs and associated sedations. In addition, with improving NGS there is potential for higher diagnosis yield. Our study provides novel insight into the charges associated with the diagnostic odyssey, and how that compares to charges of NGS. Recently, another group has also shown that NGS appears to have lower costs compared to a traditional diagnostic route in children with neurodevelopmental disorders (Soden et al., 2014). However, to our knowledge our study and that of Soden et al. are the only publications with direct information on the charges associated with diagnosis. The detailed information we have determined on the charges and diagnosis rates from the diagnostic odyssey provide important information that can be used to improve patient diagnosis, management, and care.
Acknowledgments
Grant sponsor:NIH; Grant number: DP2 MH100008; Grant sponsor: PCORI ALD Connect.
Author Contributions: Dr. Bonkowsky had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design was performed by Jackson, Taft, Vanderver, Bonkowsky. Acquisition of data was done by Jackson, Korgenski, Bonkowsky. Analysis and interpretation of data was done by all authors. Manuscript preparation was performed by all authors. JLB was supported by NIH grant DP2 MH100008, and by PCORI grant ALD Connect. NIH and ALD Connect had no role in design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication. RJT was supported by Illumina, Inc. Illumina had no role in design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication. Otherwise, the authors state that they have no conflicts of interest. Additional Contributions: We thank R. Rasmussen and T. Pysher, MD, for assistance with obtaining charge data.
Footnotes
Conflicts of interest: RJT is an employee of Illumina, Inc.
References
- [Accessed January 15, 2015]; http://www.ambrygen.com/exomenext.
- Bonkowsky JL, Nelson C, Kingston JL, Filloux FM, Mundorff MB, Srivastava R. The burden of inherited leukodystrophies in children. Neurology. 2010;75:718–725. doi: 10.1212/WNL.0b013e3181eee46b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C, Mundorff MB, Korgenski EK, Brimley CJ, Srivastava R, Bonkowsky JL. Determinants of health care use in a population-based leukodystrophy cohort. J Pediatr. 2013:162. doi: 10.1016/j.jpeds.2012.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond G, Eichler Fatemi A, Naidu S. Leukodystrophies. 1. London: Mac Keith Press; 2011. [Google Scholar]
- Soden SE, Saunders CJ, Willig LK, Farrow EG, Smith LD, Petrikin JE, LePichon JB, Miller NA, Thiffault I, Dinwiddie DL, Twist G, Noll A, Heese BA, Zellmer L, Atherton AM, Abdelmoity AT, Safina N, Nyp SS, Zuccarelli B, Larson IA, Modrcin A, Herd S, Creed M, Ye Z, Yuan X, Brodsky RA, Kingsmore SF. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffmann R, Van Der Knaap MS. Invited Article: An MRI-based approach to the diagnosis of white matter disorders. Neurology. 2009;72:750–759. doi: 10.1212/01.wnl.0000343049.00540.c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward P, Braxton A, Beuten J, Xia F, Niu Z, Hardison M, Person R, Bekheirnia MR, Leduc MS, Kirby A, Pham P, Scull J, Wang M, Ding Y, Plon SE, Lupski JR, Beaudet AL, Gibbs R, Eng CM. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]