Abstract
Triple-negative breast cancers are highly aggressive, and patients with these types of tumors have poor long-term survival. These breast cancers do not express estrogen or progesterone receptors and do not have gene amplification of human epidermal growth factor receptor 2; therefore, they do not respond to available targeted therapies. The lack of targeted therapies for triple-negative breast cancers stems from their heterogeneous nature and lack of a clear definition of driver defects. Studies have recently identified triple-negative breast cancer molecular subtypes based on gene expression profiling and representative cell lines, allowing for the identification of subtype-specific drug leads and molecular targets. We previously reported the identification of a new fungal metabolite named maximiscin (1) identified through a crowdsourcing program. New results show that 1 has selective cytotoxic efficacy against basal-like 1 MDA-MB-468 cells compared to cell lines modeling other triple-negative breast cancer molecular subtypes. This compound also exhibited antitumor efficacy in a xenograft mouse model. The mechanisms of action of 1 in MDA-MB-468 cells were investigated to identify potential molecular targets and affected pathways. Compound 1 caused accumulation of cells in the G1 phase of the cell cycle, suggesting induction of DNA damage. Indeed, treatment with 1 caused DNA double-strand breaks with concomitant activation of the DNA damage response pathways, indicated by phosphorylation of p53, Chk1, and Chk2. Collectively, these results suggest basal-like triple-negative breast cancer may be inherently sensitive to DNA damaging agents relative to other triple-negative breast cancer subtypes. These results also demonstrate the potential of our citizen crowdsourcing program to identify new lead molecules for treating the subtypes of triple-negative breast cancer.
Graphical Abstract
Triple-negative breast cancers are a highly aggressive and heterogeneous subtype of breast cancer, defined by a lack of human epidermal growth factor receptor 2 amplification and undetectable levels of estrogen/progesterone receptors.1–4 There are no approved targeted therapies for triple-negative breast cancer, so patients typically receive multimodality chemotherapy with cytotoxic agents such as taxanes and anthracyclines.5,6 The lack of targeted treatment options for triple-negative breast cancers stems from their heterogeneous nature and the diversity of their molecular drivers. However, Lehman and Bauer et al identified six molecular subtypes of triple-negative breast cancer and representative cell lines based on gene-expression profiling of tumors from triple-negative breast cancer patients.7 The molecular subtypes include two basal-like (BL1 and BL2), mesenchymal-like (ML), mesenchymal stem-like (MSL), luminal androgen receptor (LAR) and immunomodulatory (IM) subtypes. This now allows for the identification of preclinical drug leads with selective activity against these molecular subtypes of triple-negative breast cancer.
Approximately 60% of all triple-negative breast cancers are classified as basal-like according to the molecular subtyping tool TNBCtype.8 The BL1 subtype is characterized by enrichment in genes related to cell cycle progression and DNA synthesis, while the BL2 subtype shows characteristics of up-regulated growth factor signaling, glycolysis and gluconeogenesis.7,9 These molecular pathways have the potential to be useful drug targets for selectively treating these subtypes of triple-negative breast cancer. In fact, Lehman and Bauer et al demonstrated that cell lines of the BL1 subtype are particularly sensitive to DNA damaging agents in vitro.7 Interestingly, Masuda and Baggerly et al found that patients with the BL1 subtype of triple-negative breast cancer had the highest rate of pathological complete response after neoadjuvant chemotherapy, while the BL2 and LAR subtypes had the lowest rate.10 These findings demonstrate that further studies into the pharmacological sensitivities of the molecular subtypes of triple-negative breast cancer are needed to help identify new, targeted agents to effectively treat these cancers. Additionally, identification of biomarkers that predict patients’ responses will allow for optimal use of currently approved chemotherapeutic agents.
Our research group has recently focused on identifying natural products with selective cytotoxic activities against molecular subtypes of triple-negative breast cancer, with the goal of identifying new agents and molecular targets for these cancers. We previously reported the identification of a polyketide-shikimate-nonribosomal peptide synthetase-hybrid metabolite named maximiscin (1), and investigated its activity against cancer cell lines in vitro and in a xenograft mouse model of melanoma.11–13 In the current study, we report that 1 shows potent and highly selective cytotoxic activity against a cell line model of the BL1 subtype of triple-negative breast cancer compared to cells representing the other triple-negative breast cancer subtypes. The molecular mechanisms of actions of 1 were investigated in vitro to identify its potential molecular targets, which might have therapeutic value for treating the BL1 subtype. The antitumor efficacy of 1 was also evaluated in a xenograft mouse model of the BL1 subtype.
RESULTS AND DISCUSSION
Antiproliferative and Cytotoxic Activities of Compound 1
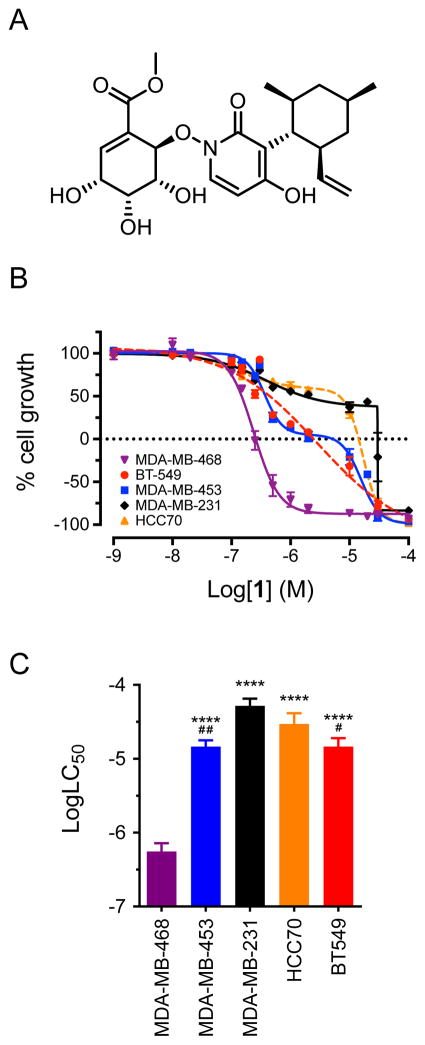
The antiproliferative and cytotoxic activities of 1 (Figure 1A) were evaluated in a panel of five triple-negative breast cancer cell lines representing five different molecular subtypes. These included MDA-MB-468, HCC70, BT-549, MDA-MB-231, and MDA-MB-453 cells, which model the BL1, BL2, ML, MSL and LAR subtypes, respectively (Table 1). Compound 1 inhibited proliferation of all five triple-negative breast cancer cell lines, but was particularly cytotoxic to MDA-MB-468 cells, which represent the BL1 subtype (Figure 1B, 1C). The concentration of 1 that caused 50% cell death (LC50) of MDA-MB-468 cells was found to be 600 nM (Table 1). This was significantly lower than the LC50 values in the four other triple-negative breast cancer cell lines, which ranged from 15 – 60 μM (Figure 1C, Table 1). For comparison, the microtubule stabilizer paclitaxel had an LC50 of 9.8 nM in MDA-MB-468 cells (Table 1). In contrast to 1, paclitaxel did not have selectivity for MDA-MB-468 cells, with LC50 values ranging from 8.2 nM to greater than 62.5 nM in the other four triple-negative breast cancer cell lines (Table 1). These results demonstrated that 1 has 25 to 100-fold cytotoxic selectivity for MDA-MB-468 cells compared to cells modeling the four other triple-negative breast cancer subtypes. Compound 1 is closely related to pyridoxatin, so we asked if pyridoxatin has similar cytotoxic potency against MDA-MB-468 cells. The concentration-response curve for pyridoxatin was nearly identical to that of 1 in MDA-MB-468 cells (Supplementary Figure S1). This suggested that pyridoxatin may be the active pharmacophore of 1. Collectively, these results suggested that MDA-MB-468 cells, and possibly BL1 cells in general, may be particularly sensitive to manipulation of the molecular target(s) of 1. This prompted further investigations into the cellular mechanisms of action of 1 in MDA-MB-468 cells.
Figure 1.
Compound 1 is selectively cytotoxic to basal-like 1 MDA-MB-468 cells. (A) Structure of 1. (B) Concentration-response curves for antiproliferative and cytotoxic effects of compound 1 (maximiscin) in five triple-negative breast cancer cell lines after 48 h treatment. Results represent mean ± SEM for at least 3 independent experiments with all concentrations tested in triplicate. (C) Comparison of log LC50 values for compound 1 in five triple-negative breast cancer cell lines. ****p < 0.0001 compared to MDA-MB-468; #p < 0.05, ##p < 0.01 compared to MDA-MB-231; one-way ANOVA with Tukey’s posthoc test. Results represent mean ± SEM for at least 3 independent experiments, each conducted with all concentrations tested in triplicate.
Table 1.
Cytotoxic Potency (LC50a) of Compound 1 and Paclitaxel (PTX) in Five Triple-Negative Breast Cancer Cell Lines
| Cell Line | Subtype | 1 (μM) | PTX (nM) |
|---|---|---|---|
| MDA-MB-468 | BL1 | 0.6 ± 0.2 | 9.8 |
| HCC70 | BL2 | 39 ± 9 | > 31.25 |
| BT-549 | ML | 15 ± 5 | 18 |
| MDA-MB-231 | MSL | 60 ± 10 | > 62.5 |
| MDA-MB-453 | LAR | 15 ± 4 | 8.2 |
LC50 is defined as the concentration causing 50% cell death compared to vehicle-treated control cells.
Effects of Compound 1 on Cell Cycle Distribution of MDA-MB-468 Cells
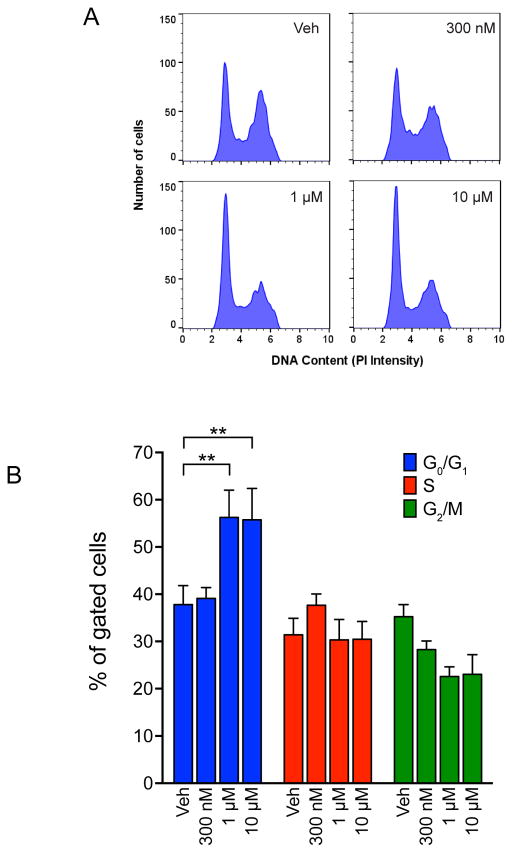
Drug-induced changes in the proportion of cells in different cell cycle phases can provide valuable information about the drug’s mechanism of action. To investigate the mechanisms of actions of 1 in MDA-MB-468 cells, its effects on cell cycle distribution were evaluated by measuring the DNA content of treated cells using flow cytometry (Figure 2). Treatment with 300 nM 1 for 18 h had no effect on the percentage of cells in each phase of the cell cycle (Figure 2). In contrast, treatment with either 1 or 10 μM 1 significantly increased the percentage of cells in the G1 phase (Figure 2). This suggested that 1 might block DNA synthesis, possibly by causing DNA damage.
Figure 2.
Compound 1 induces G1 accumulation of MDA-MB-468 cells. (A) Representative histogram of PI intensity after 18 h treatment with vehicle or indicated concentration of 1 (maximiscin). (B) Quantification of cells in each phase of the cell cycle after 18 h treatment. **p < 0.01; two-way ANOVA with Dunnett’s posthoc test; results represent mean ± SEM (n = 3).
We further investigated the cellular mechanism of action of 1 in MDA-MB-468 cells using an antibody microarray. This slide-based array measures the phosphorylation or cleavage of multiple intracellular signaling proteins in whole-cell lysates, including members of the PI3K/Akt/mTOR, MAPK/Erk, DNA damage response and apoptotic pathways. Treatment with 1 μM 1 resulted in a time-dependent increase in phosphorylation of p53 at serine 15, which began between 2 and 6 h after treatment, suggesting an accumulation of DNA damage (Supplementary Figure S2). This was consistent with the previously observed G1 accumulation. No significant changes in any other protein targets were observed with the antibody array (data not shown). These results prompted further investigations into the mechanisms of compound 1-induced changes in the cell cycle distribution of MDA-MB-468 cells.
Effects of Compound 1 on DNA Damage Response Pathways
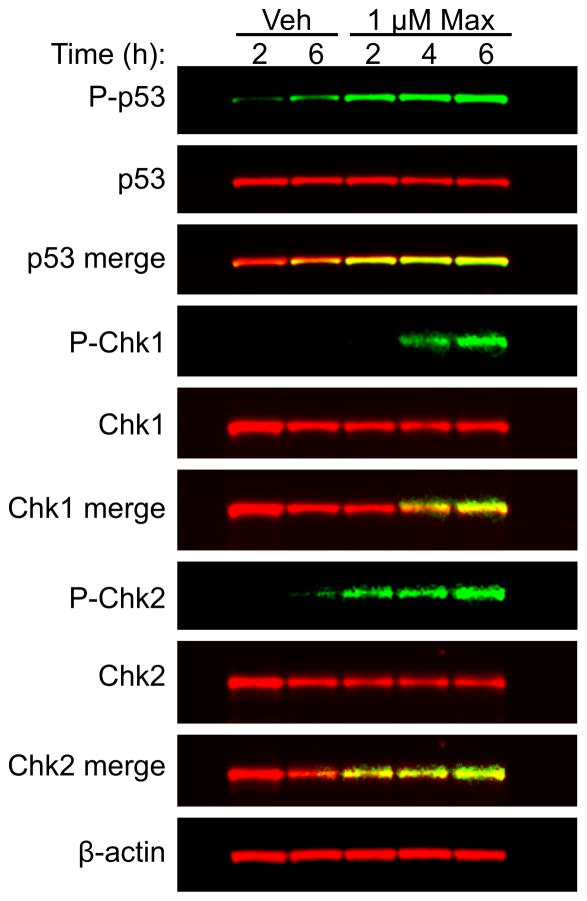
The observations that 1 induced both p53 phosphorylation and G1 accumulation suggested that 1 caused DNA damage as one of its primary mechanisms of action. The ability of 1 to activate DNA damage response signaling pathways in MDA-MB-468 cells was evaluated. We specifically evaluated early time points after treatment with 1 to observe the more direct effects of treatment, rather than secondary effects resulting from cell death. Immunoblotting of MDA-MB-468 whole-cell lysates confirmed the results of the antibody array, showing robust phosphorylation of p53 at serine 15 as soon as 2 h after treatment (Figure 3). Additionally, 1 induced phosphorylation of checkpoint kinase 1 (Chk1) and checkpoint kinase 2 (Chk2), which occurred 4 and 2 h after treatment, respectively (Figure 3). Chk1 and Chk2 are downstream effectors of ATM and ATR and are critically involved in regulating the DNA damage checkpoint.14,15 These results demonstrate that 1 activates DNA damage response pathways in MDA-MB-468 cells within 2 h of treatment.
Figure 3.
Compound 1 activates DNA damage response pathways in MDA-MB-468 cells. Immunoblotting of MDA-MB-468 whole cell lysates for total and activated (phosphorylated) p53, checkpoint kinase 1 (Chk1) and checkpoint kinase 2 (Chk2) after treatment with vehicle or compound 1 (maximiscin) for indicated time periods.
Effects of Compound 1 on Double-Strand Break Formation
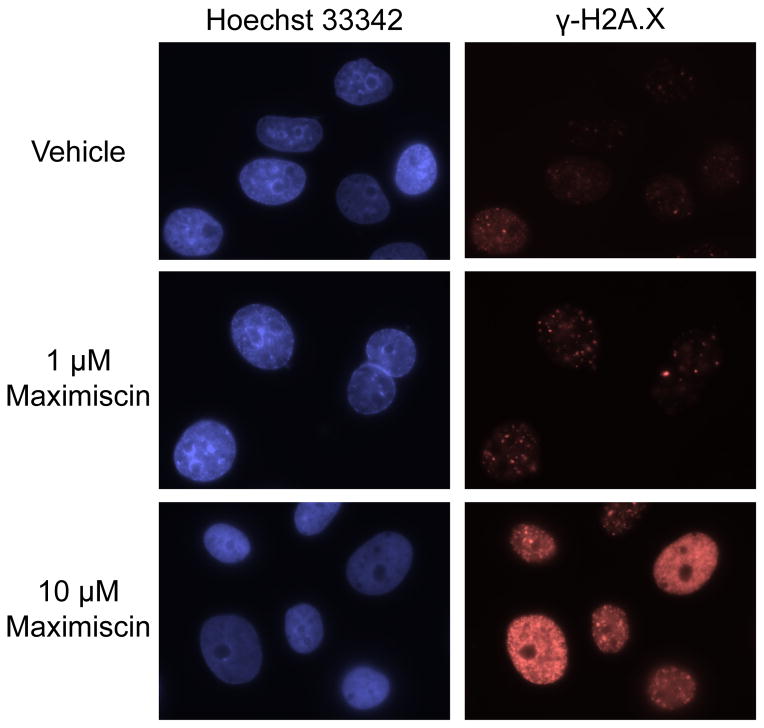
The ability of 1 to directly cause DNA damage was evaluated by determining if it induced the formation of DNA double-strand breaks. For these experiments, the accumulation of histone H2A.X phosphorylated at serine 139 (γ-H2A.X), a marker of DNA double-strand breaks,16,17 was monitored in MDA-MB-468 cells by immunofluorescence microscopy. A concentration-dependent increase in γ-H2A.X foci after treatment with 1 or 10 μM 1 for 18 h was observed, suggesting an accumulation of DNA double-strand breaks (Figure 4). Collectively these results strongly suggested that induction of DNA damage is the primary mechanism of action of 1 in MDA-MB-468 cells.
Figure 4.
Compound 1 induces DNA double-strand breaks in MDA-MB-468 cells. Immunostaining of γ-H2A.X in MDA-MB-468 cells treated with vehicle or compound 1 (maximiscin) for 18 h. Images are representative of results from two independent experiments, with each concentration tested in duplicate.
Antitumor efficacy of compound 1 in a BL1 xenograft model
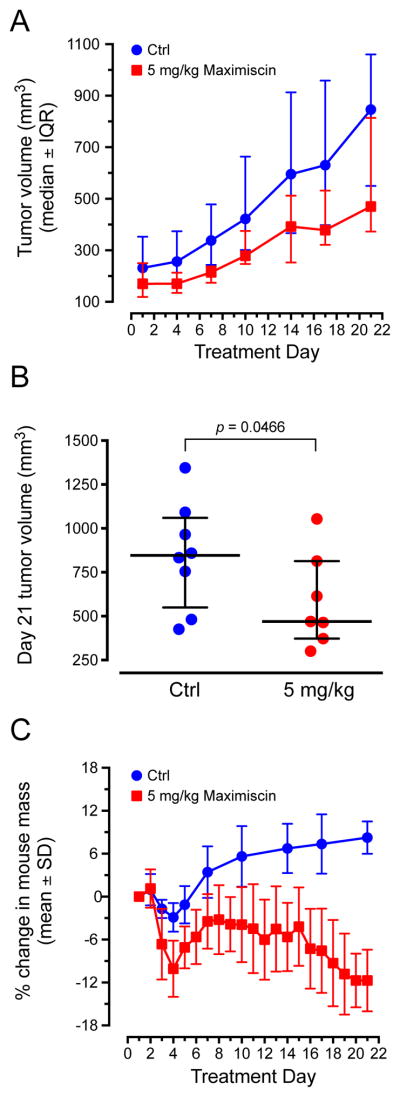
To determine if 1 has antitumor efficacy against the BL1 subtype in vivo, its efficacy against MDA-MB-468 xenografts in nude mice was evaluated. We previously established a tolerable intraperitoneal (i.p.) dose for 1 of 5 mg/kg when administered in five-day intervals, with two-day breaks from dosing between cycles.11 This dose and schedule demonstrated antitumor efficacy against a melanoma xenograft model, and suggested that frequent dosing was necessary for 1 to produce antitumor effects. In the current study, we administered 5 mg/kg 1 by i.p. injection, every day for 21 days. Compound 1 inhibited the growth of MDA-MB-468 xenografts over this time period compared to untreated controls (Figure 5A). On day 21 of treatment, tumors were significantly smaller in the mice treated with 1 than they were in control mice (p = 0.0466; Figure 5B). At this dose and schedule, mice gradually lost weight over the course of treatment, with an average weight loss of approximately 12% on day 21 (Figure 5C). These results suggested that 1 has antitumor efficacy against the BL1 subtype, but it also has a narrow therapeutic window.
Figure 5.
Compound 1 inhibits growth of MDA-MB-468 xenografts. (A) Caliper measurements of tumor volumes taken twice weekly over the course of treatment with 1 (maximiscin). Results represent median ± interquartile range (IQR) for n = 8 (Ctrl) or 7 (treated) tumors. (B) Comparison of final tumor volumes on day 21 of treatment; one-tailed Mann-Whitney rank analysis. (C) Percent change in mouse weights over the course of treatment. Results represent mean ± SD.
Conclusion
Together, these studies identified the natural product 1 as having selective cytotoxic activity against MDA-MB-468 cells modeling the BL1 subtype of triple-negative breast cancer. Mechanism of action studies suggest that induction of DNA damage is a primary mechanism of action for this compound in this sensitive cell line. Other pyridones and hydroxamic acids, such as the closely related compound pyridoxatin, have been shown to inhibit DNA synthesis,18 although the precise mechanism of action is not clear. Further supporting our hypothesis, we found that the concentration-response curve for pyridoxatin is identical to that of 1, suggesting 1 may act as a prodrug for pyridoxatin.
Gene expression profiling identified increased expression of cell cycle and DNA damage response proteins in the BL1 subtype of triple-negative breast cancer, suggesting these cancers are highly proliferative and may be particularly susceptible to DNA damage.7 Recently, Purrington et al. independently confirmed the presence of two distinct basal-like subtypes of triple-negative breast cancer with increased expression of cell cycle and DNA damage response genes.19 It has been demonstrated that BL1 cell lines are more sensitive to cisplatin and poly-ADP-ribose polymerase (PARP) inhibitors compared to cell lines representative of the other triple-negative breast cancer molecular subtypes.7 Also, BL1 xenografts are particularly sensitive to cisplatin.7 It is likely that higher proliferation rates, combined with defective DNA repair systems and higher basal levels of DNA damage, sensitize BL1 cells to DNA damaging agents.
Additional studies will be needed to identify the precise mechanisms of DNA damage induced by 1. Collectively, these results provide further evidence that the BL1 subtype of triple-negative breast cancer is particularly sensitive to DNA damaging agents, and may be better suited for treatment with classical DNA-interacting chemotherapeutic agents than other subtypes of triple-negative breast cancer. Our results also demonstrate the value of our citizen-science crowdsourcing program for identifying new lead molecules for treating triple-negative breast cancer subtypes.
EXPERIMENTAL SECTION
General Experimental Procedures
Sulforhodamine B salt, Trizma, Dulbecco’s phosphate-buffered saline (DPBS), phenylmethanesulfonyl fluoride (PMSF) and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Compounds for cell treatments were dissolved in DMSO and stored at −20°C. Maximiscin for in vivo studies was stored as a stock solution in 50:50 DMSO:Kolliphor® EL at −20°C and diluted in DPBS immediately prior to use.
Fermentation, Isolation and Purification
Maximiscin (1) was isolated from a fungal isolate Tolypocladium sp. Salcha MEA-2 and purified by column chromatography and HPLC (purity >99%) as previously described.11 The fungus was cultured in 20 L modified potato dextrose broth containing 10 g/L mashed potatoes (Great Value™) and 5 g/L glucose. Approximately 50 mg of 1 was isolated for all in vitro and in vivo experiments.
Cell Culture
MDA-MB-468, MDA-MB-453, MDA-MB-231, and HCC70 cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). The BT-549 cell line was obtained from Lombardi Comprehensive Cancer Center of Georgetown University (Washington, DC, USA) and validated by Promega (Fitchburg, WI, USA). MDA-MB-453 and MDA-MB-231 cells were cultured in improved minimum essential medium (IMEM; Gibco/Thermo Fisher Scientific, Waltham, MA, USA) containing 25 μg/mL gentamicin (Gibco) and 10% fetal bovine serum (FBS; GE Healthcare, Little Chalfont, UK). MDA-MB-468, HCC70 and BT-549 cells were cultured in RPMI-1640 medium (Sigma-Aldrich) containing 50 μg/mL gentamicin and 10 % FBS and. Cells were maintained in humidified incubators at 37°C with 5% CO2. All cell lines were initially expanded and frozen as stocks in liquid nitrogen. Cells were passaged for less than 3 months after revival.
Sulforhodamine B Assay
The antiproliferative and cytotoxic activities of maximiscin were evaluated using the sulforhodamine B (SRB) assay as previously described.20,21 Cells were treated with the compounds indicated for 48 h. The cell density at the time of treatment (T0) was measured to assess cytotoxic activity. Concentration-response curves were plotted and the concentrations causing 50% cell death compared to T0 (LC50) were interpolated from nonlinear regressions of the data using Prism 6 (GraphPad Software, La Jolla, CA, USA).
Whole-cell Lysis and Immunoblotting
MDA-MB-468 cells were treated with vehicle or maximiscin, harvested by scraping and lysed in cell extraction buffer (Life Technologies/ Thermo Fisher Scientific, Waltham, MA, USA) containing protease inhibitor cocktail, PMSF and Na3VO4 (Sigma-Aldrich). The total concentration of protein in each sample was measured with a Pierce Coomassie Plus assay kit (Life Technologies) and equal amounts of protein were separated by SDS-PAGE on NuPAGE Bis-Tris gels (Life Technologies). Proteins were transferred to PVDF membranes (EMD Millipore, Billerica, MA, USA) and probed with antibodies for Phospho-S345-Chk1, Chk1, Phospho-T68-Chk2, Chk2, Phospho-S15-p53, p53 (Cell Signaling Technology, Danvers, MA, USA) or β-actin (Sigma) diluted in Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE, USA). Membranes were incubated with appropriate IRDye 680 or IRDye 800 secondary antibodies (LI-COR Biosciences) and near-infrared fluorescence signals were captured on an Odyssey FC (LI-COR Biosciences).
Antibody Microarray
Whole-cell MDA-MB-468 lysates were prepared as described for immunoblotting and evaluated with a PathScan® intracellular signaling array (fluorescent readout; Cell Signaling Technology) according to the manufacturer’s protocol. Near-infrared fluorescent signals were captured with an Odyssey CLx (LI-COR Biosciences) and quantified with ImageStudio (LI-COR Biosciences).
Flow Cytometry
The effects of each compound on cell cycle distribution were evaluated by flow cytometry. MDA-MB-468 cells were treated with vehicle (DMSO) or predetermined concentrations of maximiscin for 18 h. Cells were then harvested, stained with Krishan’s reagent,22 and their DNA content was measured using a Muse Cell Analyzer (EMD Millipore). Data were analyzed with FlowJo 10 (FlowJo LLC, Ashland, OR, USA)
Immunofluorescence Microscopy
MDA-MB-468 cells were plated onto glass coverslips and allowed to adhere overnight. After treatment with maximiscin for 18 h, cells were fixed with paraformaldehyde and subsequently incubated with a blocking solution of 10% bovine calf serum in DPBS for 20 min at room temperature. Cells were then incubated with a primary antibody against γ-H2A.X (Cell Signaling Technology) diluted in 1% bovine serum albumin/0.3% Triton X-100 in DPBS. After incubation, cells were washed with DPBS and incubated with an Alexafluor 594-conjugated secondary antibody (Life Technologies). Coverslips were washed with PBS and stained with NucBlue live cell stain (Life Technologies) diluted in DPBS. Coverslips were mounted and visualized with an Eclipse 80i fluorescence microscope using a Plan Apo VC 60× H objective (Nikon, Tokyo, Japan). Images were captured with a CoolSNAP HQ2 camera (Photometrics, Tuscon, AZ, USA) and NIS Elements software (Nikon).
Xenograft Studies in Nude Mice
Female athymic nude mice (Envigo/Harlan Laboratories, Indianapolis, IN, USA) were injected s.c. with 2 × 106 MDA-MB-468 cells, suspended in 100 μL DPBS and 100 μL Matrigel® (BD Biosciences, San Jose, CA, USA), bilaterally into each flank. After tumors reached a minimum volume of 150 mm3, mice were assigned to one of two different groups (n = 7 or 8 tumors/group). Maximiscin (5 mg/kg) was administered daily by i.p. injection in a vehicle of 2.5% DMSO and 2.5% Kolliphor EL in DPBS. Mice were weighed and examined daily for signs of toxicity. Tumors were measured twice per week using calipers, and tumor volume (mm3) was calculated as length (mm) × width (mm) × height (mm). All mice were housed in an AAALAC-approved facility at The University of Texas Health Science Center at San Antonio and given food and water ad libitum. All procedures were IACUC approved.
Supplementary Material
Acknowledgments
Funding Sources
This study was funded by grants to S.L.M. and R.H.C. from the National Cancer Institute (UO1CA182740), the UTHSCSA President’s Council Excellence Award (S.L.M), and the Greehey Distinguished Chair in Targeted Molecular Therapeutics endowment (S.L.M.). Support of the Flow Cytometry, Macromolecular Structure and Mass Spectrometry Shared Resources of the CTRC Cancer Center Support Grant (P30 CA054174) are gratefully acknowledged. A.J.R was partially supported by the IMSD program of the NIGMS (1R25GM095480-01).
We gratefully acknowledge Ms. A. Reyor for supplying the soil sample which ultimately led to the identification of maximiscin. We also acknowledge Dr. April Risinger for thoughtful review of this manuscript.
ABBREVIATIONS
- BL1
basal-like 1
- BL2
basal-like 2
- LAR
luminal androgen receptor
- ML
mesenchymal-like
- MSL
mesenchymal stem-like
- IM
immunomodulatory
Footnotes
Author Contributions
A.J.R. and S.L.M. designed the biological experiments. L.D. and R.H.C. designed the chemical experiments. A.J.R. performed all biological experiments. L.D. isolated and identified maximiscin. A.J.R. prepared the manuscript. All authors were involved in editing and have given approval to the final version of the manuscript.
Figure S1: Concentration-response curve for pyridoxatin and compound 1. (PDF)
Figure S2: Graph of antibody array data for phospho-S15-p53. (PDF)
This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Vaz-Luis I, Ottesen RA, Hughes ME, Mamet R, Burstein HJ, Edge SB, Gonzalez-Angulo AM, Moy B, Rugo HS, Theriault RL, Weeks JC, Winer EP, Lin NU. J Clin Oncol. 2014;32:2142–2150. doi: 10.1200/JCO.2013.53.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu KD, Zhu R, Zhan M, Rodriguez AA, Yang W, Wong S, Makris A, Lehmann BD, Chen X, Mayer I, Pietenpol JA, Shao ZM, Symmans WF, Chang JC. Clin Cancer Res. 2013;19:2723–2733. doi: 10.1158/1078-0432.CCR-12-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irvin WJ, Carey LA. Eur J Cancer. 2008;44:2799–2805. doi: 10.1016/j.ejca.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 4.Abramson VG, Mayer IA. Curr Breast Cancer Reports. 2014;6:154–158. doi: 10.1007/s12609-014-0152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch A, Eroles P, Zaragoza R, Viña JR, Lluch A. Cancer Treat Rev. 2010;36:206–215. doi: 10.1016/j.ctrv.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Hirshfield KM, Ganesan S. Curr Opin Obstet & Gynecol. 2014;26:34–40. doi: 10.1097/GCO.0000000000000038. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, Pietenpol JA. Cancer Informatics. 2012;11:147–156. doi: 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehmann BD, Pietenpol JA. J Pathol. 2014;232:142–150. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, Valero V, Lehmann BD, Pietenpol JA, Hortobagyi GN, Symmans WF, Ueno NT. Clin Cancer Res. 2013;19:5533–5540. doi: 10.1158/1078-0432.CCR-13-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du L, Robles AJ, King JB, Powell DR, Miller AN, Mooberry SL, Cichewicz RH. Angew Chem Int Ed. 2014;53:804–809. doi: 10.1002/anie.201306549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du L, Robles AJ, King JB, Powell DR, Miller AN, Mooberry SL, Cichewicz RH. Angew Chem Int Ed. 2014;53:9695–9695. doi: 10.1002/anie.201306549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du L, Robles AJ, King JB, Powell DR, Miller AN, Mooberry SL, Cichewicz RH. Angew Chem Int Ed. 2015;54 doi: 10.1002/anie.201503831. [DOI] [PubMed] [Google Scholar]
- 14.Xu N, Libertini S, Zhang Y, Gillespie DA. Biochem Biophys Res Commun. 2011;413:465–470. doi: 10.1016/j.bbrc.2011.08.119. [DOI] [PubMed] [Google Scholar]
- 15.Lou Z, Minter-Dykhouse K, Wu X, Chen J. Nature. 2003;421:957–961. doi: 10.1038/nature01447. [DOI] [PubMed] [Google Scholar]
- 16.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. Nat Rev Cancer. 2008;8 doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bar-Am O, Weinreb O, Amit T, Youdim MBH. FASEB J. 2005;19:1899–1901. doi: 10.1096/fj.05-3794fje. [DOI] [PubMed] [Google Scholar]
- 18.Lee HJ, Chung MC, Lee CH, Chun HK, Kim HM, Kho YH. Korean J Microbiol Biotechnol. 1996;6:445–450. [Google Scholar]
- 19.Eccles DM, Vachon CM, Couch FJ, Purrington KS, Visscher DW, Wang C, Yannoukakos D, Hamann U, Nevanlinna H, Cox A, Giles GG, Eckel-Passow JE, Lakis S, Kotoula V, Fountzilas G, Kabisch M, Rüdiger T, Heikkilä P, Blomqvist C, Cross SS, Southey MC, Olson JE, Gilbert J, Deming-Halverson S, Kosma V-M, Clarke C, Scott R, Jones JL, Zheng W, Mannermaa A, Mannermaa Breast Cancer Res Treat. 2016 doi: 10.1007/s10549-016-3775-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyd MR, Paull KD, Rubinstein LR. Cytotoxic Anticancer Drugs: Models and Concepts for Drug Discovery and Development. Springer; US: 1992. pp. 11–34. [Google Scholar]
- 21.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 22.Krishan A. J Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.